Abstract
Male infertility is a common and complex problem affecting 1 in 20 men. Despite voluminous research in this field, in many cases, the underlying causes are unknown. Epigenetic factors play an important role in male infertility and these have been studied extensively. Epigenetic modifications control a number of processes within the body, but this review will concentrate on male fertility and the consequences of aberrant epigenetic regulation/modification. Many recent studies have identified altered epigenetic profiles in sperm from men with oligozoospermia and oligoasthenoteratozoospermia. During gametogenesis and germ cell maturation, germ cells undergo extensive epigenetic reprogramming that involves the establishment of sex-specific patterns in the sperm and oocytes. Increasing evidence suggests that genetic and environmental factors can have negative effects on epigenetic processes controlling implantation, placentation and fetal growth. This review provides an overview of the epigenetic processes (histone-to-protamine exchange and epigenetic reprogramming post-fertilization), aberrant epigenetic reprogramming and its association with fertility, possible risks for ART techniques, testicular cancer and the effect of environmental factors on the epigenetic processes.
Keywords: Epigenetics, Male infertility, ART, Chromatin remodeling, Imprinting, Protamines
Introduction
Male infertility is a complex problem where not only the genes, but also the epigenetic factors play a crucial role. There is an enormous interest in one potential cause of male infertility—the aberrant epigenetic reprogramming in male germ cells that can lead to sperm abnormalities. A number of studies have explored the causes of male infertility and now there is sufficient information supporting the idea that epigenetic changes contribute to male infertility.
What is epigenetics?
The term epigenetics refers to changes in the phenotype caused by mechanisms other than changes in DNA sequences, hence the name epi- (above or over)- genetics. Waddington [1] reintroduced the term to explain that gene action and expression that give rise to the phenotype [2]. Epigenetic changes encompass an array of molecular modifications of DNA or histones that are intimately associated with DNA. DNA wraps around histones to form nucleosomes. Nucleosomes are packaged into a higher order of structures called chromatin; modifications in chromatin control gene-expression in a spatio-temporal manner [3–6]. The genome-wide approach to studying epigenetics is defined as epigenomics.
Epigenetic mechanism of gene regulation
Two major modifications that occur in chromatin are DNA methylation and post-translational histone modifications [3, 4]. DNA methylation is a biochemical process which involves addition of a methyl group to the 5′ position of the cytosine pyrimidine ring typically occurring in a CpG dinucleotide [4]. DNA methylation occurs as a result of DNA methyltransferase (DNMT) activity. There are 3 main DNMTs: i) DNMT1 [7]—which plays a key role in maintenance of methylation; ii) DNMT 3a and iii) 3b, which are de novo methyltransferases that methylate the genomic DNA during early embryonic development [8]. The changes are acquired in a gradual rather than by an abrupt process [6, 9]. CpG islands are genomic regions that are approximately 500 base pairs long, which have a high frequency of CpG sites (CG to GC ratio >55%) [10]. These stretches of DNA are located within the promoter region of about 40% of mammalian genes which, when methylated, cause stable heritable transcriptional silencing. Hypomethylation and hypermethylation can occur simultaneously at different regions in the genome [11].
Histones are basic proteins in eukaryotic nuclei, and they package DNA into nucleosomes. H2A, H2B, H3 and H4 histones are integral part of nucleosomes. Histone modifications, such as acetylation, methylation, ubiquitylation and phosphorylation, have emerged as the main players in epigenetic regulatory mechanisms. An intricate interplay exists between modifications of the histone tails of H3 and H4, some of which act antagonistically to regulate the conversion from an active chromatin state to an inactive one termed the histone code [12]. Generally, the acetylation of histones marks active, transcriptionally competent regions, whereas hypoacetylated histones are found in transcriptionally inactive euchromatic or heterochromatic regions. In contrast, histone methylation can be a marker for both active and inactive regions of chromatin. Methylation of lysine 9 on the N terminus of histone H3 (H3-K9) is a feature of silent DNA and is globally distributed throughout heterochromatic regions. On the other hand, methylation of lysine 4 of histone H3 (H3-K4) denotes activity and is found predominantly at the promoters of active genes. H3-K9 methylation is a prerequisite for DNA methylation in fungi and plants [13, 14]. DNA methylation can also trigger H3-K9 methylation [15], as has been documented in mammals.
Epigenetic gene regulation during germ-cell development
Epigenetic mechanisms regulate DNA accessibility throughout an organism’s lifetime as specific sets of genes are active at any stage of development. Each cell type has its own epigenetic signature that reflects the developmental history and environmental influences, and is ultimately reflected in the phenotype of the cell and organism. At the time of fertilization paternal genome delivered by the mature sperm has a haploid genome and is packaged densely with protamines, whereas maternal genome arrested at metaphase II is packaged with histones. Upon fertilization, protamines are rapidly replaced by histones and oocyte completes the second metaphase, releasing the polar body. The H3 and H4 histones that associate with the paternal chromatin are more acetylated than those present in the maternal chromatin [16, 17].
