Abstract
The extremely tight binding between biotin and avidin or streptavidin makes labeling proteins with biotin a useful tool for many applications. BirA is the Escherichia coli biotin ligase that site-specifically biotinylates a lysine side chain within a 15-amino acid acceptor peptide (also known as Avi-tag). As a complementary approach to in vivo biotinylation of Avi-tag-bearing proteins, we developed a protocol for producing recombinant BirA ligase for in vitro biotinylation. The target protein was expressed as both thioredoxin and MBP fusions, and was released from the corresponding fusion by TEV protease. The liberated ligase was separated from its carrier using HisTrap HP column. We obtained 24.7 and 27.6 mg BirA ligase per liter of culture from thioredoxin and MBP fusion constructs, respectively. The recombinant enzyme was shown to be highly active in catalyzing in vitro biotinylation. The described protocol provides an effective means for making BirA ligase that can be used for biotinylation of different Avi-tag-bearing substrates.
Keywords: Biotinylation, BirA, Fusion expression, Maltose-binding protein, TEV protease cleavage, Thioredoxin
Introduction
Biotin is a small organic molecule that binds avidin and streptavidin with extremely high affinity [1]. Proteins can be covalently labeled with biotin, a process known as biotinylation, through chemical or enzymatic means. Biotinylation is one of the most commonly used protein labeling methods for easy detection, immobilization, and purification [2]. BirA, the E. coli biotin ligase, is an enzyme that can catalyze the covalent attachment of biotin to the lysine side chain within a 15-amino acid peptide termed the Avi-tag [3–6]. Many proteins have been labeled with biotin via the Avi-tag for a variety of purposes [7–10].
E. coli B strain AVB101 contains an engineered pACYC184 plasmid with an inducible birA gene. It is designed for co-expression of BirA and Avi-tagged protein of interest for in vivo biotinylation. We previously used this system to generate biotinylated proteins that can be used as immobilized ligands for interaction studies. The protein of interest, which has an Avi-tag fused to it, was expressed as a thioredoxin fusion by inserting the corresponding gene into commercial vector pET-32a. Since the AVB101 strain does not have a T7 RNA polymerase expression system and will therefore not express genes from the T7 promoter-driven pET-32a vector, we isolated the BirA encoding vector pACYC184 and co-transfected it with pET-32a harboring the target gene into E. coli strain BL21(DE3). BirA was co-expressed with the Avi-tagged target protein. While a portion of the target protein was biotinylated in vivo in this way, the biotinylation efficiency varied from batch to batch. We noticed that the expression level of BirA dropped significantly when it was co-expressed with the Avi-tagged target protein. In addition, we found that the labeled protein gradually loses its biotin during storage, which was indicated by the decreased efficiency of its binding to streptavidin-coated sensor chip. In order to more efficiently biotinylate Avi-tagged substrates and to relabel substrates whose biotin was lost during storage, we decided to produce recombinant BirA for in vitro biotinylation as a complementary approach to in vivo labeling.
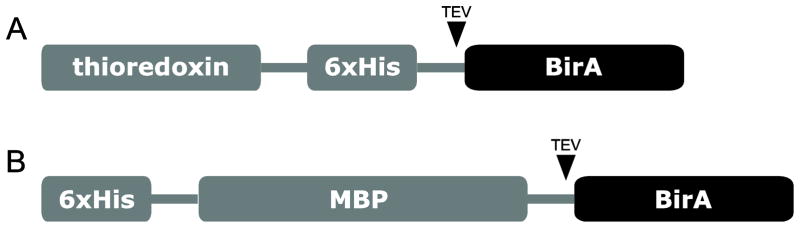
In this work, we expressed BirA biotin ligase as both thioredoxin and maltose-binding protein (MBP)1 fusions (Fig. 1). The ligase was released from its corresponding fusion by tobacco etch virus (TEV) protease cleavage, and subsequently separated from the carrier using HisTrap HP column. The purified recombinant BirA is highly active in catalyzing in vitro biotinylation. The described protocol provides an effective means for making BirA ligase that efficiently biotinylates Avi-tag-bearing substrates.
