Abstract
To prevent the accumulation of misfolded and aggregated proteins, the cell has developed a complex network of cellular quality control (QC) systems to recognize misfolded proteins and facilitate their refolding or degradation. The cell faces numerous obstacles when performing quality control on transmembrane proteins. Transmembrane proteins have domains on both sides of a membrane and QC systems in distinct compartments must coordinate to monitor the folding status of the protein. Additionally, transmembrane domains can have very complex organization and QC systems must be able to monitor the assembly of transmembrane domains in the membrane. In this review, we will discuss the QC systems involved in repair and degradation of misfolded transmembrane proteins. Also, we will elaborate on the factors that recognize folding defects of transmembrane domains and what happens when misfolded transmembrane proteins escape QC and aggregate.
Keywords: transmembrane protein, quality control, autophagy, membrane chaperone, protein folding
1. Introduction
Accumulation of misfolded proteins in a cell can lead to disruption of global protein homeostasis. To prevent the accumulation of toxic protein species, the cell has developed a variety of cellular quality control (QC) systems to recognize misfolded proteins and facilitate their refolding or degradation [1-5]. The cell contains a variety of factors, notably molecular chaperones, which aid in the folding of proteins and degradation of terminally misfolded proteins [6]. Failure of protein QC systems to manage protein loads can result in protein aggregation and formation of toxic protein species, the molecular basis for a number of diseases.
Transmembrane proteins present interesting problems for QC systems. First, transmembrane proteins have domains on both sides of a membrane and QC systems in distinct compartments must coordinate to monitor the folding status of the protein. Second, transmembrane domains can have very complex organization and QC systems must be able to monitor the assembly of transmembrane domains. Several destabilizing mutations in transmembrane domains of proteins are the basis for several diseases, including cystic fibrosis, retinitis pigmentosa, hypercholesterolemia, diabetes insipidus, and hypogonadotropic hypogonadism [7-12] Misfolded transmembrane domains will expose hydrophilic residues in the hydrophobic environment of the membrane that would normally be involved in hydrogen bonding to the hydrophobic environment of the membrane (figure 1). Here, we will discuss QC systems used for misfolded polytopic transmembrane proteins, how the cell recognizes folding defects in transmembrane domains, and what happens when the transmembrane proteins aggregate.
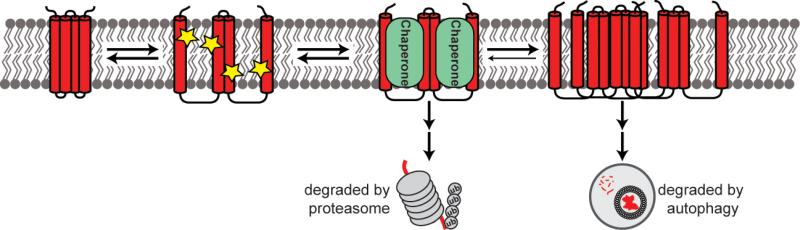
Figure 1. Quality control of a protein with a misfolded transmembrane domain.
A misfolded transmembrane protein may have improperly aligned transmembrane helices. These helices will display polar residues on the surface of the helix (indicated with stars) that can be recognized by membrane chaperones. Chaperones will prevent the misfolded protein from aggregating, potentially allowing for the refolding. Terminally misfolded proteins will be degraded by the proteasome. Proteins that escape chaperone recognition will aggregate and be degraded by autophagy.
2. Folding/misfolding of transmembrane proteins
There are two distinct types of transmembrane spanning domains in proteins: β-barrel and α-helix. α-helix transmembrane spans are common and are inserted into the ER membrane co-translationally via the Sec61 translocon complex[13]. The translocon, which binds to ribosomes [14], consists of a complex composed of the Sec61α, β, and γ subunits and translocating chain-associating membrane protein (TRAM) [15, 16]. Sec61 forms a hydrophobic tunnel in the membrane that creates a chemical environment in which translating transmembrane polypeptides can insert into the membrane and achieve proper structure [17-19]. The Sec61 tunnel can accommodate two helices at one time and facilitate interhelical bonds between them [20]. However, many proteins have a complex network of more than two transmembrane spans and the translocon must have a mechanism to prevent the co-translational aggregation of such proteins (discussed in section 2.2.1). In contrast, the less common β-barrel transmembrane domains, which are found exclusively on the outer-membrane of bacteria, chloroplast, and mitochondria, consist of a large coiled β-sheet that form a pore in the membrane [21]. Folding of the highly ordered β-barrel domain occurs in the inner membrane space in a process mediated by soluble chaperones such as Skp [22]. Once the proper folding is achieved, β-barrels are post-translationally inserted into the membrane via an energetically spontaneous process [23].