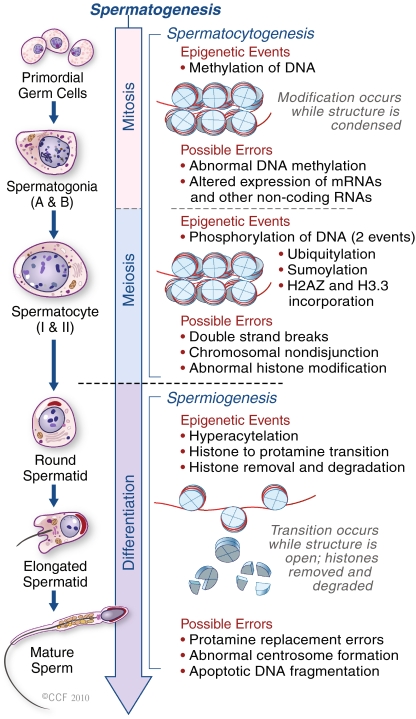
The epigenetic profile of germ cells changes dynamically during development (Fig. 1) [18–21]. In post-implantation mammalian embryos, pluripotent cells in the epiblast give rise to primordial germ cells (PGCs). In females PGCs are arrested in the prophase of meiosis-I whereas in males they enter mitotic arrest. Germ cells undergo several changes in their epigenetic profile during different stages of meiosis. For example, premeiotic PGCs and spermatogonia, exhibit unique patterns of histone modifications such as low H3K9me2 levels [22–24] but in male germ cells, these patterns change dynamically upon initiation of meiosis [25]. Changes in the composition and modification of histones could contribute to chromatin modifications that are required for proper meiosis, and for further maturation of gametes [21]. Both male and female germ cells undergo final developmental changes after meiosis. In haploid round spermatids, global nuclear remodeling occurs, although some histone marks such as H3K9me2 on the inactive X chromosome are retained [26, 27]. A testis-specific linker histone variant H1T2 appears at this stage and plays a crucial role in chromatin condensation during spermiogenesis [28]. Later, a linker histone variant HIls1 (histone-1-like protein in spermatids 1) is expressed in elongated spermatids. In the histone-protamine exchange process, nuclear histones become hyperacetylated during spermiogenesis and shortly thereafter disassemble, replaced by transition proteins (TP1 and TP2). At the final stage of spermiogenesis, transition proteins are removed and replaced by protamines [29]. The incorporation of protamines into sperm chromatin induces DNA compaction, which is important for the formation of spermatozoa and for providing a safe environment for the genome. The presence of somatic-like chromatin in the sperm nucleus could transmit different epigenetic information to the offspring. Oakes et al. [30] suggested that the genome-wide DNA methylation pattern changes little during spermiogenesis, after the pachytene spermatocyte stage. Histone methylation in spermatogenesis is carried out by the H3-K4 and H3-K9 methyltransferase.
Fig. 1.
Epigenetic events during spermatogenesis. In primodial germ cells (mitosis), DNA methylation occurs to set up the paternal specific imprints. Phosphorylation (in meiotic cell) occurs to assist in both recombination and XY body formation. Ubiquitylation, sumoylation and incorporation of H2AZ and H3.3 variants are all involved in XY body formation. Hyperacetylation occurs during spermiogenesis to assist in the Histone-Protamine exchange. Spermatocytogenesis can also give rise to chromosome non-disjunction during its meiosis I and II along with double strand breaks, abnormal histone modification and alteration in the expression on mRNA and other non-coding RNAs. DNA fragmentation is the consequence of apoptosis following double strand breaks or abnormal protamination during spermiogenesis
It has been reported that hyperacetylation of histone H4 is associated with a histone-to-protamine exchange in haploid spermatids [31–33]. Recently, Govin et al. [34] reported that the double bromodomain-containing protein, BRDT (bromodomain testis specific) binds hyperacetylated histone H4 before accumulating in condensed chromatin and helps in organizing the spermatozoon’s genome by mediating a general histone acetylation-induced chromatin compaction and maintaining a differential histone acetylation of specific regions.
A factor named BORIS (brother of regulator of imprinted sites), specifically expressed in male gonads, could be directly involved in the resetting of methylation marks during male germ cell differentiation [35]. The domains of BORIS have the same 11 Zinc Finger as CTCF (CCCTC binding factor: a somatic regulator for expression of imprinted genes), which binds to specific target DNA sequences and plays an important role in the maintenance of differential methylation patterns in somatic cells [36]. CTCF is present in both somatic and germ cells whereas BORIS is expressed specifically in the male germ line. Studies [35, 37] have shown that BORIS is linked with both methylases mediating de novo methylation and demethylases mediating erasure of imprinting marks.
Paternal impact on early embryogenesis
A growing body of evidence suggests that both genetic and epigenetic abnormalities may contribute to idiopathic male infertility, which may affect the outcome of in vitro fertilization (IVF) [38]. Advanced maternal age is one of the obvious contributors to poor fecundity [39], but little is known about the effect of paternal age. We know that advanced paternal age is associated with decreased semen volume, sperm morphology, and sperm motility, but no significant reduction in sperm concentration [40]. A number of studies have documented age-dependent changes in the testis [41, 42].