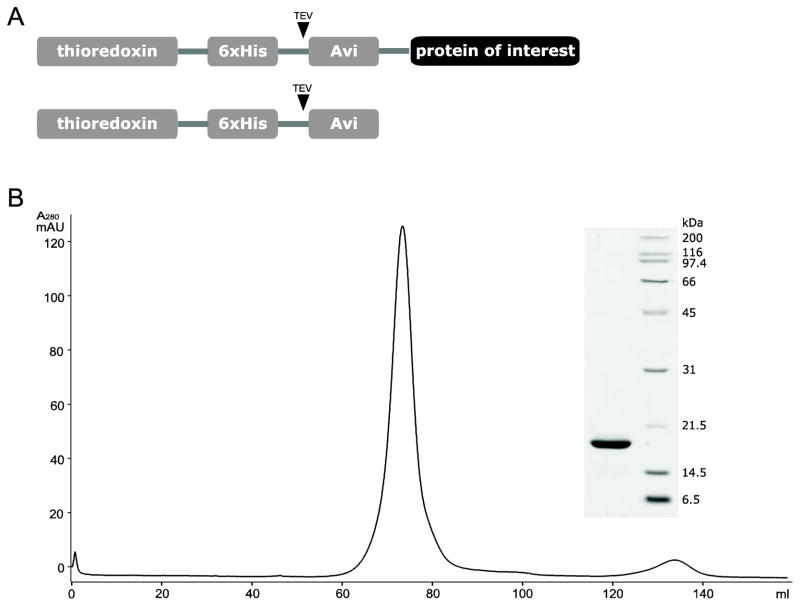
Fig. 1.
Schematic representation of the two constructs in which the target protein is expressed as a fusion protein with (A) thioredoxin or (B) MBP.
Materials and methods
Materials
One Shot BL21(DE3) competent cell and dNTP mix were obtained from Invitrogen. Escherichia coli B strain AVB101 harboring a pACYC-184 plasmid, which contains the birA gene, was purchased from Avidity. Plasmids pET-32a and pET-28a were purchased from Novagen. TEV protease expression vector pRK793 was obtained from Addgene. All oligonucleotides were synthesized at the Nucleic Acids Core Facility at the University of Texas Health Science Center at San Antonio (UTHSCSA). Vent DNA polymerase, restriction enzymes, calf intestinal alkaline phosphatase (CIP), T4 DNA ligase and DNA marker were obtained from New England Biolabs. QIAquick PCR purification kit, QIAquick gel extraction kit, QIAprep spin miniprep kit, and Ni-NTA agarose were purchased from Qiagen. Difco LB broth was purchased from BD Biosciences. Tris, glycine, SDS, 30% acrylamide/bis, and protein standards were purchased from Bio-Rad. Isopropyl β-D-1-thiogalactopyranoside (IPTG) and adenosine 5′-triphosphate (ATP) disodium salt hydrate (A7699-10G) were purchased from Gold Biotechnology and Sigma, respectively. HisTrap HP column (5 ml) and Streptavidin Sepharose were purchased from GE Healthcare. ImmunoPure D-biotin and Zeba spin desalting column (0.5 ml) were obtained from Pierce and Thermo Scientific, respectively.
Construction of fusion expression plasmids
The gene encoding BirA was amplified by PCR with primers 1 and 2 (Table 1) using pACYC-184 plasmid as template. The forward and reverse primers contain EcoRI and NotI sites, respectively. The forward primer also contains the sequence encoding a TEV protease cleavage site. The PCR amplified birA gene was digested with EcoRI and NotI, and ligated into pET-32a doubly digested with the same enzymes. The resultant recombinant plasmid, which allows the target protein to be expressed as a thioredoxin fusion (Fig. 1A), was named BirA/pET-32a.
Table 1.
Primers used for birA gene amplification and vector modification

|
Note. The restriction sites used for cloning are shown in italics, with the name of the enzyme under the sequence. In primer 1, the coding sequence for TEV protease cleavage site is underlined.