2.1. QC of polytopic membrane proteins in the ER
Transmembrane proteins can have a wide range of topologies, ranging from proteins consisting of a single transmembrane α-helix to proteins with more than 20 transmembrane helices and large soluble domains on both sides of the membrane [13]. To deal with this variety, multiple ER-membrane associated E3 ubiquitin ligases have been identified that target different types of terminally misfolded membrane proteins for degradation [24] (figure 2). The ubiquitin ligase HRD1 forms a functional QC complex with many proteins including SEL1L, OS-9, the Derlin proteins, and E2 ubiquitin-conjugating enzymes [25-27]. The HRD1 complex has been implicated in the recognition of misfolding in the transmembrane spans and the ER luminal domains of polytopic membrane proteins. SEL1L and OS-9 are believed to serve as recognition factors for misfolded ER luminal domains [24]. Another ubiquitin ligase, GP78, is homologous to HRD1 and is involved in similar QC processes [28]. The RMA1 E3 ubiquitin ligase functions in a complex with Derlin-1, the E2 ubiquitin-conjugating enzyme UBE2J1, and DNAJB12 [29, 30]. DNAJB12 is a transmembrane HSP40 chaperone with a cytosolic J-domain that co-operates with cytosolic Hsp70 to recognize misfolding in cytosolic domains of polytopic membrane proteins, allowing for RMA1 mediated ubiquitination. Little is known about an additional mammalian QC ubiquitin ligase, TEB4, but the yeast homolog, Doa10, is involved in QC of polytopic membrane proteins with cytosolic domains [31]. Recently it was shown that a diverse group of 24 transmembrane ubiquitin ligases localize to the ER membrane, suggesting that there may be many uncharacterized transmembrane ubiquitin ligases involved in the QC of membrane proteins in the ER [32]. Additionally, a host of cytosolic QC factors and ubiquitin ligase aid in the QC of polytopic membrane proteins with cytosolic domains [2, 33].
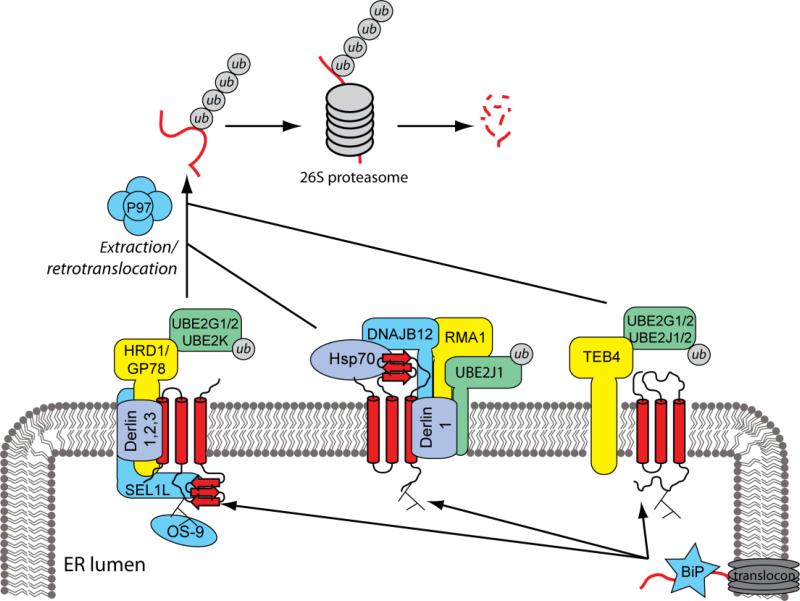
Figure 2. ER-membrane localized complexes that target misfolded membrane proteins for degradation.