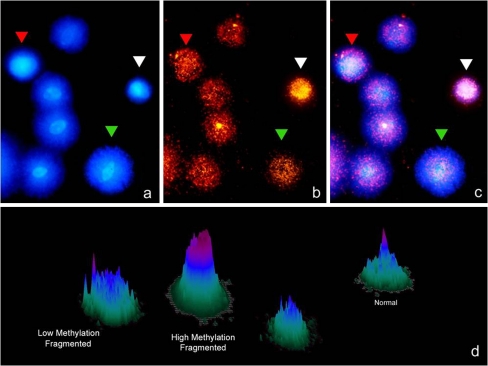
High DNA fragmentation is associated with diminished sperm count, motility, and morphology [42, 43]. Increased DNA fragmentation also decreases fertilization and implantation rates. The influence of sperm DNA methylation on pregnancy was done on mice after using methylation depletion by use of 5-aza-deoxycytidine. This molecule is base analog that when incorporated into DNA decreases the level of DNA methylation varying the gene expression [43]. One of the main problems of solving or throwing some light about the actual role of DNA modifications on fertilization is the absence of reliable techniques which allow easy and reproducible analysis of the level of DNA modification in each gamete. A simple Sperm Chromatin Dispersion (SCD) based technology which offered stable results can be used to analyze the amount of methylated DNA residues and the level of DNA damage in each sperm (Fig. 2). This technique consisted of using MABs against 5-Met-Citosine on a partially denatured DNA molecule once the SCD test was performed. The methodology is mainly based on previous results where the SCD and FISH were used to characterize the level of DNA damage and the presence of aneuploidies (CITA). Figure 2 shows a panel of some results where different levels of DNA methylation could be simultaneously correlated with the level of sperm DNA in each sperm. In some cases DNA fragmentation fully correlates with an abnormal DNA molecule while in other cases this does not occurs. This points to the fact that probably methylation and its consequence are characteristically displayed by each sperm although its variations shall be associated to certain pathological cases. Interestingly, adaptation of this technique may allow semiquantitative assesment of the level of methylation cell by cell.
Fig. 2.
Sperm DNA fragmentation using the SCD test (Halosperm) (panel a), combined with the simultaneous visualization of sperm methylation (panel b) using anti-5metyl-C. Panel 2c was produced after merging a and b. Semiquantitative representation of the differential level of sperm DNA methylation using a surface plot (panel d)
Histones are considered the best candidates for the transmission of epigenetic information because of their influence on the modification of chromatin structure, and access of transcriptional machinery to genes [44, 45]. The methyltransferases facilitate gene silencing by mono-, di- or trimethylation of lysine or arginine [3, 46]. It has yet to be determined whether modified histones play a crucial role in gene expression during early embryogenesis or if abnormal histone modifications in the sperm are associated with diminished embryo development. Alterations in methylation patterns can effect biallelic expression or repression of imprinted genes resulting in various pathologies [47]. Impaired spermatogenesis has been associated with aberrant H4 acetylation [48]. H4 hyperacetylation was also observed in infertile men exhibiting Sertoli cell only (SCO) syndrome. Sperm of patients with asthenozoospermia and teratozoospermia have decreased levels of DNA methylation [49]. Recently, researchers have begun to look at the contribution to early embryogenesis by spermatozoa beyond those of genetic factors. There is evidence of epigenetic contribution to complex diseases.
Epigenetic changes important for male gametes
Gonadal sex determination and testis development occur between embryonic days 12 and 15 (E12 to E15) in the rat (after midgestation in the human) and are initiated by the differentiation of precursor Sertoli cells in response to the testis-determining factor SRY. Aggregation of the precursor Sertoli cells, PGCs, and migrating mesonephros cells (precursor peritubular myoid cells) promotes testis morphogenesis and cord formation. The fetal testis contains steroid receptors and is a target for endocrine hormones. The androgen receptor (AR) and estrogen receptor-b (ERb) are present in Sertoli cells as well as in precursor peritubular myoid cells and germ cells at the time of cord formation (E14). Although the testis does not produce steroids at this stage of development, estrogens and androgens have the ability to influence early testis cellular functions. Treatment with endocrine disruptors vinclozolin and methoxychlor, at a critical time during gonadal sex determination (E8 to E15 in the rat), promotes an adult testis phenotype with decreased spermatogenic capacity and male infertility. This study shows that external factors can induce an epigenetic transgenerational phenotype through an apparent reprogramming of the male germ line [50]. It is not clear whether steroidal factors acting inappropriately at the time of gonadal sex determination reprogram the germ line epigenetically (altered DNA methylation) to cause the transgenerational transmission of an altered phenotype or genetic trait.
Many epigenetic modifiers, including DNA methyltransferases, histone-modification enzymes and their regulatory proteins play essential roles in germ-cell development. Some of these are specifically expressed in germ cells whereas others are more widely expressed. The crucial roles of germ-cell-specific genes such as Dnmt3L and Prdm9 were revealed by conventional knockout studies [51–53]. A recent report showed that there are numerous intra- and inter-individual differences in DNA methylation in human sperm samples [54] which could contribute to phenotypic differences in the next generation. Furthermore, it has been reported that sperm samples from oligospermic patients often contain DNA-methylation defects at imprinted loci [55, 56].