For MBP fusion expression (Fig. 1B), the commercial vector pET-28a was first modified by inserting a sequence encoding MBP (Fig. 2). This was achieved by amplifying the MBP coding sequence with primers 3 and 4 (Table 1) using vector pRK793 as the template and inserting the NdeI and BamHI double digested PCR product into similarly digested pET-28a. Next, the PCR amplified birA gene was digested with EcoRI and NotI, and ligated into the modified pET-28a plasmid doubly digested with the same enzymes. The resultant construct for MBP fusion expression was named MBP-BirA/pET-28a.
Fig. 2.
Expression/cloning region of (A) original pET-28a and (B) its modified version, in which a sequence encoding MBP was inserted between the NdeI and the BamHI sites. This modification allows the vector to be used for MBP fusion expression.
The presence and identity of the insert in both constructs was verified by diagnostic restriction digestion and DNA sequencing.
Fusion proteins expression and purification
After transformation of E. coli strain BL21(DE3) with recombinant vector BirA/pET-32a, the bacteria were grown overnight in 100 ml LB broth (with 100 μg/ml ampicillin) at 37 °C. This overnight culture was used to inoculate one liter of fresh LB medium and cells were grown at 28 °C with shaking at 250 rpm. When the culture OD600 reached 0.6, protein expression was induced by adding IPTG to a final concentration of 1 mM. The culture temperature was reduced to 18 °C following induction. After additional 22 h cultivation, cells were harvested by centrifugation at 6000 rpm for 10 min. The bacterial pellet (~6.3 g) was resuspended in 30 ml of cell lysis buffer (25 mM Tris, 200 mM NaCl, pH 8.0) and cells were lysed by sonication. The cell lysate was then shaken at 4 °C for 30 min followed by centrifugation at 20,000 rpm for 40 min. The supernatant was combined with 5 ml Ni-NTA resin suspension and shaken at 4 °C for 3 h. The resin was pelleted by spinning at 1000 rpm for 5 min and the supernatant, which contains unbound proteins, was discarded. The resin was washed twice by shaking with 35 ml of lysis buffer containing 20 mM imidazole at 4 °C for 30 min. Protein bound to the resin was eluted with 20 ml of lysis buffer containing 400 mM imidazole. The fusion protein was further purified by running through a HiLoad 16/60 Superdex 75 column equilibrated with cell lysis buffer. The MBP fusion protein encoded by MBP-BirA/pET-28a was obtained following the same procedure, except that the bacteria were grown in medium containing 50 μg/ml kanamycin.
Expression and purification of TEV protease
TEV protease was expressed and purified following the protocol developed by David Waugh’s lab [11]. In brief, E. coli strain BL21(DE3) harboring recombinant vector pRK793 was grown at 37 °C until OD600 reached 0.6, at which point IPTG was added to a final concentration of 1 mM and the culture temperature was reduced to 30°C. After 7 h, bacteria were harvested and resuspended in cell lysis buffer containing 10% glycerol and 25 mM imidazole. Cells were lysed by sonication and the cell lysate supernatant was incubated with Ni-NTA resin for 3 h with shaking at 4 °C. After washing twice with 25 mM imidazole, the protein was eluted with cell lysis buffer containing 10% glycerol and 300 mM imidazole.
Target protein release and isolation
Both thioredoxin-BirA and MBP-BirA fusion proteins were cleaved with TEV protease at a mass ratio of 100:1 (i.e., for 1 mg of purified fusion, 10 μg of protease was added) to release the target protein. The cleavage reaction was allowed to proceed at room temperature for 16 h. After cleavage, the reaction mixture was subjected to a HisTrap HP column (5 ml). The unbound portion which contains recombinant BirA was collected and stored in 50% glycerol at −20 °C for further use.
Activity test
The biological activity of recombinant BirA ligase was evaluated by applying different amounts of enzyme to 30 μM Avi-tagged substrate (in cell lysis buffer, pH 8.0) in the presence of 0.3 mM biotin and 5 mM ATP. The reaction was allowed to proceed at room temperature for 14 h. Biotinylation was confirmed by incubating the reaction mixture with Streptavidin Sepharose, which captures the biotinylated substrate. Before applying sample to the Streptavidin Sepharose, free biotin left in the solution was removed using a Zeba spin desalting column (0.5 ml).
