Abstract
Histone deacetylases (HDACs) are a family of enzymes that play significant roles in numerous biological processes and diseases. HDACs are best known for their repressive influence on gene transcription through histone deacetylation. Mapping of non-histone acetylated proteins and acetylation-modifying enzymes involved in various cellular pathways has shown protein acetylation/deacetylation also plays key roles in a variety of cellular processes including RNA splicing, nuclear transport, and cytoskeletal-remodeling. Studies of HDACs have accelerated due to the availability of small molecule HDAC inhibitors, most of which contain a canonical hydroxamic acid or benzamide that chelates the metal catalytic site. To increase the pool of unique and novel HDAC inhibitor pharmacophores, a pharmacological active compound screen was performed. Several unique HDAC inhibitor pharmacophores were identified in vitro. One class of novel HDAC inhibitors, with a central naphthoquinone structure, displayed a selective inhibition profile against HDAC6. Here we present the results of a unique class of HDAC6 inhibitors identified using this compound library screen. In addition, we demonstrated treatment of human acute myeloid leukemia cell line MV4-11 with the selective HDAC6 inhibitors decreases levels of mutant FLT-3 and constitutively active STAT5, and attenuates Erk phosphorylation, all of which are associated with the inhibitor’s selective toxicity against leukemia.
Histone deacetylases (HDACs) regulate a broad range of cellular processes by modulating protein structure and function via lysine deacetylation (1). Lysine deacetylation is a dynamic and reversible process mediated by acetyl transferases and HDACs. The balance of these dynamic post-translational modifications is theorized to play key roles in cellular signaling and homeostasis similar to protein phosphorylation (2–3). Although protein acetylation is best known for its role in transcriptional regulation via histone acetylation (4–5), recent studies have shown over 3,500 acetylation sites in approximately 1,700 acetylated proteins; in comparison to the current phosphoproteome at about 6,600 phosphorylation sites and 2,200 proteins (3). Thus, the acetylome is approaching the size of phosphoproteome and extends the mechanistic relevance and research interests in HDACs well beyond the field of transcriptional regulation and chromatin biology.
The initial classification of human HDACs is based on sequence homology and phylogenic relationships to yeast homologues. There are four distinct classes of HDACs. Class I (HDAC1, 2, 3, and 8), class IIa (HDAC4, 5, 7, and 9), class IIb (HDAC6 and 10), and class IV (HDAC11), which represent the Zn2+-dependent deacetylases (6). The class III sirtuins are mechanistically diverse NAD+-dependent deacetylases (7). Chemical inhibitors have always played a key role in the study of HDACs and their biological processes (8–9). Suberoylanilide hydroxamic acid (SAHA, Vorinostat; Merck Research Laboratories) and Romidepsin (FK-228; Gluocester Pharmaceuticals) were approved for the treatment of cutaneous T-cell lymphoma within 10 years of their discovery (10–11). HDAC inhibitors have also been shown to be promising therapies for neurodegenerative diseases and inflammatory disorders (12–14). However, how HDAC inhibitors achieve their therapeutic effects is still poorly understood. Recent studies using class-specific substrates display divergent enzymatic properties for class IIa HDACs distinguishing them from class I and IIb HDACs (15–16). The unique biochemistry associated with class IIa HDACs further complicates the mechanistic understanding of HDACs and their inhibitors. The commonly used pan-HDAC inhibitor, SAHA, appears to only inhibit Class I and IIb HDACs, with significantly lower activity against class IIa HDACs, and its inhibition profile is shared with other well known HDAC inhibitors (17–18). Proteomic studies showing that only 10% of all acetylation sites are even sensitive to SAHA treatment further emphasizes the need for novel HDAC inhibitors (3). Unique and selective HDAC inhibitors targeting specific isozymes could potentially facilitate the understanding of HDAC related molecular mechanisms. Currently four major structurally unique classes of HDAC inhibitors exist: hydroxamic acids, benzamides, short chain fatty acids, and large cyclic peptides. All these inhibitors share a common pharmacophore composed of a zinc-metal binding motif, a linker region, and a surface recognition domain (19). These commonalities of HDAC inhibitors could potentially limit the development of novel and unique HDAC inhibitors capable of inhibiting broad or selective HDACs.
Library of Pharmacologically Active Compounds (LOPAC, 1280 compounds) was screened for potential inhibitory activity against class I and IIa HDACs. We identified five potential novel HDAC inhibitors displaying different potencies and preferential selectivity against HDAC isozymes in vitro. In cells, one inhibitor induced selective hyperacetylation of tubulin indicating HDAC6 inhibition matching its in vitro profile (20–21). Several analogs were further investigated and one inhibitor, NQN-1, displayed a highly selective inhibition profile of HDAC6 in vitro and induced hyperacetylated tubulin in cells. Moreover, previous studies have shown HDAC6 controls Hsp90 chaperone activity through deacetylation of Hsp90 (22–23). We have shown that NQN-1 induces Hsp90 acetylation and causes a decrease in Hsp90 client protein FLT-3, STAT5, and phosphorylated-Erk levels in the human acute myeloid leukemia cell line, MV4-11, with associated cell death. Interestingly, hydroxamic acid selective HDAC6 inhibitors, tubacin and tubastatin A, did not induce acetylated Hsp90. In addition, the HDAC6 inhibitor caused an increase in ubiquinated protein levels in cells and promoted degradation of mutant FLT-3 through proteasome dependent pathways. It has great potential to be used as a therapeutic agent against AML or combined synergistically with other anticancer agents such as Hsp90 ATPase inhibitor, 17-AAG, or proteasome inhibitor, Bortezomib.