Epigenetics and protamine abnormalities
Marques et al. [54] suggested an association between aberrant sperm epigenetic modifications and altered spermatogenesis. During differentiation of the male gamete, the genome undergoes major changes that not only affect the DNA sequence and genetic information by homologous recombination but also alter its nuclear structure and epigenetic information. One of the challenging issues is to understand how the specific nucleoprotamine/nucleohistone structure of the sperm nucleus conveys epigenetic information and how it controls early embryonic events.
Cho et al. [57] have shown that both protamines 1 and 2 are essential for sperm function, and the haploinsufficiency of either protamine 1 (P1) or protamine 2 (P2) results in a reduced amount of the respective protein. The phosphorylation of protamines has also been shown to be very important. Indeed, mutation of the calmodulin-dependent protein kinase Camk4, which phosphorylates protamine 2, results in defective spermiogenesis and male sterility [58, 59]. The P1/P2 ratio in fertile men lies close to 1.0 [60–62] and ranges from 0.8 to 1.2 [60, 61]. Perturbation of this ratio, in either direction, is characterized by poor semen quality, increased DNA damage, and decreased fertility [63–68].
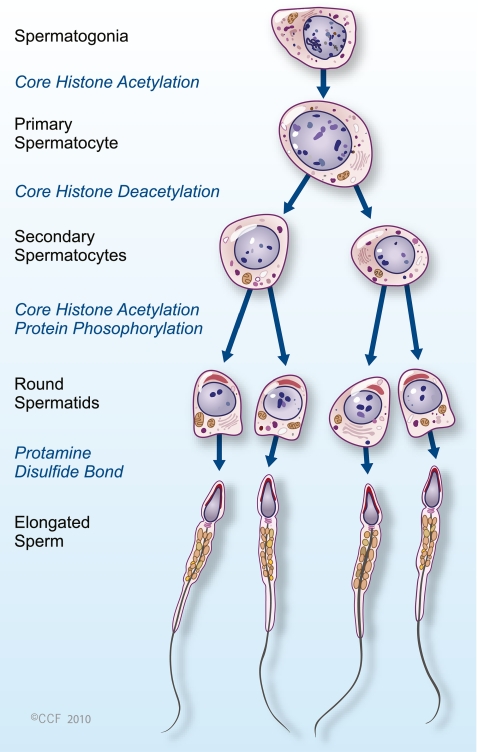
An increasing amount of data now supports the hypothesis that in mature mammalian spermatozoa, DNA is actually not homogeneously packed with protamines [29]. Some histones are retained in humans; in fact, the persistent histones could be an important epigenetic code in the sperm and may not be the result of inefficient protamine replacement [69]. Protamine replacement occurs in the elongating spermatid stage of spermatogenesis (Fig. 3), long after the completion of meiosis [70]. The elongating spermatid also undergoes other maturation events that affect its motility and fertilization ability during the period of protamine replacement; but the events have not been linked to sperm count. This has led to a hypothesis that the link between abnormal protamine replacement and generally diminished semen quality may be due to a defect in the unique system of temporal uncoupling of transcription and translation during spermatogenesis [71].
Fig. 3.
Histone to protamine exchange during spermatogenesis
The study of two relatively novel areas—sperm epigenetics and sperm transcriptome could be of particular interest in men with protamine expression abnormalities. Abnormal protamine incorporation into chromatin may affect transcription of other genes. For example, in mice, deregulation of protamines causes precocious chromatin condensation, transcription arrest, and spermatogenic failure [72]. The human sperm nucleus, which retains 10–15% of its original histone content, distributes it in a heterogeneous manner within the genome [73]. Various studies concluded that histones that are retained bind specific regions, to convey epigenetic information to the early embryo. If so, there are obvious and profound implications for sperm with abnormal protamine replacement, and for the use of such sperm, or any immature sperm, for intracytoplasmic sperm injection (ICSI). Future research will classify and characterize the role of retained histones throughout the sperm genome in mature sperm from fertile men as well as in patients with known chromatin abnormalities.