Results
Fusion protein expression and purification
BirA is a monomeric protein of 35.3 kDa. The molecular weights of thioredoxin-BirA and MBP-BirA fusions are 54.1 and 81.3 kDa, respectively. In both cases, the expressed fusion proteins were mainly found in the soluble portion and can be purified to a high degree using affinity and size-exclusion chromatography (Fig. 3).
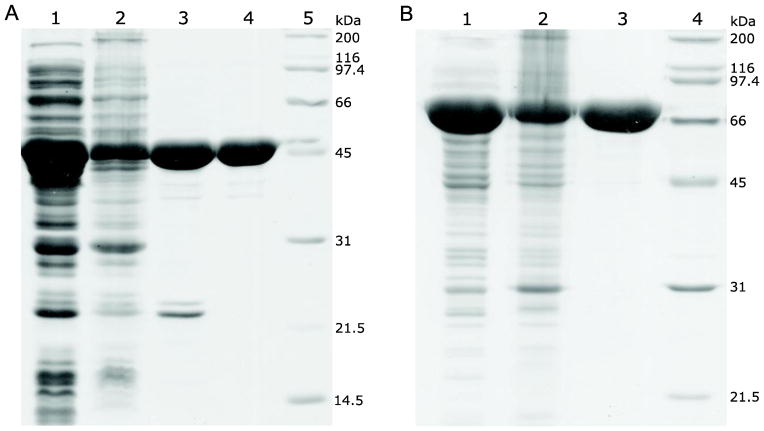
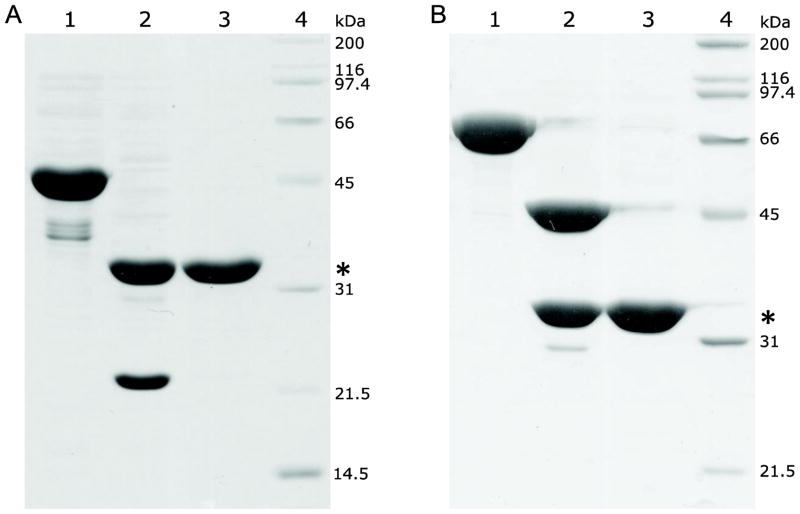
Fig. 3.
Expression and purification of BirA fusion proteins as followed by SDS-PAGE (12%). (A) The target protein was expressed as a thioredoxin fusion. Lane 1, cell lysate supernatant; Lane 2, cell lysate pellet (resuspended in 8 M urea); Lane 3, imidazole eluate (Ni-NTA resin purified protein); Lane 4, size-exclusion chromatography purified protein; Lane 5, protein standards. (B) The target protein was expressed as an MBP fusion. Lane 1, cell lysate supernatant; Lane 2, cell lysate pellet (resuspended in 8 M urea); Lane 3, affinity and size-exclusion chromatography purified fusion protein; Lane 4, protein standards.
Expression and purification of TEV protease
TEV protease was expressed as a MBP fusion, which undergoes in vivo self-cleavage and yields a soluble form of the N-terminally His-tagged enzyme [11]. The protease can be obtained in relatively pure form after a single step affinity purification using Ni-NTA resin (Fig. 4).