Misfolded membrane proteins are recognized by various ER factors, such as molecular chaperones, and directed towards ER membrane associated E3 ubiquitin-ligases. The three main mammalian ligases identified are HRD1, RMA1 and TEB4. Each ligase is part of a complex with an E2 ubiquitin-conjugating enzyme and other proteins that are involved in recognition. HRD1 substrates tend to have folding defects in luminal and transmembrane domains. RMA1 substrates are polytopic membrane proteins with large cytoplasmic domains. Substrate proteins are ubiquitinated by the respective ligase, extracted into the cytoplasm via the p97 AAA+ ATPase, and degraded by the proteasome.
2.1.1 Cystic fibrosis transmembrane conductance regulator (CFTR) as a model transmembrane QC substrate
The ion-channel cystic fibrosis transmembrane conductance regulator (CFTR) is a prominent model substrate used in the investigation of QC of membrane proteins. CFTR is a polytopic membrane protein containing twelve transmembrane spans, a cytoplasmic regulatory-domain, and two large cytoplasmic nucleotide-binding domains (NBD1/2) [34]. Mutations in CFTR are the molecular basis for cystic fibrosis (CF), a common recessive disorder [35, 36]. While over one-thousand different CF-causing mutations in CFTR have been discovered, 69% of afflicted people worldwide carry at least one copy of the ΔF508 mutation and as such ΔF508 CFTR is the by far the most studied CFTR mutant [37]. Phenylalanine-508 of CFTR is part of NBD1 and deletion of this residue creates a folding defect in the cytoplasmic domains of CFTR [38]. While, ΔF508 CFTR does appear to retain some function, virtually none of the protein reaches the cell surface. Instead, a network of quality control factors retains ΔF508 CFTR in the endoplasmic reticulum where it is targeted for proteasomal degradation.
Due to the cytoplasmic folding defect of ΔF508 CFTR, a series of checkpoints involving both ER and cytoplasmic factors are used in QC. Cytosolic Hsc70 interacts with HSP40, the U-box ubiquitin ligase CHIP, and the E2 UbcH5 to form an E3 complex that recognizes and ubiquitinates misfolded ΔF508 CFTR [33, 39, 40]. Additionally, misfolded ΔF508 CFTR is independently recognized by the ER membrane associated Hsp40 DNAJB12 [29, 30]. DNAJB12 recruits the RMA1 complex to ubiquitinate misfolded CFTR [41]. CFTR that has been ubiquitinated by RMA1 and/or CHIP is extracted from the membrane into the cytoplasm via the p97 AAA+ ATPase and degraded by the proteasome.
While the basis for misfolding and recognition of ΔF508 CFTR is an area of intense investigation, it is not known how the cell would recognize CFTR mutants that cause misfolding in the transmembrane spans. Of the 20 most common disease-causing CFTR mutations, three of them are mutations to transmembrane spans (G85E, R347P, and R334W). However, very little is known about the quality control of these CFTR mutants. G85E CFTR is degraded before it can exit the ER and none of the protein reaches the cell surface [42, 43]. Degradation of G85E is dependent on the membrane protein Derlin-1 and it is suggested that Derlin-1 is serving as a recognition factor for misfolded G85E CFTR [42]. However, much research into the QC mechanisms of misfolded CFTR and other disease-related transmembrane proteins remains.
2.2. Transmembrane domain chaperones
The key first step in a QC pathway is the recognition of a misfolded protein. Misfolded soluble proteins typically display hydrophobic patches on the surface of the protein that would normally be buried in the core. These hydrophobic patches are recognized by molecular chaperones, allowing the misfolded protein to enter a QC pathway [44]. However, less is known about the molecular factors that aid in recognition of misfolded transmembrane domains and the basis for recognition in the lipid bilayer. The proper folding of α-helix based transmembrane domains is achieved through a series of inter- and intra-helical hydrogen bonds and salt bridges [45]. A “misfolded” transmembrane domain will have misaligned helices and may expose polar residues that would normally be involved in hydrogen bonding to the hydrophobic environment of the membrane (figure 1). A plausible model is that exposed polar residues could be recognized by molecular chaperones with active transmembrane domains that monitor the folding status of client transmembrane domains. Additionally, transmembrane proteins that are unable to properly oligomerize or form complexes with binding partner will also expose polar/charged residues in the membrane that can be recognized as misfolded by transmembrane domain chaperones. While these models of recognition are plausible, no experimental evidence directly confirms them. In extreme cases of misfolding, transmembrane helices may not properly insert into the membrane. The uninserted transmembrane helix will be generally hydrophobic and exposed in an aqueous environment providing a basis for recognition by cytosolic chaperones. As an example, an unstable helix in CFTR is often not properly inserted into the membrane and contributes to improper biogenesis of CFTR [46].