Epigenetics and ART
In humans, the use of assisted reproductive technology (ART) has been shown to induce epigenetic alterations and to affect fetal growth and development. There is an open debate about the influence ART on certain pathologies, especially those that are dependent on genomic imprinting such as the Beckwith-Wiedemann or Angelman syndrome [74]. Possible epigenetic risks linked to ART techniques may result from either the use of sperm with incomplete reprogramming or from in vitro embryo procedures performed at a time of epigenetic reprogramming [75–78]. Loss of gene imprinting may occur during preimplantation under certain conditions of gamete handling [79]. Although epigenetic states are relatively stable, it has been estimated that the loss of epigenetic control (epimutation) may be one or two orders of magnitude greater than that of somatic DNA mutation [80]. Recent reports from various studies have shown that epimutations not only lead to inappropriate expression of the affected gene but may also expose hidden genetic variation [81]. It is possible that some sub-fertile couples have a genetic predisposition to epigenetic instability, which makes their offspring more susceptible to epigenetic changes, independently of whether or not they are conceived by ART. Epimutations affecting imprints can arise during imprint erasure, imprint establishment or imprint maintenance. It has been suggested by animal model studies that loss-of-function mutations of DNA methyltransferases affect all imprinted domains as well as other chromosomal regions. For example, mutations of Dnmt3a (de-novo methylase) and maintenance methylase (Dnmt) could lead to loss of imprinting and embryonic lethality [82]. Another study has shown that targeted disruption of Dnmt3L in mice caused azoospermia in homozygous mutant males, and heterozygous progeny of homozygous females died before midgestation. Another study suggested that imprinting defects and subfertility can have a common and possibly genetic cause and that super-ovulation instead of ICSI may further increase the risk factor of conceiving a child with an imprinting defect. Based on this study and the study by Chang et al. [83] it is tempting to speculate that super-ovulation leads to the maturation of epigenetically imperfect oocytes that would not have developed without treatment and may disturb the process of DNA methylation in the oocyte. It is therefore suggested that imprinting errors can lead to spontaneous abortions. The influence of sperm DNA methylation on pregnancy was documented in one study where 5-aza-deoxycytidine (5-azaC) was used to induce methylation depletion in mice. DNA methylation level decreases when 5-azaC (a base analog) is incorporated into DNA [84]. It has been proposed that the level of DNA methylation in human sperm could be linked to their ability to initiate pregnancy by assisted reproduction [85].
Epigenetics and testicular cancer
Testicular germ cells tumor (TGCT) represents approximately 98% of all testicular neoplasms and is the most common malignancy among young males [86]. Epigenetic changes that deregulate gene expression are frequently observed during the development of cancer. The epigenetic equilibrium of the normal cell is disrupted during tumorigenesis. In human neoplasms, at least two types of DNA methylation defects are found: hypomethylation, characterized by a global loss of 5-methylcytosine, and hypermethylation of regulatory regions of promoters, associated with the silencing of tumor suppressor genes. Hypomethylation was the first epigenetic abnormality to be identified in cancer cells [87–89]. Studies in mouse models have indicated a causal relation between reduced levels of 5-methylcytosine and tumor formation [90]. In contrast to the mere handful of oncogenes activated by DNA hypomethylation, a long list of tumor suppressor genes is transcriptionally silenced by DNA hypermethylation in cancer cells. TGCTs are believed to arise from primordial germ cells (PGCs)—where DNA methylation and parental imprints are erased and totipotency is restored [19, 91, 92]. A genome-wide DNA methylation study using restriction landmark genome scanning (RLGS) showed that the genome of seminomas was extensively hypomethylated and virtually completely devoid of CpG island hypermethylation [93]. In contrast, the nonseminoma group was less extensively hypomethylated and revealed variable CpG island hypermethylation levels, which were comparable with tumors of other tissues [94]. From embryonic studies in mice, a wave of demethylation immediately after fertilization has been shown to erase the majority of methylation marks in the genome, with the exception of some imprinted genes and repeat sequences [92], leading to totipotency. High expression of the de novo methyltransferases DNMT3A and DNMT3B, as well as their homologue DNMT3L, is significantly associated with the embryonal carcinoma subtype [95]. The presumptive testis-specific chromatin regulator CTCFL (BORIS) and the pluripotency marker POU5F1 (OCT3/4) have recently been proposed to share properties with the cancer/testis associated genes in being hypermethylated in somatic tissue and hypomethylated in normal testis tissue [96, 97].
The effect of epigenetic sperm abnormalities on early embryogenesis
Imprinting errors in the developing fetus have been identified and shown to cause severe pathologies. Some studies have also suggested that the use of ART increases the risk of imprinting diseases. A study by Marques et al. [56] suggested that an increase in abnormal methylation of the H19 gene in oligospermic men is associated with Beckwith-Wiedemann syndrome. A decreased genome-wide methylation pattern in sperm has also been identified with poor embryo quality in rats and decreased IVF pregnancy rates in humans [49]. Benchaib et al. [43] used 5-methyl-cytosine immunostaining as an indicator of genome-wide methylation pattern in sperm. He showed that decreased global methylation in semen samples from normospermic men is related to a poor pregnancy outcome during IVF [85] suggesting that global methylation status independently affects embryogenesis. Mitchell et al. [98] found a correlation between the frequency of P1 transcripts and pregnancy rates in men undergoing testicular sperm extraction (TESE) for ICSI. This could suggest that epigenetic regulation of DNA via nuclear packaging in the sperm is related to the function of the mature sperm. Current knowledge of genetic and epigenetic factors in sperm contributing to poor embryogenesis is limited. Both the complex path of sperm production and the delicate balance of epigenetic and genetic factors during sperm maturation contribute to the formation of a mature sperm with the ability to fertilize an oocyte and contribute to the developing embryo. A defect at any step may manifest as male infertility.