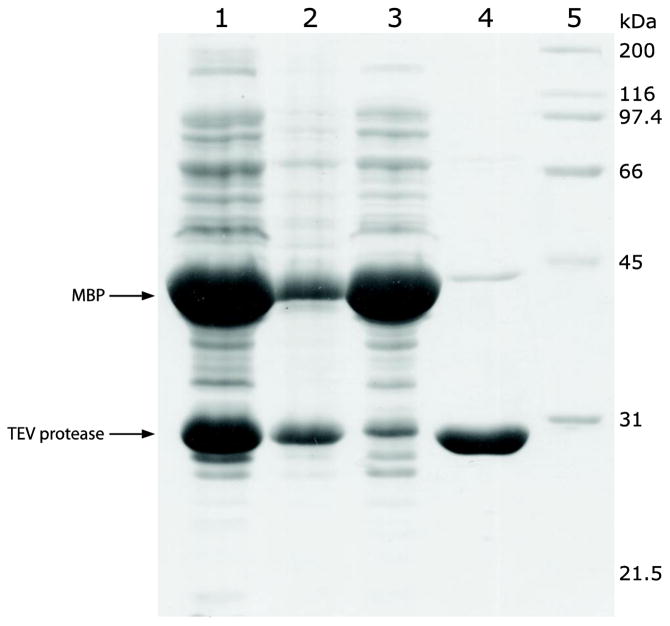
Fig. 4.
Expression and purification of TEV protease as followed by SDS-PAGE (12%). Lane 1, cell lysate supernatant; Lane 2, cell lysate pellet (resuspended in 8 M urea); Lane 3, cell lysate supernatant after binding with Ni-NTA resin (unbound proteins); Lane 4, imidazole eluate (Ni-NTA resin purified protein); Lane 5, protein standards.
Target protein release and isolation
The thioredoxin-BirA fusion protein was cleaved by TEV protease without stirring and a small scale test showed that the reaction was virtually complete in 16 h at room temperature when 10 μg TEV protease per mg fusion protein was used (Fig. 5A). TEV cleavage leaves an extra glycine at the N-terminus of BirA.
Fig. 5.
(A) Cleavage efficiency of the thioredoxin-BirA fusion protein with different amounts of TEV protease as followed by SDS–PAGE (12%). The reactions were allowed to proceed for 16 h at room temperature. Lane 1–6, 0, 2, 4, 6, 8, 10 μg of TEV protease was added to 1 mg of fusion protein, respectively; Lane 7, protein standards. (B) Separation of the target protein from the carrier thioredoxin by using a HisTrap HP column (5 ml). Lane 1–3, fractions that flowed through the column and which contain the target protein; Lane 4–5, factions that bound to the column and eluted with increasing concentration of imidazole, Lane 6, protein standards.
The released target protein was separated from the carrier using a HisTrap HP column (5 ml). The target protein will not bind to the column and comes out shortly after injection, whereas the carrier protein, which contains a His-tag, binds to the column and is eluted with increasing concentrations of imidazole (Fig. 5B).
BirA expressed as an MBP fusion was released and isolated following the same procedure. In both cases (thioredoxin-BirA and MBP-BirA), fusion cleavage and target protein isolation is highly efficient and recombinant BirA was obtained in high purity (Fig. 6). The yield for purified fusion proteins and BirA from one liter of culture is summarized in Table 2.
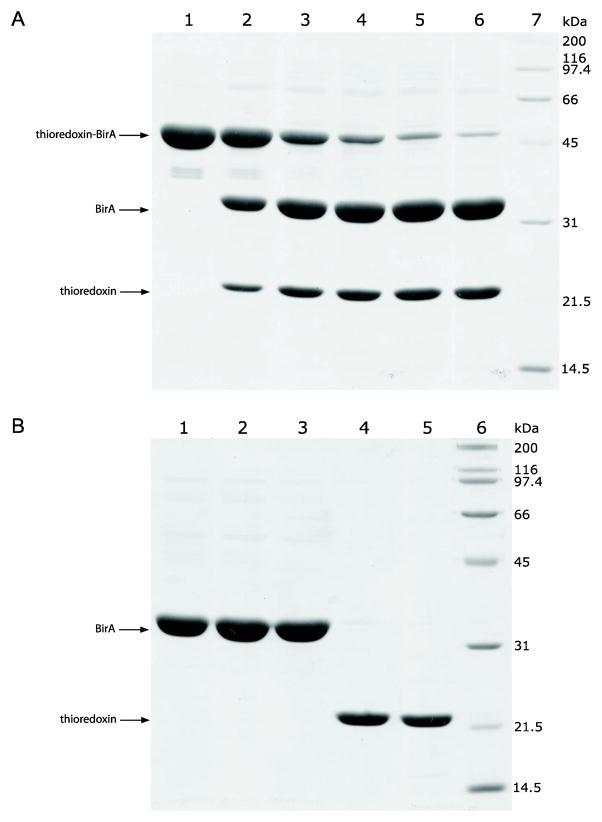
Fig. 6.