RESULTS AND DISCUSSION
Hit Identification Using Class-Specific HDAC Substrate Screening Assay
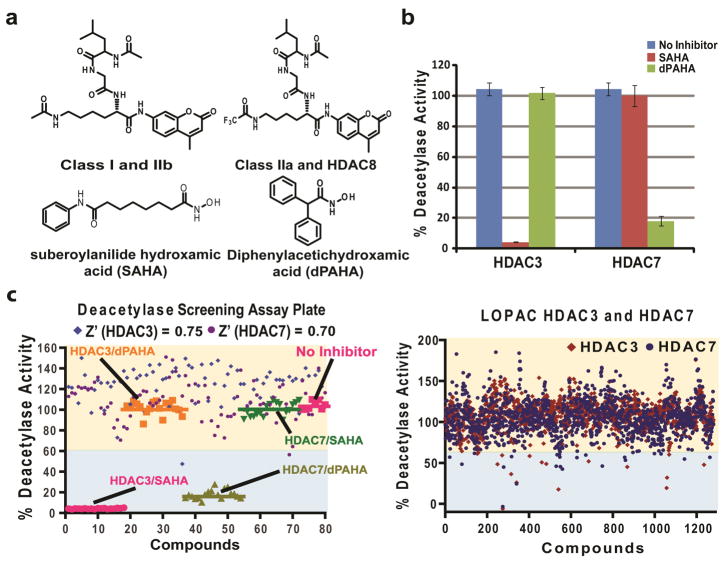
Unique small molecule inhibitors for different classes of HDACs are valuable as chemical tools or potential therapies against a variety of diseases. We have developed a high-throughput compatible HDAC screen assay using HDAC class-specific substrates. SAHA and diphenyl acetic hydroxamic acid (dPAHA) are used as positive controls for class I and class IIa respectively (Figure 1a, 1b, and 1c). We have demonstrated the positive inhibitor controls are orthogonal to each class of HDACs at 10 μM as shown in figure 1b, which is consistent with previous studies (17, 24). HDAC3 and HDAC7 have the highest activity in vitro and are used as the recombinant enzymes of choice to identify potential novel HDAC class I and II inhibitors. Although HDAC3 and HDAC7 are not identical to all other isozymes, in this study we demonstrate their utility in identifying new HDAC inhibitors that display different inhibition preferences in vitro (Figure 2).
Figure 1. HDAC class-specific substrates, inhibitors, and compound screen.
a) Structures of class I and class IIa specific substrates. SAHA and dPAHA are used for positive inhibition control of class I HDAC and class IIa respectively (10 μM). b) Orthogonal enzymatic deacetylase activities and inhibition profile of SAHA and dPAHA. c) Typical results for a 96 well plate screen (left panel) and results of 1280 LOPAC screen hits. Compounds which exhibited greater than 40% HDAC inhibition at 10 μM are consider a positive hit from the primary screen (right panel).
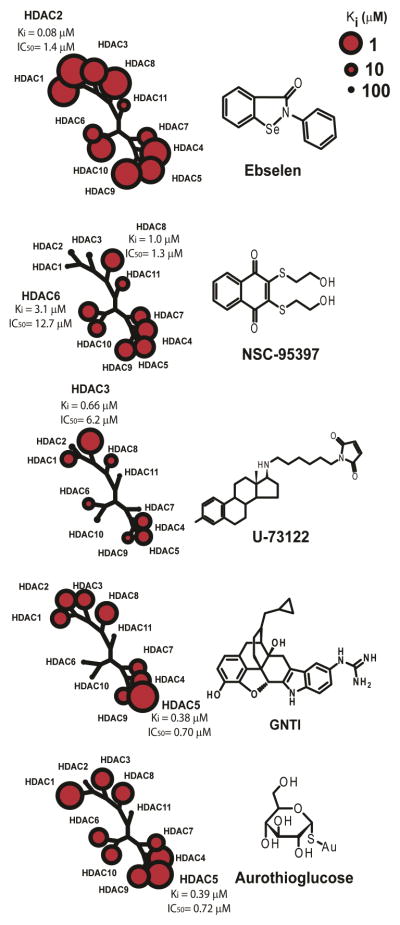
Figure 2. Identification of five positive hit HDAC inhibitors in vitro.
The five positive compounds from the screen were further tested with full panel of HDACs. The inhibition profiles are represented according to the HDAC phylogenetic classification. IC50 and Ki are labeled for the most inhibited HDAC for each compound.
The LOPAC library was screened at 10 μM concentration for each compound (Figure 1d). The average Z′ factor value for assays in both formats is 0.75 for HDAC3 plates and 0.7 for HDAC7 plates (Figure 1c). Initially, 24 compounds were selected from the primary hits for HDAC3 and HDAC7 to have >40% inhibition (standard deviations are 17.0% for HDAC3 plates and 21.7% for HDAC7 plates). Two point titrations were subsequently performed for each hit compound, and inhibitor candidates were chosen by eliminating compounds containing heavy metals, DNA chelators/intercalators, and those with poor LogP and water solubility issues. Only five compounds remained as positive hits (Figure 2). In addition, none of the five positive hits were identified as non-specific inhibitors due to in vitro aggregation verified by an aggregate formation assay against the same LOPAC compounds by assaying with/without 0.01% detergent Triton X-100 against activity of AmpC β-lactamase (a completely unrelated protein target) (25).
