CONSPECTUS
Tens of thousands of terpenoids are present in both terrestrial and marine plants as well as fungi. In the last 5–10 years, however, it has become evident that terpenes are also produced by numerous bacteria, especially soil-dwelling Gram-positive organisms such as Streptomyces and other Actinomycetes. Although some microbial terpenes, such as geosmin—the degraded sesquiterpene responsible for the smell of moist soil, the characteristic odor of the earth itself—have been known for over 100 years, few terpenoids have been identified by classical structure- or activity-guided screening of bacterial culture extracts. In fact, the majority of cyclic terpenes from bacterial species have only recently been uncovered by the newly developed techniques of “genome mining.” In this new paradigm for biochemical discovery, bacterial genome sequences are first analyzed with powerful bioinformatic tools, such as the BLASTP program or Profile Hidden Markov models, to screen for and identify conserved protein sequences harboring a characteristic set of universally conserved functional domains typical of all terpene synthases. Of particular importance is the presence of variants of two universally conserved domains, the aspartate-rich DDXX(D/E) motif and the NSE/DTE triad, (N/D)DXX(S/T)XX(K/R)(D/E). Both domains have been implicated in the binding of the essential divalent cation, typically Mg2+, that is required for cyclization of the universal acyclic terpene precursors, such as farnesyl and geranyl diphosphate.
The low level of overall sequence similarity among terpene synthases, however, has so far precluded any simple correlation of protein sequence with the structure of the cyclized terpene product. The actual biochemical function of a cryptic bacterial (or indeed any) terpene synthase must therefore be determined by direct experiment. Two common approaches are (i) incubation of the expressed recombinant protein with acyclic allylic diphosphate substrates and identification of the resultant terpene hydrocarbon or alcohol and (ii) in vivo expression in engineered bacterial hosts that can support the production of terpene metabolites. One of the most attractive features of the coordinated application of genome mining and biochemical characterization is that the discovery of natural products is directly coupled to the simultaneous discovery and exploitation of the responsible biosynthetic genes and enzymes.
Bacterial genome mining has proved highly rewarding scientifically, already uncovering more than a dozen newly identified cyclic terpenes (many of them unique to bacteria) as well as several novel cyclization mechanisms. Moreover, bioinformatic analysis has identified more than 120 presumptive genes for bacterial terpene synthases that are now ripe for exploration. In this Account, we review a particularly rich vein we have mined in the genomes of two model Actinomycetes, Streptomyces coelicolor and Streptomyces avermitilis, from which the entire set of terpenoid biosynthetic genes and pathways have now been elucidated. In addition, studies of terpenoid biosynthetic gene clusters have revealed a wealth of previously unknown oxidative enzymes, including cytochromes P450, non-heme iron-dependent dioxygenases, and flavin monooxygenases. We have shown that these enzymes catalyze a variety of unusual biochemical reactions, including 2-step ketonization of methylene groups, desaturation–epoxidation of secondary methyl groups, and pathway-specific Baeyer–Villiger oxidations of cyclic ketones.
Introduction
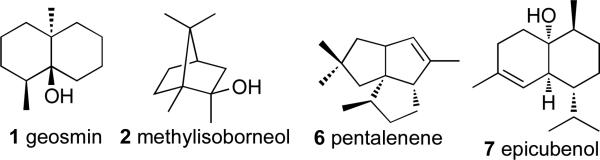
Streptomyces and other Actinomycetes are Gram-positive, soil-dwelling organisms that are prolific producers of an enormous variety of natural products. Until recently, however, reports of the isolation of terpenoid metabolites from Actinomycetes and other bacteria had been surprisingly rare. The first terpene to be isolated from Streptomyces was the degraded sesquiterpene geosmin (1), which is responsible for the characteristic odor of moist soil (Figure 1).1,2 In addition to detecting geosmin in 17 different species of Streptomyces, a cyanobacterium and a myxobacterium, Gerber also isolated another common metabolite with a characteristic musty, camphor-like odor, identified as 2-methylisoborneol (2). In 1981, Bentley reported the first evidence for the likely isoprenoid origin of both geosmin and 2-methylisoborneol produced by Streptomyces antibioticus.3
FIGURE 1.
Bacterial terpenes.
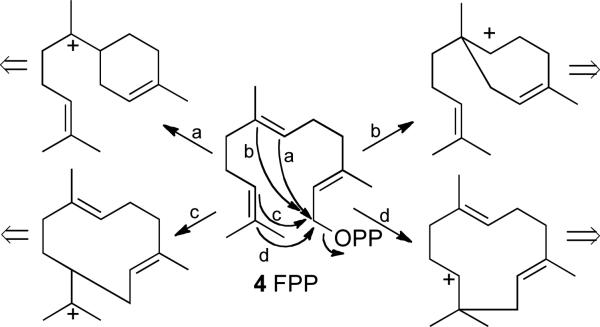
The several hundred parent cyclic monoterpene, sesquiterpene, and diterpene hydrocarbons and alcohols are all formed by variations of a common cyclization mechanism initiated by enzyme-catalyzed ionization of the universal acyclic precursors geranyl (3, GPP), farnesyl (4, FPP), and geranylgeranyl diphosphate (GGPP), respectively (Scheme 1). Intramolecular attack of the resultant allylic cation on the central or distal double bonds followed by well-precedented cationic transformations and eventual quenching of the positive charge by deprotonation or by capture of water can account for the formation of the enormous variety of cyclic terpenes.
SCHEME 1.
Cyclization of farnesyl diphosphate (4).
Bacterial Terpene Synthases
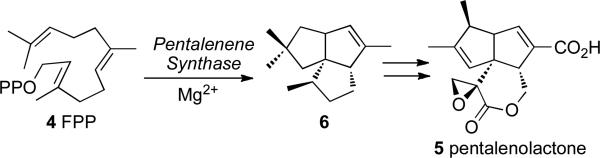
Pentalenolactone (5) is a sesquiterpenoid antibiotic that has been isolated from more than 30 species of Streptomyces.4,5 Pentalenene synthase, which we first isolated from Streptomcyes exfoliatus UC5319 and subsequently cloned and expressed in Escherichia coli,6 catalyzes the cyclization of farnesyl diphosphate (4) to the parent sesquiterpene hydrocarbon pentalenene (6) (Scheme 2). Extensive experiments with stereospecifically labeled FPP have established the detailed mechanism and stereochemistry of the cyclization.7 The crystal structure of pentalenene synthase has also been determined.8 The only other bacterial terpene synthase to have been isolated by analogous biochemical assay-guided methods is epicubenol (7) synthase from Streptomyces sp. LL-B7.9
SCHEME 2.
Pentalenene synthase and the biosynthesis of pentalenolactone.