Epigenetic regulation and nutrition
Epigenetic programming is tightly regulated, both temporally and spatially, during fetal development and lactation [19, 20, 99]. Dietary supplementation with a methyl donor during pregnancy increases the proportion of pups carrying a methylated IAP (Intracisternal-A particles) sequence [100, 101]. There are many environmental and metabolic factors that can influence patterns of histone acetylation and DNA methylation, two major epigenetic marks. Metabolic factors influencing these epigenetic modifications include intranuclear levels of acetyl-CoA for HAT activity, NAD+ for Sir2 deacetylases, ATP for the deacetylation of chromatin substrates by at least some HDACs and methyl donors of SAM provided by the folate-methionine pathway. Once the specific epigenetic patterns corresponding to ‘labile’ and ‘locked’ situations are identified, these patterns could be useful for diagnosis and prognosis [102]. They may also represent new types of targets for the development of novel diets and drugs to prevent or to abolish aberrant gene silencing, which may be associated with treatment failure [103, 104].
Most of these studies have looked at modifications of the pattern of DNA methylation, as it is the easiest epigenetic mark to study. Nutrition during early development can influence DNA methylation because one-carbon metabolism is dependent on dietary methyl donors and on co-factors such as methionine, choline, folic acid and vitamin B-12 [104]. The availability of dietary methyl donors and cofactors is therefore very critical during development. The epigenetic change, caused by a decrease in DNMT1 activity, [105] can be prevented by folate supplementation [106]. Although not directly regulated by nutrition, maternal behavior also programs the epigenetic regulation (DNA methylation and histone acetylation) of the gluco-corticoid receptor gene in the hippocampus and determines the stress responses of the offspring [107, 108].
It was recently reported that a methyl-donor-deficient diet in postnatal life can permanently affect the expression of IGF2, resulting in growth retardation [109]. This suggests that the effects of nutrition are not only limited to the fetal stage but nutrition during postnatal development can also permanently alter the epigenetic regulation of imprinted genes. In humans, diet has been shown to affect the DNA methylation status of patients with hyperhomocysteinaemia. This disease is characterized by the accumulation of S-adenosylhomocysteine (an inhibitor of DNA methyltransferases). The impact of diet, nutrients or drugs on early epigenetic programming must be seriously considered to achieve a directed epigenetic regulation in spermatogenesis.
The role of environmental factors in epigenetic modifications
Epigenetic modifications provide a putative link between the environment and alterations in gene expression that might lead to disease phenotype. An increasing body of evidence from animal studies support the role of environmental epigenetics in disease susceptibility. Environmental exposures to nutritional, chemical and physical factors have the potential to alter gene expression and therefore, modify adult disease susceptibility in various ways through changes in the epigenome. These genomic targets contain CpG islands and other DNA sequences, although in some cases the status of histone modifications in the same region, determine levels of gene expression.
Monozygotic twins provide an interesting model for studying the role of environmental factors in epigenetic modifications [110]. A large epigenetic study on monozygotic twins [111] recently showed that twins are epigenetically concordant at birth in most cases, and that epigenetic differences (DNA methylation and histone modifications) accumulate with age in monozygotic twins. Remarkably, the twins displaying the greatest epigenetic differences were found to be those who had lived together for the smallest amount of time. This finding underlines the relative importance of environmental factors in addition to intrinsic factors.
Future perspectives
Whether common diseases have an epigenetic basis is still open to speculation, but if they do, this holds great promise for medicine. Knowledge of genetic and epigenetic modifications of germ cells is necessary for the production of functional gametes and for overcoming infertility. Categorization of infertile men using a more detailed analysis of DNA methylation patterns might reveal a new level of reduced fertilization, implantation or pregnancy rates. Epigenetic studies offer an important window to understanding the role of environmental interactions with the genome in causing disease, and in modulating those interactions to improve human health. Our increasing knowledge over last 10 years is beginning to be translated into new approaches to molecular diagnosis and targeted treatments across the clinical spectrum.
Footnotes
Capsule
Epigenetic mechanisms regulate germ cell development and differentiation. Sperm epigenome is critical for optimal embryogenesis. Sperm from infertile men are prone to epigenetic instability and this may lead to increased incidence of imprinting defects in children conceived by advanced assisted reproductive procedures.