Confirmation of In Vitro HDAC Inhibition and Selectivity Using HDAC Deacetylase Biochemical Assay
The five compounds identified with the LOPAC library screen were further characterized with a HDAC deacetylase homogenous assay with class-specific substrates (acetylated substrate: HDAC1, 2, 3, 6, and 10; triflouroacetylated substrate: HDAC4, 5, 7, 8, 9 and 11) (17). The median inhibition concentration (IC50), was measured through a 12 point titration of inhibitor concentrations using full panel of human recombinant HDAC enzymes. Each compound displayed a different inhibition potency and selectivity toward different HDAC isozymes (Figure 2, Table 1, and Supporting Information). In vitro, ebselen (2-phenyl-1,2-benzisoselenazol-3(2H)-one), an antioxidant, and aurothioglucose (1-thio-D-glucopyronaose gold salt), an anti-inflammatory compound, displayed inhibitory activity for both class I and IIa HDACs. U-73122 (1-[6-[[(17beta)-3-methoxyestra-1,3,5(10)-trien-17-yl]amion]hexyl]-1H-pyrrole-2,5-dione), an aminosteroid, showed an isozyme selective preference toward HDAC3. NSC-95397 (2,3-bis[(2-Hydroxyethyl)thio]-1,4-naphthoquinone), a dual-specificity phosphatase inhibitor, displayed preferences toward HDAC6 and class IIa HDACs. GNTI ditrifluoroacetate (guanidinyl-naltrindole di-trifluoroacetate) is an opioid antagonist used in research which is highly selective for the κ opioid receptor, and displays inhibition preference toward HDAC5 (Figure 2 and Table 1).
Table 1. HDAC inhibition IC50 and Ki for the five LOPAC hit compounds.
The IC50 values lower than 100 μM were determined from at least two independent titrations (Supporting Information). The IC50 values were converted to Ki using Cheng-Prusoff equation assuming classical inhibitory mechanism.
| Ebselen | NSC-95397 | U-73122 | GNTI | Aurothioglucose | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 | Ki | IC50 | Ki | IC50 | Ki | IC50 | Ki | IC50 | Ki | |
| HDAC1 | 1.5 ± 0.4 | 0.15 | >180 | N/A | 38.4 ± 15.5 | 4.11 | 30.2 ± 7.3 | 3.24 | 4.4 ± 1.1 | 0.47 |
| HDAC2 | 1.4 ± 0.3 | 0.08 | >100 | N/A | >100 | N/A | 26.2 ± 7.6 | 1.48 | >180 | N/A |
| HDAC3 | 2.1 ± 1.5 | 0.23 | >100 | N/A | 6.2 ± 4.0 | 0.66 | 22.7 ± 3.1 | 2.43 | 11.5 ± 3.2 | 1.23 |
| HDAC4 | 1.2 ± 0.2 | 0.20 | 7.2 ± 2.8 | 1.23 | 31.2 ± 0.2 | 5.33 | 13.4 ± 1.6 | 2.29 | 4.1 ± 2.1 | 0.7 |
| HDAC5 | 0.78 ± 0.1 | 0.42 | 3.1 ± 0.2 | 1.68 | 10.5 ± 7.4 | 5.68 | 0.70 ± 0.1 | 0.38 | 0.72 ± 0.5 | 0.39 |
| HDAC6 | 12.8 ± 2.6 | 3.10 | 12.7 ± 2.5 | 3.07 | 45.2 ± 12.6 | 10.96 | >180 | N/A | 14.4 ± 5.8 | 3.49 |
| HDAC7 | 10.9 ± 2.2 | 3.09 | 13.1 ± 4.3 | 3.72 | >180 | N/A | 18.9 ± 1.0 | 5.36 | 22.4 ± 4.4 | 6.35 |
| HDAC8 | 0.35 ± 0.2 | 0.11 | 1.3 ± 0.1 | 1.03 | 10.9 ± 3.1 | 8.63 | 2.0 ± 0.4 | 1.58 | 1.4 ± 0.3 | 1.11 |
| HDAC9 | 0.97 ± 0.5 | 0.20 | 4.8 ± 1.4 | 2.04 | 29.8 ± 5.6 | 12.67 | 12.3 ± 1.7 | 5.23 | 2.8 ± 0.1 | 1.19 |
| HDAC10 | 0.95 ± 0.4 | 0.42 | 14.5 ± 1.5 | 6.40 | >100 | N/A | >180 | N/A | 4.9 ± 1.5 | 2.16 |
| HDAC11 | 22.5 ± 2.4 | 4.41 | 37.8 ± 1.4 | 7.41 | >180 | N/A | >180 | N/A | >180 | N/A |
All five compounds have unique structural motifs and display differential inhibition activity against different HDAC isozymes in vitro. The IC50s for the five compounds are in the μM to high nM range and apparent Ki was calculated using Cheng-Prusoff equation, which is consistent with the measurements from the initial library screens (26). Further structure-activity relationship analysis could potentially yield novel and selective HDAC inhibitors for different HDAC isozymes and increases the collection of HDAC inhibitor pharmacophores.
In Vivo Activity of Selected Compounds
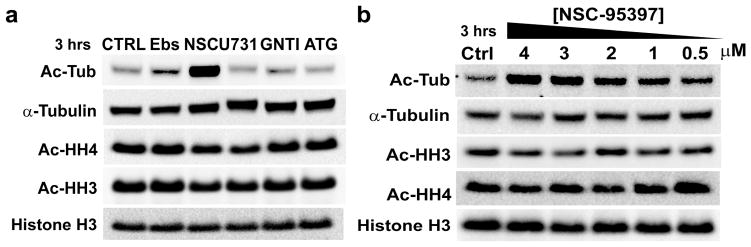
Activity in vitro does not necessarily represent how these compounds will act in vivo. HDACs are commonly known to function in large complexes with other functional proteins including other HDACs, and inhibitors affecting recombinant HDAC enzymes in vitro might not affect the HDAC complex activity (27). Furthermore, the special biochemistry of class IIa HDACs and the lack of an identified class IIa cellular substrate necessitate further investigation in regards to the relevance of class IIa HDAC enzymatic activity. Regardless, the five compounds were examined for their ability to induce histone H3, histone H4, and tubulin acetylation in MV4-11 cells. Only one out of the five compounds induced tubulin acetylation at 10 μM (Figure 3a). NSC-95397 induces hyperacetylated tubulin without an increase in acetylated histone levels. Concentration dependent induction of acetylated tubulin by NSC-95397 was observed indicating concentration dependent inhibition (Figure 3b).