“Genome mining” has proved to be a powerful paradigm for the discovery and characterization of natural product biosynthetic genes.10 The actual biochemical function of a candidate terpene synthase can be assigned by incubation of the recombinant cyclase with the GPP, FPP, or GGPP and identification of the resulting monoterpene, sesquiterpene, or diterpene product. This in vitro approach, which simultaneously provides enhanced levels of protein for mechanistic, mutational, and structural studies, can be powerfully complemented by in vivo expression of individual genes or clusters of terpenoid biosynthetic genes in suitably engineered host bacteria, followed by identification of the newly generated metabolic products. In this Account, we review our recent application of genome mining to the discovery of new bacterial terpene synthases and the elucidation of terpenoid biosynthetic pathways, focusing on two prototypical Actinomycetes, S. coelicolor and S. avermitilis.
Terpene Synthase Bioinformatics
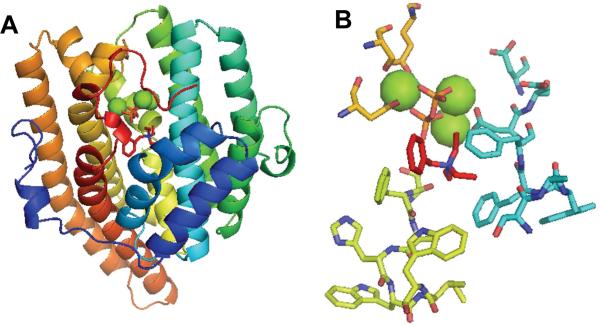
Within the more than 20 Actinomycete genome sequences reported to date are more than 100 presumed or confirmed terpene synthases. Bioinformatic analysis alone has been incapable of assigning a specific biochemical function to most newly recognized terpene synthase genes. In spite of the very substantial differences in overall primary amino acid sequence, terpene synthases typically display two highly conserved Mg2+-binding domains: an aspartate-rich motif, DDXX(D/E) or DDXXX(D/E), usually found ~80-120 amino acids (aa) downstream of the N-terminus of microbial synthases, and a second “NSE/DTE” triad, (N/D)DXX(S/T)XX(K/R)(D/E), located 140±5 aa downstream of the aspartate-rich motif.11 Crystallographic studies of nearly a dozen monoterpene and sesquiterpene synthases from bacterial, fungal, and plant sources have established that these two motifs, which are located at opposite sides of the rim of the deep active site cavity, are responsible for cooperative binding of three divalent cations and the pyrophosphate moiety of the substrate, precisely positioning the acyclic allylic diphosphate substrate and activating it for the ionization that triggers the cyclization cascade (Figure 2).8,12,13 Despite the significant differences in primary sequence, these terpene cyclases also all share a common α-helical fold. The detailed contour of the cyclase active site is thought to play a major role in the proper folding of the allylic diphosphate substrate and chaperoning the highly reactive carbocationic intermediates so as to control the formation of the characteristic cyclic terpene product.12
FIGURE 2.
Structure of a typical terpene synthase. A. Epi-isozizaene synthase in complex with three Mg2+ ions (green spheres), inorganic pyrophosphate (orange), and the benzyltriethylammonium cation (red), showing universal Type I all-α-helical terpene synthase fold. B. Detail of active site, including conserved 99DDXXD (cyan) and 240NXXXSXXXE (yellow) motifs as well as aromatic side chains (cyan and lemon) lining the active site cavity
While local sequence alignment searches using the BLAST algorithm can recognize many presumptive terpene synthases, they may still miss matches of low overall sequence similarity. Hidden Markov Models (HMMs) provide a powerful means of codifying the underlying pattern of functional domains and searching for proteins of similar function even in the absence of significant levels of primary sequence similarity.14,15 The HMM parameters are first estimated from a training set of protein sequences that can include multiple alignments that incorporate three-dimensional structural information. The resulting profile HMM can distinguish members of the relevant protein functional families from non-members with a high degree of accuracy.16-18
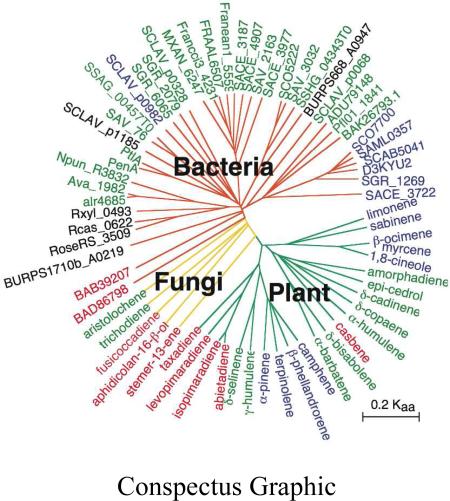
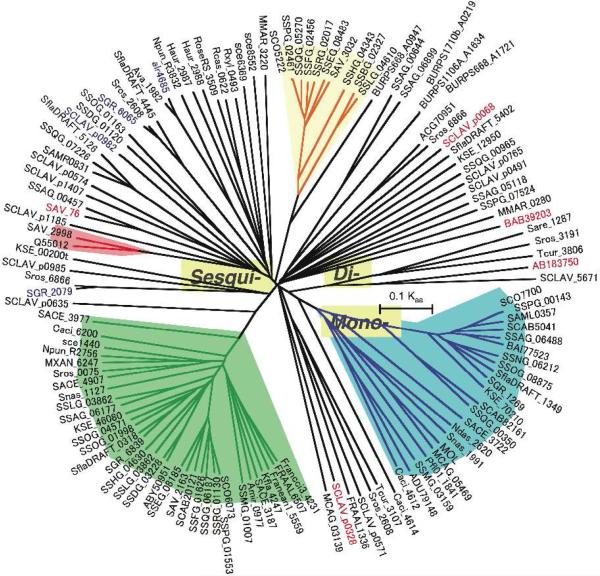
We first used a profile Hidden Markov Model from the Pfam database (Pfam entry PF03906), based on the two universally conserved terpene synthase family metal-binding domains, to search the 2008 NCBI database of bacterial genome sequences in order to harvest all the presumptive terpene cyclase sequences.19 From 1,922,990 predicted proteins, 41 proteins initially selected as strong matches were classified into three distinct groups on the basis of phylogenetic analysis. Group I contained 12 protein sequences provisionally assigned as monoterpene cyclases. Group II, by far the largest group of 27 protein sequences, included the known sesquiterpene cyclases pentalenene synthase, germacradienol/geosmin synthase, and epi-isozizaene synthase (see below) and close orthologues as well as several presumptive sesquiterpene synthases of unknown biochemical function. Group III consisted of two diterpene synthase sequences, including a cyclase implicated in terpentecin biosynthesis. These first-pass harvested synthases have since been used to generate a second-generation model specific for bacterial terpene synthases. A phylogenetic tree illustrating the grouping of over 120 candidate bacterial sequences is shown in Figure 3.