While no transmembrane domain chaperones have been extensively characterized, several proteins have been identified that could function as transmembrane domain chaperones in various QC pathways.
2.2.1. The Translocon and TRAM
The Sec61 translocon contains a hollow hydrophobic tunnel in the membrane that creates a chemical environment in which translating membrane polypeptides can insert into the membrane and achieve proper folding [17-19]. The translocon tunnel can accommodate two helices at one time and facilitate interhelical bonds between them [20]. However, some proteins have a complex network of more than twenty transmembrane helices and the translocon channel would be unable to facilitate the organization of such proteins. It has been suggested that translocating chain-associating membrane protein (TRAM), an essential component of the translocon complex [15, 16], could act as a transmembrane domain chaperone [47, 48]. Chemical crosslinking experiments show that TRAM interacts with translocated helices containing charged residues in translating polypeptides [49, 50]. TRAM contains four relatively hydrophilic transmembrane helices that could be sites for recognition of exposed charges in client transmembrane helices [48]. A general model for TRAM as a chaperone would be that it binds exposed polar/charged residues in already inserted transmembrane helices of translating polypeptides, preventing aggregation until all the protein has been completely synthesized so that proper folding can be achieved.
2.2.2. E. coli YidC
In E.coli, the protein YidC has been highly implicated as a key player in membrane protein biogenesis and shown to be an interactor of the E.coli translocon [51-53]. Notably, YidC is required for the proper folding of the lactose permease LacY [52]. As LacY contains a very complex 12-membrane spanning domain, with no significant cytosolic or periplasmic domains, this suggests that YidC is a chaperone that helps organize complex transmembrane domains. A distantly related mammalian homolog of YidC (Oxa1L) localizes to the mitochondria and interacts with mitochondrial ribosomes, suggesting that Oxa1L could have a similar function to YidC in the mitochondria [54]. However, no proteins with any significant homology to YidC are found outside of the mitochondria and thus there are no YidC-like proteins that could act as a transmembrane domain chaperone in the ER, where the bulk of membrane protein synthesis takes place.
2.2.3. Yeast Shr3p
Shr3p is an 4 transmembrane spanning protein in yeast that recognizes, prevents aggregation of, and facilitates refolding of transmembrane domains of general amino-acid permease 1 (Gap1) [55, 56]. Gap1 contains 12 transmembrane helices with no significant extracellular or cytoplasmic domains suggesting that Shr3p acts as a chaperone to fold and organize transmembrane domains. There are no obvious mammalian homologs of Shr3p.
2.2.4. HRD1
The ER membrane associated ubiquitin-ligase HRD1 is involved in the degradation of numerous proteins with transmembrane folding defects [24, 57]. Mutations to the transmembrane domain of HRD1 prevent proteins with misfolded transmembrane domains, but not ER luminal domains, from being degraded [58]. Additional in vitro studies suggest that the transmembrane domain of HRD1 directly recognizes misfolded membrane proteins. A reasonable model is that the transmembrane domain of HRD1 binds misfolded transmembrane domains of client proteins, bringing the client protein into a complex with the cytosolic RING-finger domain, allowing for ubiquitination and proteasomal degradation.