Figure 3. Induction of acetylated protein in more complex cellular context.
a) AML cells, MV4-11, were treated with 10 μM concentration of each compound. The acetylation levels were assessed for acetylated tubulin (Ac-Tub), acetylated histone H3 (Ac-HH3), and acetylated histone H4 (Ac-HH4). Selective induction of hyperacetylated tubulin was observed indicating inhibition of HDAC6. b) Concentration dependent hyperacetylation of acetylated tubulin without increases in acetylated histone H3 and H4.
Selective HDAC6 Inhibitors from the LOPAC Library Screen
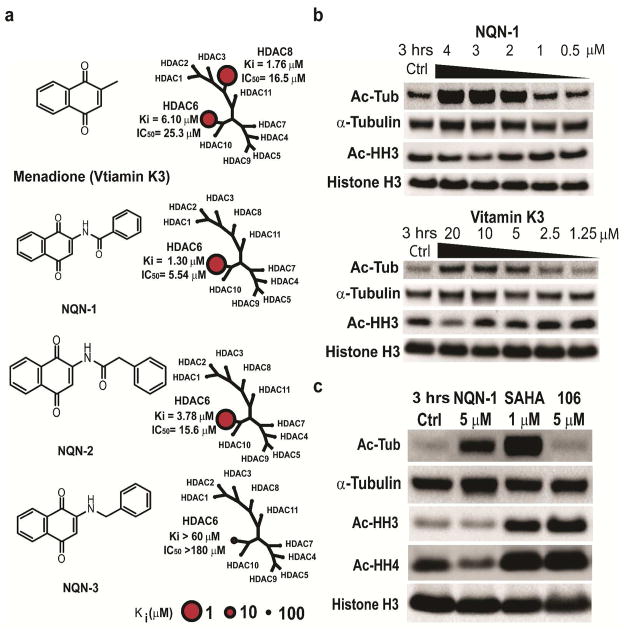
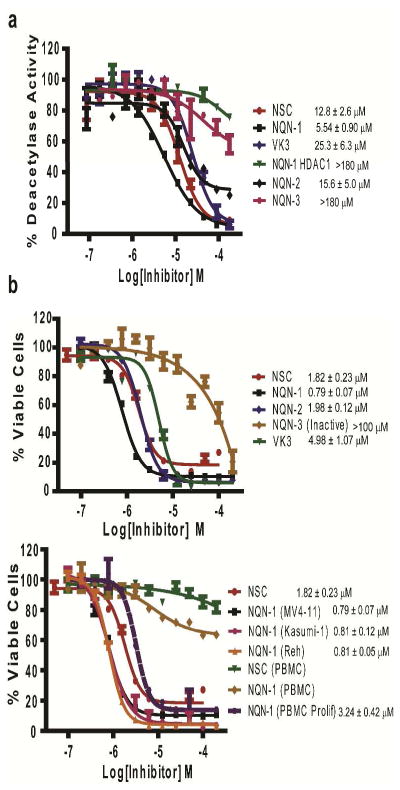
NSC-95397 contains a central naphthoquinone structure. Naphthoquinone is the central structural motif of many natural products. Most notably it is the central structure for vitamin K. We purchased and synthesized analogs of NSC-95397 to assess their HDAC6 inhibition including vitamin K1/K2/K3, NQN-1 (PPM-18), NQN-2, and NQN-3. Vitamin K1 and K2 only inhibited HDAC6 above 200 μM (data not shown), while vitamin K3, 2-methyl-naphthoquinone, selectively inhibited HDAC6 and HDAC8 at low μM (Figure 4a). NQN-1 (2-benzyl-amino-naphthoquinone) a known anti-inflammatory agent, displayed a highly selective inhibitory profile for HDAC6 and a slight increase in inhibition potency (IC50 of 5.54 ± 0.90 μM and Ki of 1.30 ± 0.22 μM) and did not inhibit other HDACs in our panel (Figure 4a, 5a, and Supporting Information). NQN-2, an analog of NQN-1 with one carbon bond extension, also selectively inhibited HDAC6 with a slight loss in its HDAC6 inhibition potency. NQN-3 is the least active analog of NQN-1 with the carbonyl functional group removed. NQN-3 has little HDAC6 inhibitory activity with IC50 > 180 μM and little cellular toxicity (Figure 4a and 5a). Simple acetylated 2-acetamido-1,4-naphthoquinone (NQN-4) and 1,4-naphthoquione were also examined for their HDAC6 inhibition and cellular toxicity (Supporting Information). Figure 5, panel b shows the concentration-dependent toxicity of MV4-11 cells by the NQN compounds as measured by MTS cell proliferation/toxicity assay. The NQN compounds that show μM HDAC6 inhibition displayed cell killing activity consistent with their HDAC6 inhibitory abilities. NQN-3 loses both its HDAC6 inhibition and cell killing abilities with a single key carbonyl functional group removed from NQN-1 suggesting HDAC6’s role in selective leukemia toxicity. Although the exact compound concentration and its distribution in cells is unknown, the cell killing concentration over which the compounds are active coincides with the Ki values measured in vitro. All active compounds are able to induce concentration dependent hyperacetylated tubulin consistent with inhibition of HDAC6 (Figure 4b and 5a). The specific acetylation of tubulin and histones are also assessed in comparison SAHA (HDAC1,2,3, and 6 inhibitor) and inhibitor 106 (HDAC3 selective inhibitor) (Figure 4c) (28). As expected, NQN-1 selectively induced hyperacetylated tubulin without an increase in histone H3 and H4 acetylation. However, NQN-1 unexpectedly caused a decrease in acetylated histone H4, and similar results were also observed for tubastatin A and tubacin with decreases in acetylated histones (Supporting Information). One possible hypothesis for this is that HDAC6 might control the activity of other HDACs or histone acetyltransferases (HATs), which have also been reported to be regulated through acetylation (29). Further studies of how HDAC6 and HDAC6 inhibitors affect histone acetylation are currently underway. Due to the selectivity and potency of NQN-1, we chose NQN-1 as a representative for more detailed follow-up assays.