FIGURE 3.
Phylogenetic tree of bacterial terpene synthases. Synthases are each labeled by their proton-coding/gene names. The green-colored area indicates a clade of germacradienol/geosmin synthases, the red-colored area corresponds to pentalenene synthases, beige-shaded regions are epi-isozizaene synthases and the blue-colored zone contains 2-methylisoborneol synthases or 2-methylenebornane synthases. SAV_76 is avermitilol synthase, SCLAV_p0068 (SSCG_03688) is (+)-T-muurolol synthase, SCLAV_0328 (SSCG_02150) is (-)-δ-cadinene synthase, BAB39203 is terpentecin synthase and AB183750 is pimara-9(11),15-diene synthase
In vitro and in vivo investigations of terpene synthases and terpenoid biosynthetic pathways
The majority of Streptomyces proteins, including terpene synthases, can be well-expressed in E. coli using either native or synthetic genes. Incubation of the purified recombinant terpene synthase with FPP, GPP or GGPP generates a cyclized terpene hydrocarbon or alcohol whose structure can be assigned by GC-MS comparison with known standards or by detailed NMR analysis. A complementary in vivo approach is to use a metabolically engineered E. coli host designed for the efficient expression of terpenoid metabolites.20 We have also developed engineered strains of S. avermitilis from which ~1 MB of DNA harboring the majority of natural product biosynthetic genes have been deleted.21-23 These so-called “SUKA” mutants are robust natural product factories that retain the capability of the parent industrial S. avermitilis strain to synthesize biological building blocks and export biosynthetic end-products. Indeed, the SUKA mutants are excellent hosts for the introduction of individual biosynthetic genes or even entire biosynthetic gene clusters from Streptomyces or even eukaryotic sources, allowing determination of their biochemical function. The resultant transformants can also be used for the production of multi-mg quantities of otherwise inaccessible, complex biosynthetic intermediates.
Case Studies
I. Terpene Synthases of Streptomyces coelicolor A3(2)
Geosmin synthase
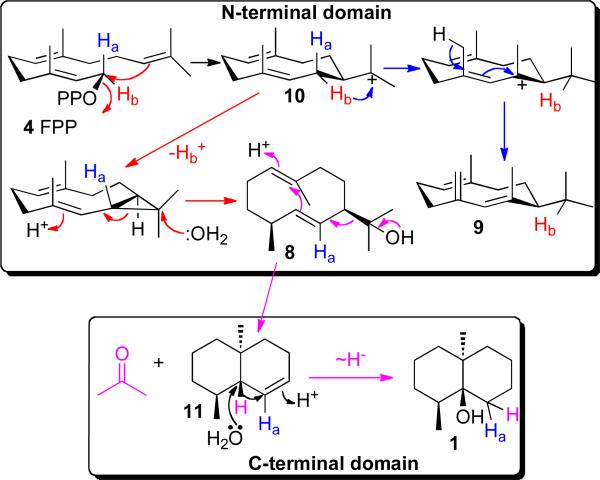
The 8.6-Mb linear genome of S. coelicolor A3(2), usually considered the model Streptomyces organism, harbors 7,825 predicted genes,24 of which three correspond to predicted terpene synthases. The 2181-bp sco6073 gene encodes an unusually large 726-aa protein in which both the N-terminal (366 aa) and C-terminal (339 aa) halves showed ~30% identity to pentalenene synthase. The N-terminal domain harbors variants of the characteristic conserved Mg2+-binding domains 86DDHFLE and 229NDLFSYQRE, while the C-terminal half displays an unusual 455DDYYP motif as well as a canonical 598NDVFSYQKE sequence. We initially found that the full-length recombinant protein expressed in E. coli catalyzed the Mg2+-dependent cyclization of FPP to an 85:15 mixture of (4S,7R)-germacra-1(10)E,5E-diene-11-ol (8) and (7S)-germacrene D (9) (Scheme 3).25,26 Incubations with chirally deuterated FPP established that both sesquiterpene products resulted from partitioning of a common germacradienyl cation intermediate 10.27
SCHEME 3.
Cyclization of farnesyl diphosphate by the bifunctional geosmin synthase, SCO6073.
Independently, Gust et al reported that in-frame deletion of the entire sco6073 gene resulted in complete loss of production of geosmin.28 The combined in vitro biochemical and in vivo molecular genetic evidence thus established that the sco6073 gene encodes a germacradienol synthase and that formation of germacradienol must be an essential step in the formation of geosmin. We then unexpectedly discovered that incubation of FPP with germacradienol synthase also produced geosmin.29 Using both the recombinant C-terminal domain of SCO6073 protein as well as the full-length protein with site-specific mutations in either of the two Mg2+-binding motifs of the C-terminal domain, we demonstrated that geosmin synthase is in fact a bifunctional protein (Scheme 3).30 The N-terminal domain catalyzes the cyclization of FPP to germacradienol (8) and germacrene D (9) while the C-terminal domain is responsible for the proton-initiated sequential retro-Prins fragmentation of germacradienol (8) with loss of the 2-propanol side-chain as acetone to give the intermediate octalin 11 that is then converted to geosmin (1).25,31,32
The geosmin synthase gene is highly conserved, with more than 50 orthologues found in the genomes of a variety Actinomycetes, myxobacteria, and cyanobacteria that display 45-99% identity to SCO6073.33,34 Incubation of recombinant SAV_2163 protein (GeoA) from S. avermitilis with FPP produced a mixture of germacradienol and geosmin while the geoA mutant no longer produced geosmin.35 Curiously, the erythromycin producer Saccharopolyspora erythraea harbors three distinct geosmin synthase orthologs. The production of geosmin and 2-methylisoborneol by cyanobacteria is responsible for episodes of unpleasant taste and odor in public water supplies as well as an off-taste in fish and other products of aquaculture. We have identified the geosmin synthase of the model cyanobacterium Nostoc punctiforme by incubation of the recombinant protein with FPP.36 In spite of the ubiquity of geosmin-producing microorganisms and a variety of imaginative suppositions as to the possible role of this volatile organic metabolite (for example in attracting various species, from earthworms to camels),37 there has been only limited experimental support for these speculations. For example glass eels (Anguila anguila), which migrate from salt water to fresh water, are strongly attracted by geosmin which is thus thought to serve as an important inland water marker (although this would have no obvious evolutionary benefit to Streptomcyes).38
2-Methylisoborneol synthase
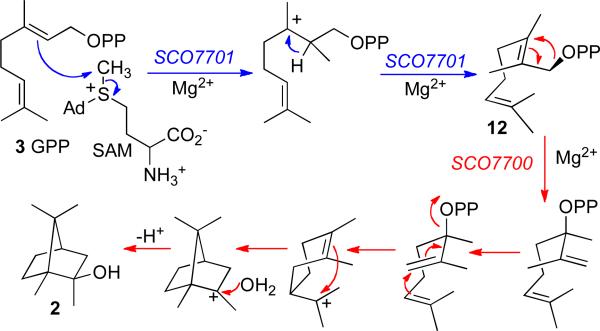
The 1323-bp sco7700 gene encodes a 440-aa protein with <20% identity to pentalenene synthase that incorporates variants of the two characteristic terpene synthase Mg2+-binding motifs, an unusual 197DDCYCED acidic motif and a more conventional downstream NSE triad 345NDLYSYTKE. Intriguingly, the 3′-end of the sco7700 gene is separated by only 16-bp from the downstream coding sequence, sco7701, which had been annotated only as a generic C-methyltransferase. Recalling that Bentley had reported that 2-methylisoborneol is labeled by [14C-methyl]methionine,3 it appeared that we might be dealing with a 2-gene biosynthetic operon for the formation of 2-methylisoborneol (2). Indeed, incubation of a reconstituted mixture of recombinant SCO7700 and SCO7701 with GPP (3) and S-adenosyl-L-methionine (SAM) gave 2-methylisoborneol (2) as the major product (Scheme 4).39 Incubation of GPP and SAM with recombinant SCO7701 gave exclusively the previously unknown acyclic substrate (E)-2-methylgeranyl diphosphate (12, 2-MeGPP). Finally, direct incubation of synthetic 12 with SCO7700 gave 2-methylisoborneol (2). These results are consistent with the reported incorporation of labeled mevalonate and methionine into 2-methylisoborneol by the myxobacterium Nannocystis exedens as well as the isolation of 2-methylgeraniol from this organism.40
SCHEME 4.