2.2.5. Derlin-1
DerlineIt has been suggested that the known QC factor Derlin-1 can act as a transmembrane domain chaperones [30]. Derlin-1 forms complexes with membrane spanning domain 1 (MSD1) of CFTR, which lacks any cytoplasmic structure, and Derlin-1 promotes ER retention and degradation of CFTR [41, 42]. Degradation of G85E CFTR, a transmembrane domain mutant, is dependent on Derlin-1 and it is suggested that Derlin-1 can serve as a recognition factor for misfolded G85E CFTR [42]. Derlin-1 interacts with the ER transmembrane ubiquitin ligases RMA1 and HRD1 [24, 41]. An interesting model is that Derlin-1 provides an additional level of recognition of substrates for these ubiquitin ligases, in a manner similar to the transmembrane domain of HRD1. While, Derlin-1 is predicted to have 4 transmembrane spans, recent evidence suggest that it actually has 6 transmembrane spans which contain a number of polar and charged residues [59].
2.2.6. BAP29/BAP31
BCR-associated proteins 29/31 (BAP29/BAP31) are ubiquitously expressed transmembrane proteins that form a high molecular weight complex that likely functions as transmembrane domain chaperones involved in the QC of transmembrane proteins in the ER. BAP29/BAP31 is involved in the recognition and ER retention of membrane-bound immunoglobulins (mIgs) with unstable transmembrane domains[60]. However, BAP31 does not interact with and retain an mIg that contains very hydrophobic and stable transmembrane domains [60]. BAP29 and BAP31 have multiple exposed polar and charged residues in their transmembrane spans. It is proposed that these polar/charged residues can interact with exposed hydrophilic sequences in unstable transmembrane spans of mIgs, providing a molecular basis for recognition and retention [60]. Similarly, BAP31 has been implicated in the ER retention of several other transmembrane proteins with known unstable transmembrane domains, including CFTR, cellubrevin, and cytochrome P450 2C2 [61, 62]. The fact that BAP29/BAP31 can regulate the ER retention of proteins with diverse functions and a wide range of topologies suggest that it may be a general ER transmembrane domain chaperones, rather than a regulator of a specific biological process. An analogous high molecular weight complex found in the mitochondria, consisting of prohibitin and BAP37, is involved in the stabilization of membrane proteins in the mitochondria [63].
3. Aggregation and autophagy of transmembrane proteins
Misfolded proteins that escape recognition by molecular chaperones are prone to aggregation [64]. Aggregates can be toxic to the cell and need to be degraded [65, 66]. Aggregates cannot be degraded by the proteasome as they are physically too large to enter the narrow proteasomal barrel [67]. While protein aggregates can be disaggregated by certain molecular chaperones, this does not appear to be the primary mechanism of aggregate clearance in mammalian cells [68]. Instead, aggregates are often degraded by macroautophagy [69]. Macroautophagy (simply referred to as autophagy) is a process in which parts of the cell, such as aggregated or damaged organelles, are packaged into double-membrane autophagosome and sent to the lysosome for degradation [70]. Unlike the proteasome, the lysosome is easily capable of degrading large protein aggregates (figure 3). Recently, there has been a significant amount of investigation into how the cell recognizes and targets protein aggregates for autophagic degradation.
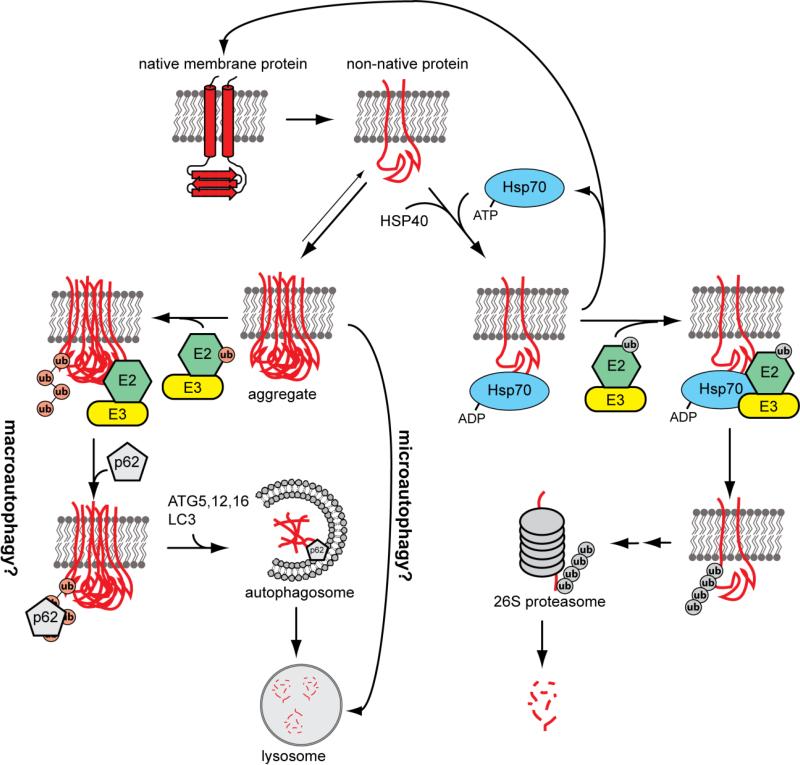
Figure 3. Model for the partitioning of misfolded membrane proteins between autophagy and proteasomal degradation.