Figure 4. Selective HDAC6 inhibition of naphthoquinone analogs.
a) Inhibition profiles of naphthoquinone analogs. b) Concentration dependent induction of acetylated tubulin in AML MV4-11 cells. c) NQN-1 selectively induces hyperacetylated tubulin without an increase in acetylated histone level. SAHA (class I HDAC and HDAC6 inhibitor) induces both acetylated tubulin and histone, HDAC inhibitor 106 (HDAC3 selective inhibitor) only induces hyperacetylated histone H3/H4.
Figure 5. HDAC inhibition titration and selective toxicity against leukemia cells.
a) NQN-1 and analogs display selective inhibition against HDAC6. Minimal inhibitory activity was observed for other HDACs (HDAC1 shown and Supporting Information). b) NQN analogs cause selective toxicity against leukemia cells consistent with their ability to inhibit HDAC6 in vitro. The least potent inhibitor of HDAC6, NQN-3, did not inhibit MV4-11 proliferation (>100 μM). Resting PBMC viability was not affected by either NQN-1 or NSC treatments. PBMC proliferation was inhibited at 3.24 μM which is 4-fold higher than the toxicity against AML cells ex vivo.
Selective Toxicity in Acute Myeloid Leukemia
The exact molecular basis for cell line specificity of different anticancer agents is not well understood. SAHA is particularly selective against cutaneous T-cell lymphoma. HDAC6 inhibition and knock-down has been shown to be effective against multiple myeloma and enhancing the toxicity of the Hsp90 inhibitor, 17-AAG, against acute myeloid leukemia (30–31). To determine whether our hit compounds can selectively kill leukemia cells over normal peripheral blood mononuclear cells (PBMC), we compared the cell-killing activity of NSC-95397 and NQN-1 in MV4-11 (AML), Kasumi (AML), Reh (ALL), and PBMC from healthy donors. Figure 5, panel b, shows NSC-95397 and NQN-1 are inactive against non-proliferating PBMC (no mitogen stimulation), while NQN-1 inhibits proliferating PBMC at 3.2 μM. NQN-1 showed selectivity killing of MV4-11 and leukemia cells, but this effect is almost non-existent against non-proliferating PBMC and 4-fold over mitogen stimulated proliferating PBMC. This result suggests proliferation is one of the factors that contribute to the selective cellular toxicity of our inhibitors. However, many factors can also contribute to the differences in cell-line toxicity of a given compound such as differences in cellular uptake, metabolism differences, or potential off target interaction with specific cellular components in cells. Optimization of NQN-1 to improve its leukemia cell specificity will be of great interest for further studies.
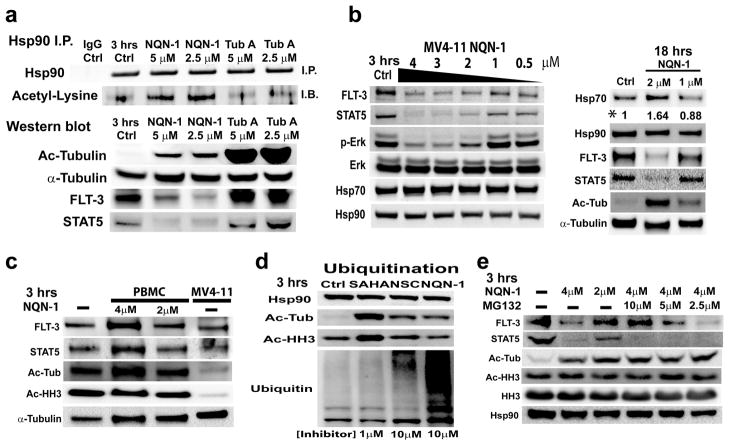
Effect of Active NQN-1 on the Levels Hsp90 acetylation, Hsp70, FLT-3, STAT5, and Erk phosphorylation
To be more explicit in the mechanism of action, it is critical to explore cellular pathways affected by the inhibitors. HDAC6 is known to control Hsp90 chaperone function through acetylation of its lysine residues and affects its client protein stabilities i.e. FLT-3, BCR-ABL, Raf-1, Akt, etc. (22, 31–32). Inhibition of HDAC6 and selective knock-down of HDAC6 have shown to reduce mutant FLT-3, Erk phosphorylation, and increase total ubiquitinated protein in cells. We compared our selective HDAC6 inhibitor, NQN-1, with recently reported selective HDAC6 inhibitor, tubastatin A, with >1000 selectivity over HDAC1 using immunoprecipitation (I.P.) and immunoblot techniques (33). NQN-1 inhibits HDAC6 and induces Hsp90 acetylation 3 hr following treatment. No induction of Hsp90 acetylation was observed in tubastatin A treated cells, although higher induction of tubulin acetylation was observed for tubastatin A than NQN-1 (Figure 6a). Western blots on total cell extracts of MV4-11 cell, containing the mutant FLT-3 and constitutively active STAT5 (a downstream effector of tyrosine activation with a key role in AML) (34–35). Figure 6, panel a and b, shows FLT-3 and STAT5 protein levels decrease within 3 hr following NQN-1 treatment but not for tubastatin A. This is consistent with the inhibitor’s ability to induce Hsp90 acetylation. Similar results have also been reported by Bali et al. of another selective HDAC6 inhibitor, tubacin, which showed tubacin does not significantly increase Hsp90 acetylation even after 16 hrs post inhibitor treatments (22, 36). We have also examined the ability of other HDAC inhibitors to induce Hsp90 acetylation 3 hr post-treatment. Surprisingly, SAHA, TSA, and inhibitor 106 did not induce Hsp90 acetylation or reduce FLT-3 and STAT5 levels suggesting a direct link to Hsp90 acetylation and FLT-3 and STAT5 depletions (Supporting Information). Contrary to previous proteomic studies, SAHA was reported to induce acetylation of some residues of Hsp90 after 24 hrs in MV4-11 cells (3). HDAC inhibitors are known to down-regulate HDAC expression through transcriptional controls (37–39); therefore, it is possible that prolonged treatment of SAHA would decrease the level of HDAC6 in MV4-11 resulting in Hsp90 hyperacetylation and inhibition after 24 hrs. Other thiolated HDAC6 inhibitors, which substitute a thiol for the hydroxamic acid moiety, only have modest preference for HDAC6, thus were not compared in this study (33, 40–41). In PBMC, little hyperacetylated tubulin was observed 3 hr post-treatment, and normal FLT-3 and STAT5 protein levels were not affected by the treatment (Figure 6c), which is consistent with a previous study using tubacin (30). The authors attribute the lack of tubacin-induced tubulin hyperacetylation on the low expression of HDAC6 in PBMC. This is consistent with our observation that acetylated tubulin and acetylated histone H3 in normal PBMC are several folds higher than the malignant AML MV4-11 cells indicating either PBMC has a low expression of HDAC6 or HDAC6 is inactive in PBMC population from health donors (Figure 6c). This result suggests that HDAC6 activity and inhibition affects normal cells differently than leukemia cells, and it offers an explanation to the selective nature of the inhibitor. Moreover, the inhibition of Hsp90 chaperone activity due to HDAC6 inhibition increases Hsp70 protein level after 6 hrs (Figure 6b and Supporting Information), which is consistent with previous reports of increased Hsp70 by HDAC6 siRNA knock-down.