Biosynthesis of 2-methylisoborneol.
In the meantime, the Kitasato group had carried out an independent analysis of bacterial 2-methylisoborneol biosynthesis.19 Significantly, the majority of the 12 genes for the Group I proteins tentatively identified as monoterpene synthases by the profile HMM search (Figure 3) were flanked by a gene encoding a predicted SAM-dependent C-methyltransferase. Indeed, five of the strains harboring these gene pairs produced 2-methylisoborneol, while a sixth, Micromonospora olivasterospora, generated the corresponding dehydrated homo-monoterpene, 2-methylenebornane (13, 2-MB). Furthermore, when each of the two-gene segments from Streptomyces ambofaciens, Streptomyces lasaliensis, and Sac. erythraea were inserted into S. avermitilis SUKA16 downstream of the strong, constitutive rpsJ (sav4925) promoter, the resultant exconjugants each produced 2-methylisoborneol. Finally, incubation of recombinant S. lasaliensis 2-methylisoborneol synthase and the 2-MeGPP synthase with GPP and SAM gave 2-methylisoborneol (2), while incubation of the same substrates in the absence of the monoterpene cyclase, followed by phosphatase treatment gave (E)-2-methylgeraniol. The preliminary X-ray crystallographic analysis of the S. lasaliensis 2-MeGPP synthase has recently been reported.41 A new profile HMM for the region around two conserved metal-binding motifs of microbial terpene synthases also identified a single open reading frame of 397 aa within the genome sequence of the cyanobacterium, Pseudanabaena limonetica.42 Heterologous expression of the corresponding gene product has confirmed the identity of the cyanobacterial 2-methylisoborneol synthase as well as the coupled 2-MeGPP synthase.
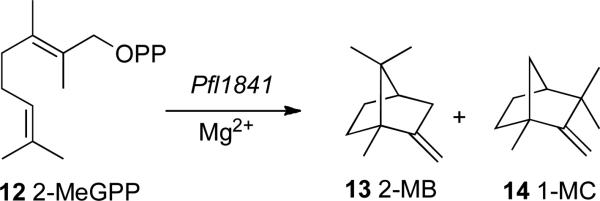
We have recently found that the Pfl_1841 protein of Pseudomonas fluorescens PfO-1 catalyzes the cyclization of 2-MeGPP (12) to a mixture of 2-methylenebornane (13) and 1-methylcamphene (14) (Scheme 5).43 A truncated recombinant form of M. olivasterospora 2-methylenebornane synthase, which is clustered in the phylogenetic tree with Pfl_1841 (Figure 3), also catalyzes the formation of 2-methylenebornane from 2-MeGPP (H. Ikeda, unpublished).
SCHEME 5.
Biosynthesis of 2-methylenebornane (13) and 1-methylcamphene (14)
Epi-isozizaene synthase and the biosynthesis of albaflavenone
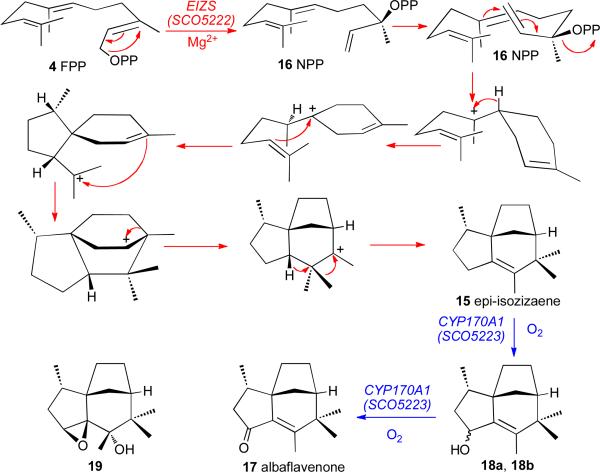
The S. coelicolor A3(2) sco5222 gene encodes a 361-aa protein with only 24% identity to S. exfoliatus pentalenene synthase. The encoded SCO5222 protein harbors two simple variants of the conserved Mg2+-binding domains, an aspartate-rich 99DDRHD and the downstream triad 240NDLCSLPKE. Incubation of the recombinant protein with FPP gave as the major product a novel tricyclic sesquiterpene hydrocarbon identified as (+)-epi-isozizaene (15) (Scheme 6).44 Using stereospecifically deuterated FPP showed that the cyclization to give 15 takes place with net retention of configuration at C-1 of FPP through the demonstrated intermediacy of (3R)-nerolidyl diphosphate (16, NPP) (Scheme 6).44,45 The 1.60 Å resolution X-ray crystal structure of recombinant S. coelicolor A3(2) epi-isozizaene synthase in complex with three Mg2+ ions, inorganic pyrophosphate, and the carbocation analogue benzyltriethylammonium cation (BTAC) showed the universal Class I α-helical terpene synthase fold in a closed conformation with an active site contour that closely complements the natural epi-isozizaene product (Figure 2).46
SCHEME 6.