Native proteins are prone to unfolding. Unfolded membrane proteins with cytosolic domains can be recognized by molecular chaperones (i.e. HSP70), preventing aggregation. The unfolded protein will be refolded or targeted for proteasomal degradation by ubiquitin ligases (i.e. CHIP) via K48-linked poly ubiquitin chains. Additionally, Misfolded/unfolded proteins are prone to aggregation. Aggregated proteins can be resolubilized by disaggregates or degraded by autophagy. Aggregates may be conjugated with K63-linked polyubiquitin chains. The ubiquitin binding protein p62 will bind K63-linked polyubiquitin chains and facilitate the packaging of aggregates into autophagosomes. Autophagosomes will fuse with the lysosome and aggregates will be degraded. Additionally, it is possible that membrane protein aggregates could be degraded by microautophagy, an autophagosome independent form of autophagy.
When the proteasome is inhibited various soluble and integral membrane proteins, including CFTR, are sent to the aggresome, an inclusion body formed at the microtubule organizing center [71]. Some proteins targeted to the aggresome are degraded by autophagy, and it has been suggested that the aggresome may be a sequestration compartment for proteins prior to autophagic degradation [72, 73]. Additionally, yeast with impaired proteasomal degradation accumulate CFTR in an aggresome-like compartment, and eventually degrade it via autophagy [74]. However, in mammalian cells, CFTR that accumulates during proteasome inhibition does not appear to be degraded by autophagy suggesting that the aggresome is a holding compartment for misfolded/aggregated membrane proteins [75].
Very few examples of membrane proteins being degraded by autophagy are found in the literature. A mutant form of dysferlin, a single transmembrane spanning protein with a large (>200 amino acids) cytoplasmic domain, has been shown to be partitioned between the proteasome and the lysosome [76]. Similarly, mutant mouse olfactory receptors have been shown to accumulate in the presence of lysosome inhibitors, suggesting that they are degraded by autophagy [77]. While it is clear that membrane proteins can be selected for autophagy, the mechanism for how membrane proteins are selected for and degraded by autophagy is entirely unknown.
It is known that many aggregates and inclusion bodies in the cytosol are ubiquitin positive and that inactivation of autophagy causes a buildup of ubiquitinated aggregates [64, 69, 78]. Specifically, it has been suggested that certain types of polyubiquitin chains will target proteins to different fates (figure 3) [79]. Soluble proteins being targeted for autophagy will have K63-linked polyubiquitin chains, while those being targeted for the proteasome will have primarily K48-linked polyubiquitin chains. Several ubiquitin binding proteins that have a high affinity for K63-linked polyubiquitin chains, such as p62, NBR1, and HDAC6, have been implicated in selection of substrates for autophagy [80-82]. This led to the model that protein aggregates are conjugated with K63-linked polyubiquitin chains and selected for autophagy by these ubiquitin binding proteins [78, 79]. This model is controversial as it has been shown that in autophagy deficient mice, all types of polyubiquitin linkages accumulate and not just K63-linkages [83]. This seems to suggest that ubiquitination of aggregated protein is merely an indirect consequence. However, as there is tight crosstalk between proteasomal and autophagic degradation pathways [84, 85] it is unclear exactly what the expected results would be in these experiments.