(A) thioredoxin and (B) MBP fusion cleavage and target protein purification as followed by SDS-PAGE (12%). Lane 1, corresponding fusion proteins purified by affinity and size-exclusion chromatography; Lane 2, reaction mixture after TEV protease cleavage; Lane 3, purified BirA; Lane 4, protein standards. The position of bands corresponding to the released BirA is indicated by an asterisk.
Table 2.
Yields of fusion proteins and BirA from the two different constructs
| Thioredoxin-BirA (65.2%)a | MBP-BirA (43.4%) | |
|---|---|---|
| Purified fusion protein (mg/L) | 45.9 | 79.2 |
| Purified BirA (mg/L) | 24.7 (29.9)b | 27.6 (34.4) |
Note. In both cases, roughly 6.3 g wet cells were obtained from 1 liter of culture.
Mass percentage of the target protein in the corresponding fusion.
Theoretical yield calculated based on the amount of fusion protein and the corresponding mass percentage of the target protein in the two fusions.
Activity test
We previously made a construct for producing Avi-tagged target protein expressed as a thioredoxin fusion. By introducing a stop codon at the end of the Avi-tag coding sequence, this construct allows the expression of C-terminally Avi-tagged thioredoxin (Fig. 7A). Expression of this Avi-tagged thioredoxin was induced with 1 mM IPTG at 37 °C. The protein was highly soluble and was purified by affinity and size-exclusion chromatography (Fig. 7B). This Avi-tagged thioredoxin was used as a substrate for BirA catalyzed in vitro biotinylation.
Fig. 7.
(A) Schematic representation and (B) size-exclusion chromatography elution profile of the Avi-tagged thioredoxin. Inset shows SDS-PAGE analysis of the major peak component.
The substrate was treated with different amounts of recombinant BirA. SDS-PAGE analysis suggested that the biotinylated substrate migrates slightly slower than its unlabeled counterpart (Fig. 8A). The lanes corresponding to low enzyme concentration (Fig. 8A, Lane 2 and 3) showed two bands, indicating the samples contain a mixture of biotinylated and non-biotinylated forms of the substrate. The enzyme is highly active and at 1:100 enzyme to substrate molar ratio the reaction was complete in 4 h at room temperature. This is indicated by the vanishing of non-biotinylated substrate starting from Lane 3 in Fig. 8B.
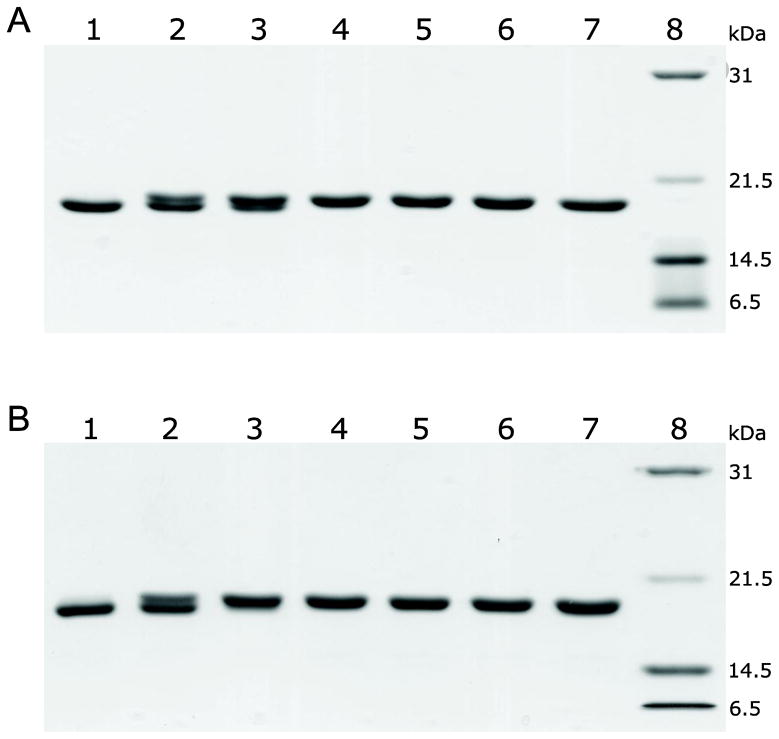
Fig. 8.