Biosynthesis of epi-isozizaene (15) and albaflavenone (17)
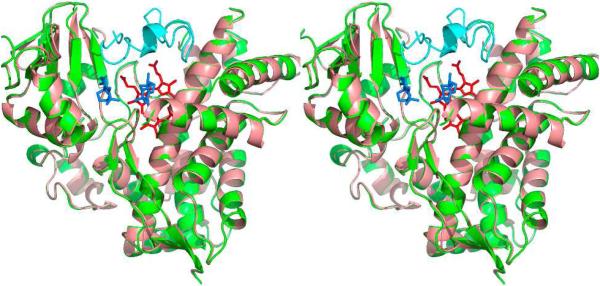
The S. coelicolor A3(2) sco5222 gene shares a four-nucleotide ATGA transcriptional overlap at its 3′-end with the sco5223 gene encoding the cytochrome P450 CYP170A1. Purified recombinant CYP170A1 catalyzed an unusual series of two consecutive allylic oxidations to convert epi-isozizaene (15) to the camphoraceous antibiotic albaflavenone (17) by way of an epimeric mixture of albaflavenols (18a, 18b) (Scheme 6).47 GC-MS analysis of S. coelicolor A3(2) cultures established the time-dependent appearance of epi-isozizaene (15), both albaflavenols (18a, 18b), and albaflavenone (17), while disruption of the CYP170A1 gene abolished formation of all three oxidation products. The crystal structure of CYP170A1 revealed that epi-isozizaene binds in the active site in two orientations with respect to the heme (Figure 4).48
FIGURE 4.
Stereoview of overlaid ligand-free CYP170A1 (SCO5223) (salmon) and ligand-bound (epi-isozizaene - blue) structures (marine) (heme in red). The closed BC loop in the epi-isozizaene complex structure is highlighted in cyan. (Reproduced with permission from Figure 1, ref. 48)
The orthologous SAV_3032 protein of S. avermitilis also catalyzed the expected cyclization of FPP to epi-isozizaene.23 Importantly, sav3032 also shares a 4-nucleotide ATGA overlap with the downsteam sav3031 gene encoding a 456-aa cytochrome P450, CYP170A2. When this sav3032 gene was placed under control of the rpsJ promoter in S. avermitilis SUKA16, the resultant transformants accumulated epi-isozizaene. Coexpression of the sav3031 gene for CYP170A2 generated both (4R)- and (4S)-albaflavenol (18a, 18b) and albaflavenone (17), as well as a previously unknown, epoxy-alcohol 19.
The two-gene operon for albaflavenone biosynthesis is highly conserved and apparently widely distributed, with orthologous pairs of transcriptionally coupled synthase/P450 genes with 56-100% identity evident in ten species of Streptomyces, nearly half of all those with reported genome sequences.23
II. Terpene Synthases of Streptomyces avermitilis MA-4680
Streptomyces avermitilis is responsible for the production of the widely used anthelmintic polyketide avermectins. The 9.03-Mb linear genome of S. avermitilis harbors four terpene synthases among its complement of 7575 predicted proteins.49 In addition to geosmin synthase (SAV_2163)40 and epi-isozizaene synthase (SAV_3032),23 these include a newly discovered cyclase, avermitilol synthase (SAV_76), and a pentalenene synthase (PtlA, SAV_2998).
Avermitilol synthase
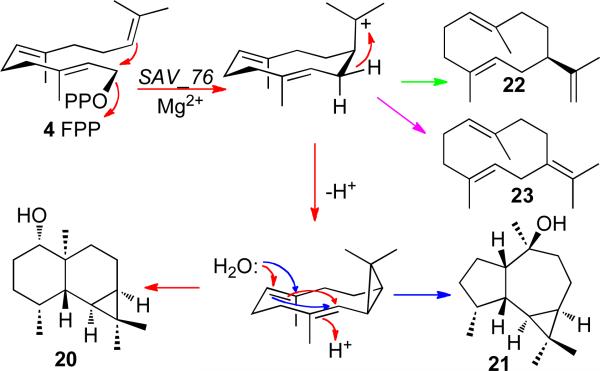
The sav76 gene of S. avermitilis encodes a 335-aa protein with 35% identity to S. exfoliatus pentalenene synthase harboring the conserved asparate-rich 80DDQFD and the triad 239NDVYSLEKE. Other than a single predicted ortholog from Streptomyces sp. Mg1 (78% identity), there are no other significant matches among predicted bacterial gene products. Incubation of the purified recombinant SAV76 protein with FPP gave a mixture consisting of a novel sesquiterpene alcohol, avermitilol (20, 85%), accompanied by the known isomer viridiflorol (21, 3%) as well as germacrenes A (22, 1%) and B (23, 5%) (Scheme 7).22 The mechanism and stereochemistry of this unusual cyclization has been experimentally established. Cultures of S. avermitilis SUKA17 harboring sav76 under control of the rpsJ promoter produced avermitilol (15%) along with viridiflorol (2%) and the oxidized derivative of 20, avermitilone (67%).
SCHEME 7.
Cyclization of FPP to avermitilol (20).
Pentalenene synthase and discovery of the neopentalenolactone biosynthetic pathway
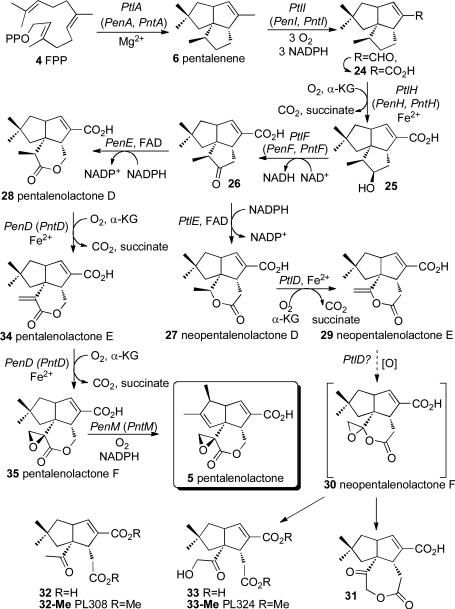
The sav_2998 gene of S. avermitilis encodes a 336-aa protein (PtlA) with 76% identity to the well-characterized pentalenene synthase of S. exfoliatus UC5319. Incubation of recombinant PtlA with FPP gave pentalenene (6).50 The ptlA gene is itself located within a 13.4-kb ptl gene cluster encoding 13 predicted protein coding sequences (CDSs)that we initially thought might represent the set of biosynthetic genes for pentalenolactone itself (Figure 5). Indeed, we established that sav2990 (gap1), located at the 5′-end of the cluster, is a resistance gene that encodes a pentalenolactone-insensitive glyceraldehyde-3-phosphate dehydrogenase.50 By systematic expression of each of the individual CDSs of the ptl gene cluster, we demonstrated that PtlI (SAV_2999) is a cytochrome P450 catalyzing the 3-step oxidation of pentalenene to pentalenal and, almost certainly, to 1-deoxypentalenic acid (24),51 that PtlH (SAV_2991) is an α-ketoglutarate- and non-heme iron-dependent dioxygenase that mediates the 11-β-hydroxylation of 24 to give 25,52,53 and that PtlF (SAV_2993) catalyzes the NAD+-dependent dehydrogenation of 25 to the cyclopentanone 26 (Scheme 8).54
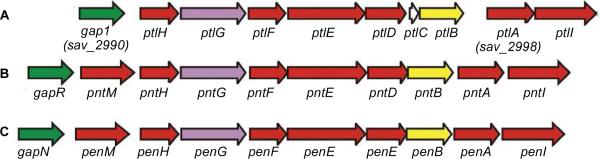
FIGURE 5.