A disease-causing mutant of the soluble protein α1-antitrypsin is known to be degraded by autophagy in an ubiquitin independent manner. When overexpressed, the Z-variant of α1-antitrypsin (ATZ) will aggregate in the ER lumen and sections of the ER containing these aggregates are selectively packaged into autophagosomes [86, 87]. While, the exact mechanism for autophagic degradation of ATZ is not clear, it is possible that aggregated transmembrane proteins in the ER could be degraded by the same mechanism as luminal ATZ aggregates.
4. Concluding remarks
Many major questions remain about how the cell deals with misfolded membrane proteins. First, what QC factors are responsible for the recognition of misfolded transmembrane spans? As discussed in section 2.2, several proteins have been implicated as transmembrane domain chaperones, however the mechanism by which most of these proteins act and the diversity of their substrates remains poorly understood. Also, how are aggregated membrane proteins selected for autophagy? Is this via ubiquitin dependant autophagy, or perhaps a process similar to autophagy of ATZ aggregates in the ER lumen? As only several examples of transmembrane proteins that are degraded by autophagy have been described, the ability to answer this question will depend on the identification and characterization of more transmembrane proteins that are degraded by autophagy. Answers to these questions will give insight into the fundamental biological mechanisms that are the basis for a diverse set of diseases.
Highlights.
We discuss the basis for unfolding of transmembrane proteins.
We discuss the quality control pathways responsible for recognition of misfolded transmembrane.
We discuss what happens when transmembrane proteins aggregate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goeckeler JL, Brodsky JL. Molecular chaperones and substrate ubiquitination control the efficiency of endoplasmic reticulum-associated degradation. Diabetes Obes Metab. 2010;12(Suppl 2):32–38. doi: 10.1111/j.1463-1326.2010.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Maattanen P, Gehring K, Bergeron JJ, Thomas DY. Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Tatsuta T. Protein quality control in mitochondria. J Biochem. 2009;146:455–461. doi: 10.1093/jb/mvp122. [DOI] [PubMed] [Google Scholar]
- 5.Arvan P, Zhao X, Ramos-Castaneda J, Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- 6.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 7.Beglova N, Blacklow SC. The LDL receptor: how acid pulls the trigger. Trends Biochem Sci. 2005;30:309–317. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 9.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 10.O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 11.Bichet DG. Nephrogenic diabetes insipidus. Adv Chronic Kidney Dis. 2006;13:96–104. doi: 10.1053/j.ackd.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Janovick JA, Maya-Nunez G, Conn PM. Rescue of hypogonadotropic hypogonadism-causing and manufactured GnRH receptor mutants by a specific protein-folding template: misrouted proteins as a novel disease etiology and therapeutic target. J Clin Endocrinol Metab. 2002;87:3255–3262. doi: 10.1210/jcem.87.7.8582. [DOI] [PubMed] [Google Scholar]
- 13.Shao S, Hegde RS. Membrane Protein Insertion at the Endoplasmic Reticulum. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 15.Gorlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 17.Skach WR. The expanding role of the ER translocon in membrane protein folding. J Cell Biol. 2007;179:1333–1335. doi: 10.1083/jcb.200711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapoport TA, Goder V, Heinrich SU, Matlack KE. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Hegde RS, Lingappa VR. Membrane protein biogenesis: regulated complexity at the endoplasmic reticulum. Cell. 1997;91:575–582. doi: 10.1016/s0092-8674(00)80445-6. [DOI] [PubMed] [Google Scholar]
- 20.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delcour AH. Structure and function of pore-forming beta-barrels from bacteria. J Mol Microbiol Biotechnol. 2002;4:1–10. [PubMed] [Google Scholar]
- 22.Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 23.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 25.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 26.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto YH, Kimura T, Momohara S, Takeuchi M, Tani T, Kimata Y, Kadokura H, Kohno K. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct Funct. 2010;35:107–116. doi: 10.1247/csf.10023. [DOI] [PubMed] [Google Scholar]