References
- 1.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert SF, Sarkar S. Embracing complexity: organicism for the 21st century. Dev Dyn. 2000;219(1):1–9. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1036>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 3.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 4.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7(10):793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 5.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132(8 Suppl):2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 7.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 8.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 9.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 10.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 12.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 13.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414(6861):277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 14.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21(24):6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A. 2003;100(15):8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124(22):4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 17.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 18.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 20.Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129(2):137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 21.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434(7033):583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 22.McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262(1):1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 23.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278(2):440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Payne C, Braun RE. Histone lysine trimethylation exhibits a distinct perinuclear distribution in Plzf-expressing spermatogonia. Dev Biol. 2006;293(2):461–472. doi: 10.1016/j.ydbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 26.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16(7):660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 27.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10(4):521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102(8):2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, Khochbin S. Establishment of male-specific epigenetic information. Gene. 2005;345(2):139–153. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Oakes CC, Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci U S A. 2007;104(1):228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev. 1992;31(3):170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- 32.Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol. 2000;79(12):950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- 33.Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34(6):384–390. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- 34.Govin J, Lestrat C, Caron C, Pivot-Pajot C, Rousseaux S, Khochbin S. Histone acetylation-mediated chromatin compaction during mouse spermatogenesis. Ernst Schering Res Found Workshop. 2006;57:155–172. doi: 10.1007/3-540-37633-x_9. [DOI] [PubMed] [Google Scholar]
- 35.Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12(5):399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 36.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003;33(1):66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 37.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, Cui H, Niemitz EL, Rasko JE, Docquier FM, Kistler M, Breen JJ, Zhuang Z, Quitschke WW, Renkawitz R, Klenova EM, Feinberg AP, Ohlsson R, Morse HC, 3rd, Lobanenkov VV. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99(10):6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emery BR, Carrell DT. The effect of epigenetic sperm abnormalities on early embryogenesis. Asian J Androl. 2006;8(2):131–142. doi: 10.1111/j.1745-7262.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- 39.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 40.Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertil Steril. 2001;75(2):237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- 41.Kuhnert B, Nieschlag E. Reproductive functions of the ageing male. Hum Reprod Update. 2004;10(4):327–339. doi: 10.1093/humupd/dmh030. [DOI] [PubMed] [Google Scholar]
- 42.Wyrobek AJ, Eskenazi B, Young S, Arnheim N, Tiemann-Boege I, Jabs EW, Glaser RL, Pearson FS, Evenson D. Advancing age has differential effects on DNA damage, chromatin integrity, gene mutations, and aneuploidies in sperm. Proc Natl Acad Sci U S A. 2006;103(25):9601–9606. doi: 10.1073/pnas.0506468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benchaib M, Ajina M, Lornage J, Niveleau A, Durand P, Guerin JF. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: a preliminary study. Fertil Steril. 2003;80(4):947–953. doi: 10.1016/s0015-0282(03)01151-8. [DOI] [PubMed] [Google Scholar]
- 44.Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr Opin Cell Biol. 2007;19(3):257–265. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 45.Ito T. Role of histone modification in chromatin dynamics. J Biochem. 2007;141(5):609–614. doi: 10.1093/jb/mvm091. [DOI] [PubMed] [Google Scholar]
- 46.Biermann K, Steger K. Epigenetics in male germ cells. J Androl. 2007;28(4):466–480. doi: 10.2164/jandrol.106.002048. [DOI] [PubMed] [Google Scholar]
- 47.Schaefer CB, Ooi SK, Bestor TH, Bourc’his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316(5823):398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- 48.Faure AK, Pivot-Pajot C, Kerjean A, Hazzouri M, Pelletier R, Peoc’h M, Sele B, Khochbin S, Rousseaux S. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod. 2003;9(12):757–763. doi: 10.1093/molehr/gag101. [DOI] [PubMed] [Google Scholar]
- 49.Benchaib M, Braun V, Lornage J, Hadj S, Salle B, Lejeune H, Guerin JF. Sperm DNA fragmentation decreases the pregnancy rate in an assisted reproductive technique. Hum Reprod. 2003;18(5):1023–1028. doi: 10.1093/humrep/deg228. [DOI] [PubMed] [Google Scholar]
- 50.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(5551):2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 52.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 53.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438(7066):374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 54.Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, Petronis A. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79(1):67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16(21):2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 56.Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363(9422):1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 57.Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28(1):82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- 58.Wu JY, Means AR. Ca(2+)/calmodulin-dependent protein kinase IV is expressed in spermatids and targeted to chromatin and the nuclear matrix. J Biol Chem. 2000;275(11):7994–7999. doi: 10.1074/jbc.275.11.7994. [DOI] [PubMed] [Google Scholar]
- 59.Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25(4):448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- 60.Balhorn R, Cosman M, Thornton K, Krishnan VV, Corzett M, Bench G, Kramer C, Lee J, IV, Hud NV, Allen M, Priety M, Meyer-IIse W, Brown J, Kirz J, Zhang X, Bradbury E, Maki G, Braun R, Breen W. Protamine mediated condensation of DNA in mammalian sperm. In: Gagnon C, editor. The male gamete: from basic knowledge to clinical applications. Vienna: Cache River Press; 1999. pp. 55–70. [Google Scholar]