Neopentalenolactone and pentalenolactone biosynthetic gene clusters. (A) S. avermitilis ptl gene cluster for neopentalenolactone biosynthesis. (B) Streptomyces arenae pnt gene cluster for pentalenolactone biosynthesis. (C) S. exfoliatus pen gene cluster for pentalenolactone biosynthesis. Biosynthetic genes in red, pentalenolactone-insensitive gapdh in green, FPP synthase in yellow.
SCHEME 8.
Biosynthesis of pentalenolactone (5) and neopentalenolactone F (30).
Unexpectedly, however, incubation of 26 with the flavin-dependent Baeyer-Villiger monooxygenase PtlE (SAV_2994) gave the previously unknown metabolite neopentalenolactone D (27), an isomer of the expected product pentalenolactone D (28).55 We have also established that PtlD (SAV_2995) is an α-ketoglutarate- and non-heme iron-dependent dioxygenase that catalyzes the desaturation of 28 to a second novel metabolite, neopentalenolactone E (29) (Scheme 8).56 The S. avermitilis ptl gene cluster is thus likely responsible for biosynthesis of a previously unknown metabolite, predicted to be neopentalenolactone F (30). In fact, cultures of S. avermitilis SUKA5 were found to produce neopentalenoketolactone (31), most likely resulting from rearrangement of 30.55 When the ptl gene cluster was placed under control of the strong constitutive ermE promoter, the resulting cultures of SUKA16 accumulated two new metabolites, 32 and 33, characterized as the derived methyl esters PL308 (32-Me) and PL324 (33-Me), that are presumably formed by hydrolysis of 29 and 30, respectively. Notably, the S. avermitilis ΔptlD mutant was blocked in formation of 29, 32, and 33, but still produced neopentalenolactone D (27), while the corresponding double deletion mutant, S. avermitilis ΔptlEΔptlD, lacked 27, accumulating instead its precursor, ketone 26 (Scheme 8).
III. Pentalenolactone Biosynthesis in S. exfoliatus UC5319 and Streptomyces arenae TU469
We have cloned and sequenced the complete pentalenolactone biosynthetic gene clusters from two established producers of the antibiotic, S. exfoliatus UC5319 and S. arenae TU469.56 Each of the two clusters harbors a homologous set of 10 unidirectionally transcribed CDSs, including close orthologs of each of the previously characterized gene products in the S. avermitilis ptl gene cluster (Figure 5).
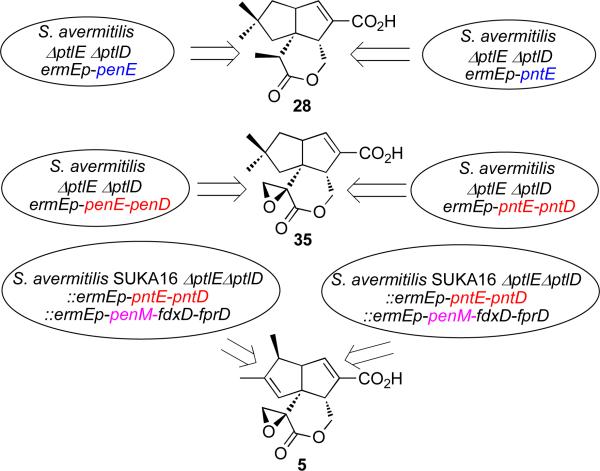
The recombinant enzymes PenE and PntE from S. exfoliatus and S. arenae, respectively, each catalyze the flavin-dependent, Baeyer-Villiger oxidation of 26 exclusively to pentalenolactone D (28). Introduction of the corresponding penE and pntE genes into the double deletion mutant, S. avermitilis ΔptlEΔptlD, resulted in heterologous production of 28 (Figure 6). Recombinant PenD and PntD each catalyzed the α-ketoglutarate- and non-heme iron-dependent, 2-step desaturation/epoxidation of 28 to pentalenolactones E (34) and F (35) (Scheme 8). The corresponding S. exfoliatus ΔpenD and S. arenae ΔpntD deletion mutants both accumulated 28 but were unable to produce either 34, 35, or 5, while complementation of the S. avermitilis ΔptlEΔptlD double deletion mutant with either penE plus penD or pntE plus pntD led to heterologous production of pentalenolactone F (35) (Figure 6).
FIGURE 6.
Engineered biosynthesis of pentalenolactone metabolites by complementation of S. avermitilis SUKA ΔptlEΔptlD mutants.
The recombinant cytochromes P450, PenM and PntM, each catalyzed the unusual oxidative rearrangement of 35 to pentalenolactone (5) (Scheme 8).57 In support of these observations, the S. exfoliatus ΔpenM and S. arenae ΔpntM deletion mutants both accumulated 35 and were blocked in the formation of pentalenolactone, which could be restored by complementation with either penM or pntM. Similarly, complementation of the S. avermitilis ΔptlEΔptlD::ermE-pntE-pntD mutant with penM or pntM also resulted in the production of pentalenolactone (5) (Figure 6).
Conclusions
Mining of the rapidly emerging number of bacterial genome sequences has already uncovered a treasure trove of new terpene synthases,58-63 previously unknown terpene metabolites, and novel variations on the universal terpene cyclization mechanisms while providing powerful genetic and structural tools to investigate the biological roles of these ubiquitous yet still poorly understood natural products. The combined application of bioinformatic analysis, microbial genetics, mechanistic enzymology, and structural biology should continue to uncover the interrelationships among terpene synthase primary sequence, functional domain organization, protein structure, and cyclization mechanism and specificity, for not only bacterial, but fungal, and plant terpene synthases as well.