- 30.Grove DE, Fan CY, Ren HY, Cyr DM. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTR{Delta}F508. Mol Biol Cell. 2011;22:301–314. doi: 10.1091/mbc.E10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neutzner A, Neutzner M, Benischke AS, Ryu SW, Frank S, Youle RJ, Karbowski M. A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J Biol Chem. 2011;286:8633–8643. doi: 10.1074/jbc.M110.197459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 34.Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 35.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 36.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 37.Bobadilla JL, Macek M, Jr., Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 38.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 39.Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, Cyr DM. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 42.Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- 43.Grove DE, Rosser MF, Ren HY, Naren AP, Cyr DM. Mechanisms for rescue of correctable folding defects in CFTRDelta F508. Mol Biol Cell. 2009;20:4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 45.Bowie JU. Membrane protein folding: how important are hydrogen bonds? Curr Opin Struct Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tector M, Hartl FU. An unstable transmembrane segment in the cystic fibrosis transmembrane conductance regulator. EMBO J. 1999;18:6290–6298. doi: 10.1093/emboj/18.22.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cross BC, High S. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J Cell Sci. 2009;122:1768–1777. doi: 10.1242/jcs.046094. [DOI] [PubMed] [Google Scholar]
- 48.Tamborero S, Vilar M, Martinez-Gil L, Johnson AE, Mingarro I. Membrane insertion and topology of the translocating chain-associating membrane protein (TRAM) J Mol Biol. 2011;406:571–582. doi: 10.1016/j.jmb.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 50.Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 51.Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 52.Nagamori S, Smirnova IN, Kaback HR. Role of YidC in folding of polytopic membrane proteins. J Cell Biol. 2004;165:53–62. doi: 10.1083/jcb.200402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhn A, Stuart R, Henry R, Dalbey RE. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 2003;13:510–516. doi: 10.1016/j.tcb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Haque ME, Spremulli LL, Fecko CJ. Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem. 2010;285:34991–34998. doi: 10.1074/jbc.M110.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kota J, Ljungdahl PO. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J Cell Biol. 2005;168:79–88. doi: 10.1083/jcb.200408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kota J, Gilstring CF, Ljungdahl PO. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J Cell Biol. 2007;176:617–628. doi: 10.1083/jcb.200612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehara K, Xie W, Ng DT. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol. 2010;188:707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schamel WW, Kuppig S, Becker B, Gimborn K, Hauri HP, Reth M. A high-molecular-weight complex of membrane proteins BAP29/BAP31 is involved in the retention of membrane-bound IgD in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2003;100:9861–9866. doi: 10.1073/pnas.1633363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Annaert WG, Becker B, Kistner U, Reth M, Jahn R. Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J Cell Biol. 1997;139:1397–1410. doi: 10.1083/jcb.139.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szczesna-Skorupa E, Kemper B. BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J Biol Chem. 2006;281:4142–4148. doi: 10.1074/jbc.M509522200. [DOI] [PubMed] [Google Scholar]
- 63.Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 65.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 66.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 67.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6' proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 68.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 70.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15:47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chin LS, Olzmann JA, Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans. 2010;38:144–149. doi: 10.1042/BST0380144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu L, Sztul E. ER-associated complexes (ERACs) containing aggregated cystic fibrosis transmembrane conductance regulator (CFTR) are degraded by autophagy. Eur J Cell Biol. 2009;88:215–226. doi: 10.1016/j.ejcb.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gelman MS, Kannegaard ES, Kopito RR. A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2002;277:11709–11714. doi: 10.1074/jbc.M111958200. [DOI] [PubMed] [Google Scholar]
- 76.Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 77.Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 2003;4:416–433. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- 78.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 79.Yao TP. The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer. 2010;1:779–786. doi: 10.1177/1947601910383277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 82.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 83.Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, Yamamoto M, Tanaka K, Mizushima N, Komatsu M, Kopito RR. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 85.Kawaguchi T, Miyazawa K, Moriya S, Ohtomo T, Che XF, Naito M, Itoh M, Tomoda A. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol. 2011;38:643–654. doi: 10.3892/ijo.2010.882. [DOI] [PubMed] [Google Scholar]
- 86.Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perlmutter DH. Alpha-1-antitrypsin deficiency: importance of proteasomal and autophagic degradative pathways in disposal of liver disease-associated protein aggregates. Annu Rev Med. 2011;62:333–345. doi: 10.1146/annurev-med-042409-151920. [DOI] [PubMed] [Google Scholar]