Figure 6. NQN-1 effects on Hsp90 acetylation, mutant FLT-3, STAT5, and Erk phosphorylation.
a) NQN-1 induces Hsp-90 acetylation, and causes degradation of FLT-3, STAT5, and a decrease in the level of phos-Erk 3 hr post inhibitor treatment in MV4-11 cells. b) NQN-1 decreases FLT-3, STAT5, and phos-Erk in a concentration dependent manner. Increase in Hsp70 level was only observed after prolonged treatment, 18 hrs (*quantification with spot-density). c) Tubulin acetylation level did not increase, and normal FLT-3 and STAT5 did not decrease when PBMC was treated with NQN-1. Significantly higher acetylated tubulin and histone H3 levels were observed in PBMC (n=4 normal donors) in comparison to malignant MV4-11 cells. d) NQN-1 and NSC treatment resulted in an increase in total ubiquitinated protein level in MV4-11 cells. e) Proteasome inhibitor, MG132, rescued the depletion of mutant FLT-3 when co-administrated at 10 μM. However, the proteasome inhibitor did not prevent depletion of STAT5 indicating a proteasome independent degradation pathway that is separate from the Hsp90 function.
Previous studies have also shown Hsp90 client protein degradation is mainly mediated through proteasomal pathways with increased cellular ubiquitinated protein levels (22, 30–31). NQN inhibitors also increase the level of ubiquitinated proteins within 3 hr following treatment (Figure 6d). To further elucidate the role HDAC6 inhibition and proteasomal pathways play in FLT-3 (a known client protein of Hsp90) and STAT5 degradation, proteasome inhibitor MG132 was used to rescue FLT-3 degradation caused by HDAC6 inhibition and loss of Hsp90 chaperone function. As expected, FLT-3 degradation is rescued by 10 μM MG132 co-treatment. This is consistent with our hypothesis of HDAC6 inhibition affecting Hsp90 client protein through proteasome degradation (Figure 6e). However, MG132 did not prevent STAT5 degradation, indicating that HDAC6 inhibition might also play a role in controlling the function of other proteases, which will be investigated further.
CONCLUSION
In this study, we have shown that it is possible to identify novel HDAC inhibitors with potential for improved isozyme selectivity that do not conform to the canonical structural motifs of known HDAC inhibitors (19). We demonstrated that HDAC class-specific substrates and highly active recombinant HDAC enzymes can be used to screen a compound library in high-throughput compatible format. Using this assay to screen 1280 compounds, we have identified, for the first time, a class of small molecule inhibitors of HDAC6 with IC50s in the μM range that do not contain either hydroxamic acid or benzamide metal chelating groups.
In human AML cells, the prototype compound NQN-1 induced hyperacetylation of tubulin and Hsp90 within 3 hrs following treatment, which the known hydroxamic acid based inhibitor, tubastatin A, does not. It also caused degradation of the Hsp90 client protein, mutant FLT-3, as well as constitutively active STAT5, which are known to be critical for leukemia cell proliferation (32, 34). The degradation of mutant FLT-3 is proteasome-dependent and can be rescued with the proteasome inhibitor, MG132, while STAT5 degradation is proteasome-independent and requires further investigation. Additionally, the compound shows moderately selective toxicity for leukemia cells versus normal proliferative PBMC and is inactive against non-proliferative PBMC. Similarly, the HDAC6 selective inhibitor, tubacin, has been shown to selectively induce apoptotic cell death in multiple myeloma and acute lymphoblastic leukemia cells with a lesser effect on normal PBMC (30, 42). Namdar et al. recently reported that tubacin only slows the growth but does not induce cell death of normal human foreskin fibroblast and transformed human prostate and breast cancer cells (43). These studies suggest HDAC6 inhibition offers potential selective toxicity against cancers of leukemic or plasma origins.