Acknowledgments
The work carried out in each of our laboratories was supported by NIH grant GM30301 (D.E.C.) and by a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT Japan, from JSPS 20310122 and from the Institute for Fermentation, Osaka, Japan (H.I.). We would each like to thank the dedicated coworkers who carried out the studies that are described, as well as Prof. Satoshi Omura for his advice and encouragement in the study of the S. avermitilis terpene synthases.
Biography
David E. Cane was born in New York in 1944. After undergraduate study at Harvard (B. A. 1966), he earned his Ph. D. in 1971 from Harvard University for research in synthetic organic chemistry with Prof. E. J. Corey. After postdoctoral study in natural products biosynthesis and bioorganic chemistry with Prof. Duilio Arigoni at the Eidgenössiche Technische Hochschule in Zürich, he joined the faculty of Brown University in 1973 where he is now Vernon K. Krieble Professor of Chemistry and Professor of Biochemistry. His research interests span the fields of organic chemistry and biochemistry and are focused on the biochemical origins of naturally occurring substances, including antibiotics, vitamins, and common flavor and odor compounds. He is particularly interested in unraveling the mechanistic and stereochemical details of multistep biochemical transformations catalyzed by a single enzyme (for example a terpene synthase) or a multifunctional enzyme (such as a modular polyketide synthase) in which the reaction intermediates remain sequestered in the enzyme active site or sites, either non-covalently or covalently. His awards include the Ernest Guenther Award (1985), the Cope Scholar Award (2000), and the Repligen Award (2005) of the American Chemical Society. He is a Fellow of the American Association of the Advancement of Science.
Haruo Ikeda was born in Tokyo, Japan (1954). He received his B.S. (1977) and M.S. (1979) in pharmaceutical sciences at Kitasato University. He obtained his Ph.D. (1982) in pharmaceutical sciences from Kitasato University, where he studied biosynthesis of 16-membered macrolide antibiotics produced by Streptomyces spp. with Professor Satoshi Omura. He began studying Streptomyces genetics with Professors Sir David A. Hopwood and Keith F. Chater at the Department of Genetics, John Innes Institute, UK as a postdoctoral fellow working on the development of actinophage vectors and characteristics of the gene involving catabolite regulation in Streptomyces. He then took a faculty position in the School of Pharmaceutical Sciences, Kitasato University (1983-2002) and is a now afull Professor (2002-present) at Kitasato Institute for Life Sciences and the Graduate School of Infection Control Sciences, Kitasato University. His research interests are in the study of the biosynthesis of microbial secondary metabolites using bioinformatics and in engineering and designing the Streptomyces chromosome for the evaluation and optimization of secondary metabolism. He received the Japan Award for Scientific Promotion (1991) and the Japan Award (1999) from the Society for Actinomycetes and the Sumiki-Umezawa Memorial Award of theJapan Antibiotics Research Association (2000).
References
- 1.Berthelot M, André G. Sur l'odeur propre de la terre. Compt. Rend. 1891;112:598–599. [Google Scholar]
- 2.Gerber NN. Geosmin, from microorganisms, is trans-1,10-dimethyl-trans-9-decalol. Tetrahedron Lett. 1968:2971–2974. [Google Scholar]
- 3.Bentley R, Meganathan R. Geosmin and methylisoborneol biosynthesis in streptomycetes. Evidence for an isoprenoid pathway and its absence in non-differentiating isolates. FEBS Lett. 1981;125:220–222. doi: 10.1016/0014-5793(81)80723-5. [DOI] [PubMed] [Google Scholar]
- 4.Martin DG, Slomp G, Mizsak S, Duchamp DJ, Chidester CG. The structure and absolute configuration of pentalenolactone (PA 132). Tetrahedron Lett. 1970:4901–4904. doi: 10.1016/s0040-4039(00)99739-9. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi S, Ogawa Y, Yonehara H. The Structure of pentalenolactone (PA-132). Tetrahedron Lett. 1969:2737–2740. doi: 10.1016/s0040-4039(01)88256-3. [DOI] [PubMed] [Google Scholar]
- 6.Cane DE, Sohng J-K, Lamberson CR, Rudnicki SM, Wu Z, Lloyd MD, Oliver JS, Hubbard BR. Pentalenene synthase. Purification, molecular cloning, sequencing and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 7.Cane DE, Oliver JS, Harrison PHM, Abell C, Hubbard BR, Kane CT, Lattman R. The Biosynthesis of pentalenene and pentalenolactone. J. Am. Chem. Soc. 1990;112:4513–4524. [Google Scholar]
- 8.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 9.Cane DE, Tandon M, Prabhakaran PC. Epicubenol synthase and the enzymic cyclization of farnesyl diphosphate. J. Am. Chem. Soc. 1993;115:8103–8106. [Google Scholar]
- 10.Corre C, Challis GL. New natural product biosynthetic chemistry discovered by genome mining. Nat. Prod. Rep. 2009;26:977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- 11.Felicetti B, Cane DE. Aristolochene synthase: mechanistic analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 2004;126:7212–7221. doi: 10.1021/ja0499593. [DOI] [PubMed] [Google Scholar]
- 12.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 13.Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 14.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 15.Eddy SR. Profile Hidden Markov models. Bioinformatics (Oxford, England) 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 16.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucl. Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucl. Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucl. Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl. Acad. Sci. U S A. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada H, Yu F, Okamoto S, Kuzuyama T, Utsumi R, Misawa N. Efficient synthesis of functional isoprenoids from acetoacetate through metabolic pathway-engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2009;81:915–925. doi: 10.1007/s00253-008-1724-7. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U S A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou WK, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE. Genome Mining in Streptomyces avermitilis: Cloning and Characterization of SAV_76, the Synthase for a New Sesquiterpene, Avermitilol. J. Am. Chem. Soc. 2010;132:8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamatsu S, Lin X, Nara A, Komatsu M, Cane DE, Ikeda H. Characterization of a silent sesquiterpenoid biosynthetic pathway in Streptomyces avermitilis controlling epi-isozizaene and albaflavenone biosynthesis and isolation of a new oxidized epi-isozizaene metabolite. Microb. Biotech. 2011;4:184–191. doi: 10.1111/j.1751-7915.2010.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O-Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 25.Cane DE, Watt RM. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc. Natl. Acad. Sci. U S A. 2003;100:1547–1551. doi: 10.1073/pnas.0337625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gansser D, Pollak FC, Berger RG. A sesquiterpene alcohol from Streptomyces citreus CBS 109.60. J. Nat. Prod. 1995;58:1790–1793. [Google Scholar]