- 61.Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl. 2001;22(4):604–610. [PubMed] [Google Scholar]
- 62.Corzett M, Mazrimas J, Balhorn R. Protamine 1: protamine 2 stoichiometry in the sperm of eutherian mammals. Mol Reprod Dev. 2002;61(4):519–527. doi: 10.1002/mrd.10105. [DOI] [PubMed] [Google Scholar]
- 63.Chevaillier P, Mauro N, Feneux D, Jouannet P, David G. Anomalous protein complement of sperm nuclei in some infertile men. Lancet. 1987;2(8562):806–807. doi: 10.1016/s0140-6736(87)92547-5. [DOI] [PubMed] [Google Scholar]
- 64.Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44(1):52–55. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- 65.Belokopytova IA, Kostyleva EI, Tomilin AN, Vorob’ev VI. Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev. 1993;34(1):53–57. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- 66.Carrell DT, Emery BR, Liu L. Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil Steril. 1999;71(3):511–516. doi: 10.1016/s0015-0282(98)00498-1. [DOI] [PubMed] [Google Scholar]
- 67.Razavi S, Nasr-Esfahani MH, Mardani M, Mafi A, Moghdam A. Effect of human sperm chromatin anomalies on fertilization outcome post-ICSI. Andrologia. 2003;35(4):238–243. doi: 10.1046/j.1439-0272.2003.00566.x. [DOI] [PubMed] [Google Scholar]
- 68.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20(5):1298–1306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 69.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 70.Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, Grootegoed JA. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol. 1999;207(2):322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- 71.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update. 2007;13(3):313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 72.Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103(3–4):217–224. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- 73.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236(4804):962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 74.Gosden R, Trasler J, Lucifero D, Faddy M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361(9373):1975–1977. doi: 10.1016/S0140-6736(03)13592-1. [DOI] [PubMed] [Google Scholar]
- 75.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13(8):839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 76.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74(4):599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson JR, Williams CJ. Genomic imprinting and assisted reproductive technology: connections and potential risks. Semin Reprod Med. 2005;23(3):285–295. doi: 10.1055/s-2005-872457. [DOI] [PubMed] [Google Scholar]
- 78.Horsthemke B, Buiting K. Imprinting defects on human chromosome 15. Cytogenet Genome Res. 2006;113(1–4):292–299. doi: 10.1159/000090844. [DOI] [PubMed] [Google Scholar]
- 79.Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131(15):3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 80.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165(4):2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33(1):70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 82.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 83.Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83(2):349–354. doi: 10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelly TL, Li E, Trasler JM. 5-aza-2′-deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl. 2003;24(6):822–830. doi: 10.1002/j.1939-4640.2003.tb03133.x. [DOI] [PubMed] [Google Scholar]
- 85.Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, Guerin JF. Influence of global sperm DNA methylation on IVF results. Hum Reprod. 2005;20(3):768–773. doi: 10.1093/humrep/deh684. [DOI] [PubMed] [Google Scholar]
- 86.Bosl GJ, Motzer RJ. Testicular germ-cell cancer. N Engl J Med. 1997;337(4):242–253. doi: 10.1056/NEJM199707243370406. [DOI] [PubMed] [Google Scholar]
- 87.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 88.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 89.Ehrlich M. The controversial denouement of vertebrate DNA methylation research. Biochemistry (Mosc) 2005;70(5):568–575. doi: 10.1007/s10541-005-0150-z. [DOI] [PubMed] [Google Scholar]
- 90.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 91.Bestor TH. Cytosine methylation and the unequal developmental potentials of the oocyte and sperm genomes. Am J Hum Genet. 1998;62(6):1269–1273. doi: 10.1086/301891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 93.Lind GE, Skotheim RI, Lothe RA. The epigenome of testicular germ cell tumors. APMIS. 2007;115(10):1147–1160. doi: 10.1111/j.1600-0463.2007.apm_660.xml.x. [DOI] [PubMed] [Google Scholar]
- 94.Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomaki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21(24):3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 95.Almstrup K, Hoei-Hansen CE, Nielsen JE, Wirkner U, Ansorge W, Skakkebaek NE, Rajpert-De Meyts E, Leffers H. Genome-wide gene expression profiling of testicular carcinoma in situ progression into overt tumours. Br J Cancer. 2005;92(10):1934–1941. doi: 10.1038/sj.bjc.6602560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoffmann MJ, Muller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol. 2006;72(11):1577–1588. doi: 10.1016/j.bcp.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 97.Jong J, Looijenga LH. Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: history and future. Crit Rev Oncog. 2006;12(3–4):171–203. doi: 10.1615/critrevoncog.v12.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 98.Mitchell V, Steger K, Marchetti C, Herbaut JC, Devos P, Rigot JM. Cellular expression of protamine 1 and 2 transcripts in testicular spermatids from azoospermic men submitted to TESE-ICSI. Mol Hum Reprod. 2005;11(5):373–379. doi: 10.1093/molehr/gah169. [DOI] [PubMed] [Google Scholar]
- 99.Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129(8):1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 100.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100(5):2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20(1):63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 102.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 103.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 104.MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, Hsu JL, Janke SM, Pham TD, Lane RH. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics. 2004;18(1):43–50. doi: 10.1152/physiolgenomics.00042.2004. [DOI] [PubMed] [Google Scholar]
- 105.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97(6):1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135(6):1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 107.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 108.Meaney MJ, Szyf M, Seckl JR. Epigenetic mechanisms of perinatal programming of hypothalamic-pituitary-adrenal function and health. Trends Mol Med. 2007;13(7):269–277. doi: 10.1016/j.molmed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 109.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15(5):705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 110.Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61(5 Pt 2):38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- 111.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]