All the active compounds derived from the screen targeting HDAC 6 contain the central 1,4-naphthoquinone ring system. In the recombinant enzyme and cellular assays performed to date, NQN-1 displayed the strongest potency and selectivity at inducing AML cell death, HDAC6 inhibition, and Hsp90 acetylation. Interestingly, the carbonyl linking the naphthoquinone and the phenyl groups is required for its HDAC6 inhibitory activity and toxicity against leukemia cells, while vitamin K3, with a simple 2-methyl-1,4-napthoquione structure, inhibits HDAC6 with IC50 of 25 μM. We theorize that although the phenyl group from NQN-1 improves the specificity towards HDAC6, the larger phenyl group might not be ideal. This is supported by the potency decrease imparted by a single methylene linker extension in NQN-2. The lack of toxicity of NQN-3 and the attenuation of the NQN-1 toxic effects by over-expression of HDAC6 (Supporting Information) suggest that the toxicity is target specific and is unlikely due to non-specific toxicity from the 1,4-naphthoquione core. Further structural investigation will be done to improve both the affinity and specificity of this unique class of HDAC6 inhibitors.
Using compounds identified and derived from the novel high-throughput screen developed in this study, we demonstrated that inhibition of HDAC6 and interference with Hsp90 chaperone function results in the degradation of critical proteins and attenuation of signaling cascades that are known to promote leukemia cell growth. This data, combined with demonstrated selective toxicity of the compounds towards AML cells, provides a strong rationale for the development of novel small molecule inhibitors of HDAC6. This unique class of HDAC6 inhibitor already shows microM activity in various in vitro and ex vivo assays. Future generations of inhibitors with increased potency and selectivity for HDAC6 will not only be useful chemical probes for investigations seeking to elucidate the isozyme’s involvement in various pathologies, but also have the potential to be developed into novel leukemia and cancer therapeutics.
METHODS
Materials
Recombinant human HDAC1, HDAC2, HDAC3/N-CoR2, HDAC5, HDAC6, HDAC8, HDAC9 and HDAC10 expressed in baculovirus, were purchased from BPS Bioscience. Recombinant human HDAC4 and HDAC7 were purchased from Millipore, and HDAC11 was purchased from Enzo Life Sciences. Acetylated-Lys(acetyl)-7-amino-methylcoumarin (acetyl-Lys(Ac)-AMC) was purchased from MP Biomedicals. Acetyl-Lys(trifluoroacetyl)-7-amion-methylcoumarin (acetyl-Lys(TFA)-AMC) was purchased from Bachem. Leucine, glysine, lysine acetyl and trifluoroacetyl substrates (Ac-KGL and TFA-KGL substrates) were synthesized as previously described (Supporting Information). Suberoylanilide hydroxamic acid (SAHA) and Tubastatin A were purchased from Cayman Chemical, and Tubacin was purchased from Enzo Life Science. Diphenyl acetic hydroxamic acid was synthesized as previously described (24). All LOPAC chemicals were purchased from Sigma-Aldrich and dissolve with DMSO to a minimum of 10 mM stock solution. All other chemicals were purchased from Sigma-Aldrich. NQN-1 was purchased from Sigma-Aldrich and synthesized in our laboratory (Supporting Information). NQN-2, NQN-3, and NQN-4 were synthesized as described and characterized with H1-NMR and LC-MS/MS (Supporting Information).
Cells and Antibodies
MV4-11, Kasumi-1, and Reh leukemia cells were purchased from American Type Culture Collection (ATCC). Normal PBMCs were purchased from Cellular Technology Inc. MV4-11 is a human AML cell line expressing mutant FLT-3. MV4-11 was cultured in recommended media supplemented with 10% heat-inactivated fetal bovine serum. PMBC was thawed with serum free wash buffer and cultured in standard culture media. All cells were grown in a humidified atmosphere with 5% CO2 at 37°C. Acetylated tubulin, tubulin, acetylated-HH4, acetylated HH3, FLT-3, STAT5, Erk, phos-Erk, ubiquitin, Hsp90, and horseradish peroxide (HRP) secondary antibodies used for immunoprecipitation and Western blot were purchased from Santa Cruz Biotechnology. Hsp70 and acetylated-lysine antibodies were purchased from Cell Signaling Technology. Histone H3 antibody was purchased from Abcam, and acetylated-Hsp90 lysineK69 antibody was purchased from Alpha Diagnostic International.
Primary Hit Identification Using Substrate Specific HDAC Assays and Hit Identification
The LOPAC library was obtained from Sigma-Aldrich. HDAC3 and HDAC7 were pre-incubated with screening compounds for 2 hr and deacetylate reactions were initiated with addition of HDAC substrates (acetyl-Lys(Ac)-AMC for HDAC3 and acetyl-Lys(TFA)-AMC for HDAC7). The reactions were allowed to proceed for 2 hr and developed with 25 μL trypsin peptidase at 10 mg/mL. Z′-factor, a quality parameter used to judge high-throughput performance, is determined using known HDAC inhibitors as positive control for both HDAC3 and HDAC7.
Biochemical HDAC Deacetylase Inhibition Assay
Individual IC50 values for each HDAC isozymes were measured with the homogenous fluorescence release HDAC deacetylase assay. Purified recombinant enzymes were incubated with serial diluted inhibitors at the indicated concentration. The deacetylase activities of class I HDACs 1, 2, 3, 6, and 10 were measured by assaying enzyme activity using AMC-K(Ac)GL substrate and AMC-K(TFA)GL substrate for class IIa HDACs 4,5,7, 8, 9 and 11 as previously described (15, 17). Deacetylated AMC-KGL was sensitive toward lysine peptidase and free fluorogenic 4-methylcoumarin-7-amide (MCA) was generated which can be excited at 355 nm and observed at 460 nm. The data was analyzed on a plate to plate basis in relationship to the control and imported into analytical software (Graphpad Prism).
Compound Libraries and Analogs
LOPAC library was obtained from Sigma-Aldrich. NQN analogs were synthesized as described, characterized with 1H and 13C NMR, mass-spectrometry, and analytical HPLC (Supporting Information).
MTS Colorimetric Cell Proliferation/Toxicity Assay
The inhibitor’s effect on cell proliferation and the ability to initiate apoptosis were determined using Promega CellTiter 96 Aqueous One Cell Proliferation assay. The effective dose 50 (EC50) was determine from the inhibitor against cell proliferation, and the data were analyzed with Graphpad Prism.