- 27.He X, Cane DE. Mechanism and stereochemistry of the germacradienol/germacrene D synthase of Streptomyces coelicolor A3(2). J. Am. Chem. Soc. 2004;126:2678–2679. doi: 10.1021/ja039929k. [DOI] [PubMed] [Google Scholar]
- 28.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, He X, Cane DE. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J. Am. Chem. Soc. 2006;128:8128–8129. doi: 10.1021/ja062669x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, He X, Cane DE. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 2007;3:711–715. doi: 10.1038/nchembio.2007.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J, Cane DE. Geosmin biosynthesis. Mechanism of the fragmentation-rearrangement in the conversion of germacradienol to geosmin. J Am Chem Soc. 2008;130:428–429. doi: 10.1021/ja077792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawrath T, Dickschat JS, Muller R, Jiang J, Cane DE, Schulz S. Identification of (8S,9S,10S)-8,10-dimethyl-1-octalin, a key intermediate in the biosynthesis of geosmin in bacteria. J. Am. Chem. Soc. 2008;130:430–431. doi: 10.1021/ja077790y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh B, Oh TJ, Sohng JK. Exploration of geosmin synthase from Streptomyces peucetius ATCC 27952 by deletion of doxorubicin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2009 doi: 10.1007/s10295-009-0605-0. [DOI] [PubMed] [Google Scholar]
- 34.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 35.Cane DE, He X, Kobayashi S, Omura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. (Tokyo) 2006;59:471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- 36.Giglio S, Jiang J, Saint CP, Cane DE, Monis PT. Isolation and characterization of the gene associated with geosmin production in cyanobacteria. Environ. Sci. Technol. 2008;42:8027–8032. doi: 10.1021/es801465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons P. Camels act on a hump. The Guardian. 2003 Mar 6; [Google Scholar]
- 38.Tosi L, Sola C. Role of geosmin. a typical inland water odour, in guiding glass eels Anguilla anguilla (L.) migration. Ethology. 1993;95:177–185. [Google Scholar]
- 39.Wang CM, Cane DE. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor. J. Am. Chem. Soc. 2008;130:8908–8909. doi: 10.1021/ja803639g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickschat JS, Nawrath T, Thiel V, Kunze B, Muller R, Schulz S. Biosynthesis of the off-flavor 2-methylisoborneol by the myxobacterium Nannocystis exedens. Angew. Chem. Int. Ed. Engl. 2007;46:8287–8290. doi: 10.1002/anie.200702496. [DOI] [PubMed] [Google Scholar]
- 41.Ariyawutthiphan O, Ose T, Tsuda M, Gao Y, Yao M, Minami A, Oikawa H, Tanaka I. Crystallization and preliminary X-ray crystallographic study of a methyltransferase involved in 2-methylisoborneol biosynthesis in Streptomyces lasaliensis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:417–420. doi: 10.1107/S1744309110051523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giglio S, Chou WK, Ikeda H, Cane DE, Monis PT. Biosynthesis of 2-methylisoborneol in cyanobacteria. Environ. Sci. Technol. 2011;45:992–998. doi: 10.1021/es102992p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou WKW, Ikeda H, Cane DE. Cloning and characterization of Pfl_1841, a 2-methylenebornane synthase in Pseudomonas fluorescens PfO-1. Tetrahedron. 2011;67:6627–6632. doi: 10.1016/j.tet.2011.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X, Hopson R, Cane DE. Genome mining in Streptomyces coelicolor: molecular cloning and characterization of a new sesquiterpene synthase. J. Am. Chem. Soc. 2006;128:6022–6023. doi: 10.1021/ja061292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin X, Cane DE. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor. Mechanism and stereochemistry of the enzymatic formation of epi-isozizaene. J. Am. Chem. Soc. 2009;131:6332–6333. doi: 10.1021/ja901313v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aaron JA, Lin X, Cane DE, Christianson DW. Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry. 2010;49:1787–1797. doi: 10.1021/bi902088z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J. Biol. Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR. Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J. Biol. Chem. 2009;284:36711–36719. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quaderer R, Omura S, Ikeda H, Cane DE. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlI, a cytochrome P450 of Streptomyces avermitilis. J. Am. Chem. Soc. 2006;128:13036–13037. doi: 10.1021/ja0639214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You Z, Omura S, Ikeda H, Cane DE. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlH, a non-heme iron dioxygenase of Streptomyces avermitilis. J. Am. Chem. Soc. 2006;128(20):6566–6567. doi: 10.1021/ja061469i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You Z, Omura S, Ikeda H, Cane DE, Jogl G. Crystal structure of the non-heme iron dioxygenase PtlH in pentalenolactone biosynthesis. J. Biol. Chem. 2007;282:36552–36560. doi: 10.1074/jbc.M706358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You Z, Omura S, Ikeda H, Cane DE. Pentalenolactone biosynthesis: Molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Arch. Biochem. Biophys. 2007;459:233–240. doi: 10.1016/j.abb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. Genome mining in Streptomyces avermitilis: A biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry. 2009;48:6431–6440. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo MJ, Zhu D, Endo S, Ikeda H, Cane DE. Genome Mining in Streptomyces. Elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry. 2011;50:1739–1754. doi: 10.1021/bi1019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu D, Seo MJ, Ikeda H, Cane DE. Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J. Am. Chem. Soc. 2011;133:2128–2131. doi: 10.1021/ja111279h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J. Bacteriol. 2008;190:6084–6096. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu Y, Chou WKW, Hopson R, Cane DE. Genome mining in Streptomyces clavuligerus: Expression and biochemical characterization of two new cryptic sesquiterpene synthases. Chem. Biol. 2011;18:32–37. doi: 10.1016/j.chembiol.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakano C, Kim H-K, Ohnishi Y. Identification and Characterization of the Linalool/Nerolidol Synthase from Streptomyces clavuligerus. ChemBioChem. 2011 doi: 10.1002/cbic.201100501. DOI: 10.1002/cbic.201100501, 0000. [DOI] [PubMed] [Google Scholar]
- 61.Nakano C, Kim HK, Ohnishi Y. Identification of the First Bacterial Monoterpene Cyclase, a 1,8-Cineole Synthase, that Catalyzes the Direct Conversion of Geranyl Diphosphate. Chembiochem. 2011;12:1988–1991. doi: 10.1002/cbic.201100330. [DOI] [PubMed] [Google Scholar]
- 62.Nakano C, Horinouchi S, Ohnishi Y. Characterization of a novel sesquiterpene cyclase involved in (+)-caryolan-1-ol biosynthesis in Streptomyces griseus. J. Biol. Chem. 2011;286:27980–27987. doi: 10.1074/jbc.M111.265652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano C, Kudo F, Eguchi T, Ohnishi Y. Genome Mining Reveals Two Novel Bacterial Sesquiterpene Cyclases: (–)-Germacradien-4-ol and (–)-epi-α-Bisabolol Synthases from Streptomyces citricolor. ChemBioChem. 2011;12:2271–2275. doi: 10.1002/cbic.201100418. [DOI] [PubMed] [Google Scholar]