Immunoprecipitation and Western Blotting
Treated cells were washed with cold Hanks’ Balanced Salt Solution (Invitrogen) follow by lysis with a low salt lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5 Triton X-100, 1X protease and phosphatase inhibitor cocktail, and 1 μM TSA) for 30 min, and followed by a 15s sonication pulse at 30 watts. For immunoprecipitation, the lysates were incubated with Hsp90 conjugated Dynabeads (Invitrogen) for 1 hr according to the manufacture instruction. The immunoprecipitates were immunoblotted and test against Hsp90 (loading control), anti-acetylated lysine (acetylation level), and acetylated-Hsp90K69. For Western blot, the cell lysates were denatured with LDS loading buffer (Invitrogen) and run on 4–12% Tris-Glysine gradient gels after centrifugation. Total histone H3, acetylated histone H3, acetylated H4, FLT-3, STAT5, Hsp70, Hsp90, and acetylated α-tubulin were assessed with primary antibody, follow by rabbit or mouse IgG-HRP secondary antibody. The blot was developed using Pierce Thermo Dura-ECL reagent (Thermo-Fisher) and visualized using Alpha Innotech gel imager (Cell Biosciences).
Data Analysis
The inhibition of HDAC activity by small-molecule inhibitor candidates was measured as described above. Data were analyzed by non-linear regression with determination of IC50 and standard deviation using GraphPad Prism. Ki values were calculated based on the Cheng-Prusoff equation for standard fast-on/off inhibitor, Ki = IC50/(1+([S]/Km)), with IC50 values derived from the nonlinear curve fit of the dose response curves. Ki, inhibition constant; [S], substrate concentration; Km, Michaelis constant.
Supplementary Material
Acknowledgments
We thank C. Smith for his assistance in HDAC compound screens; R. Schnellmann and C. Beeson for the LOPAC library and valuable discussion. This work was supported by MUSC and the South Carolina Clinical & Translational Research Institute in MUSC CTSA, NIH/NCRR Grant Number UL1RR029882.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 6.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 9.Sternson SM, Wong JC, Grozinger CM, Schreiber SL. Synthesis of 7200 small molecules based on a substructural analysis of the histone deacetylase inhibitors trichostatin and trapoxin. Org Lett. 2001;3:4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 10.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci U S A. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 19.Miller TA, Witter DJ, Belvedere S. Histone deacetylase inhibitors. J Med Chem. 2003;46:5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 20.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 23.Kekatpure VD, Dannenberg AJ, Subbaramaiah K. HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. J Biol Chem. 2009;284:7436–7445. doi: 10.1074/jbc.M808999200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Tessier P, Smil DV, Wahhab A, Leit S, Rahil J, Li Z, Deziel R, Besterman JM. Diphenylmethylene hydroxamic acids as selective class IIa histone deacetylase inhibitors. Bioorg Med Chem Lett. 2009;19:5684–5688. doi: 10.1016/j.bmcl.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 27.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J Biol Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandl A, Heinzel T, Kramer OH. Histone deacetylases: salesmen and customers in the post-translational modification market. Biol Cell. 2009;101:193–205. doi: 10.1042/BC20080158. [DOI] [PubMed] [Google Scholar]
- 30.Hideshima T, Bradner JE, Wong J, Chauhan D, Richardson P, Schreiber SL, Anderson KC. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci U S A. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao R, Fiskus W, Yang Y, Lee P, Joshi R, Fernandez P, Mandawat A, Atadja P, Bradner JE, Bhalla K. HDAC6 inhibition enhances 17-AAG--mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 32.Lin TY, Fenger J, Murahari S, Bear MD, Kulp SK, Wang D, Chen CS, Kisseberth WC, London CA. AR-42, a novel HDAC inhibitor, exhibits biologic activity against malignant mast cell lines via down-regulation of constitutively activated Kit. Blood. 2010;115:4217–4225. doi: 10.1182/blood-2009-07-231985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003;17:120–124. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 35.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse JF, Capiod JC, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 36.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dokmanovic M, Perez G, Xu W, Ngo L, Clarke C, Parmigiani RB, Marks PA. Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol Cancer Ther. 2007;6:2525–2534. doi: 10.1158/1535-7163.MCT-07-0251. [DOI] [PubMed] [Google Scholar]
- 38.Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T, Wurst W, Minucci S, Gottlicher M. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 39.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki T, Kouketsu A, Itoh Y, Hisakawa S, Maeda S, Yoshida M, Nakagawa H, Miyata N. Highly potent and selective histone deacetylase 6 inhibitors designed based on a small-molecular substrate. J Med Chem. 2006;49:4809–4812. doi: 10.1021/jm060554y. [DOI] [PubMed] [Google Scholar]
- 41.Itoh Y, Suzuki T, Kouketsu A, Suzuki N, Maeda S, Yoshida M, Nakagawa H, Miyata N. Design, synthesis, structure--selectivity relationship, and effect on human cancer cells of a novel series of histone deacetylase 6-selective inhibitors. J Med Chem. 2007;50:5425–5438. doi: 10.1021/jm7009217. [DOI] [PubMed] [Google Scholar]
- 42.Aldana-Masangkay GI, Rodriguez-Gonzalez A, Lin T, Ikeda AK, Hsieh YT, Kim YM, Lomenick B, Okemoto K, Landaw EM, Wang D, Mazitschek R, Bradner JE, Sakamoto KM. Tubacin suppresses proliferation and induces apoptosis of acute lymphoblastic leukemia cells. Leuk Lymphoma. 2011;52:1544–1555. doi: 10.3109/10428194.2011.570821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namdar M, Perez G, Ngo L, Marks PA. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc Natl Acad Sci U S A. 2010;107:20003–20008. doi: 10.1073/pnas.1013754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.