(A) Biotinylation at different enzyme to substrate molar ratios. Reactions were allowed to proceed for 14 h at room temperature. Lanes 1–7: 0, 0.05, 0.1, 0.2, 0.3, 0.4 and 0.5 μM of recombinant BirA was added to 30 μM of substrate, respectively; Lane 8, protein standards. (B) Biotinylation reaction monitored as a function of time (enzyme to substrate molar ratio was 1:100). Lane 1–7, aliquots taken from the reaction at 0, 2, 4, 6, 8, 10 and 12 h, respectively; Lane 8, protein standards.
In addition to the different migration on SDS-gel, biotinylation of the substrate was confirmed by its ability to interact with streptavidin. After incubation with Streptavidin Sepharose, Avi-tagged thioredoxin from the control reaction (non-biotinylated form) remained in the solution (Fig. 9, Lane 3) whereas the substrate from biotinylation reaction was captured and disappeared from the solution (Fig. 9, Lane 4). The captured substrate can be recovered by applying biotin to the resin (Fig. 9, Lane 5).
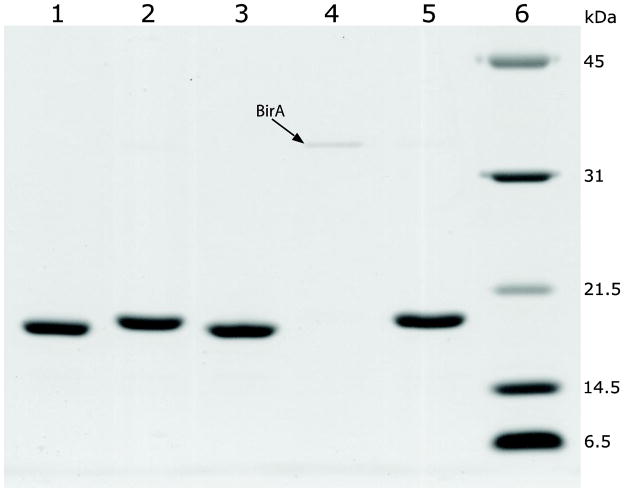
Fig. 9.
Capture of biotin-labeled thioredoxin by Streptavidin Sepharose to confirm biotinylation. Lane 1, unlabeled thioredoxin; Lane 2, labeled thioredoxin; Lane 3, solution of unlabeled thioredoxin after incubation with Streptavidin Sepharose; Lane 4, solution of biotin-labeled thioredoxin after incubation with Streptavidin Sepharose (the faint band on the gel is BirA as indicated); Lane 5, biotin eluate that recovers the biotin-labeled thioredoxin captured by the resin; Lane 6, protein standards.
The recombinant BirA derived from the two fusion constructs are identical in sequence. Whereas the BirA used in the assay shown in Figure 8 was generated from the thioredoxin fusion, the ligase generated from MBP fusion was equally active in catalyzing biotinylation (data not shown). In fact, both fusion proteins exhibit similar catalytic capacity as the released biotin ligase (Fig. 10). Thus, the fusion proteins can be directly used for in vitro biotinylation. This actually has an advantage. As both fusions contain His-tag, they can be removed after biotinylation by running the reaction mixture through a Ni-NTA column if the substrate does not contain a His-tag.
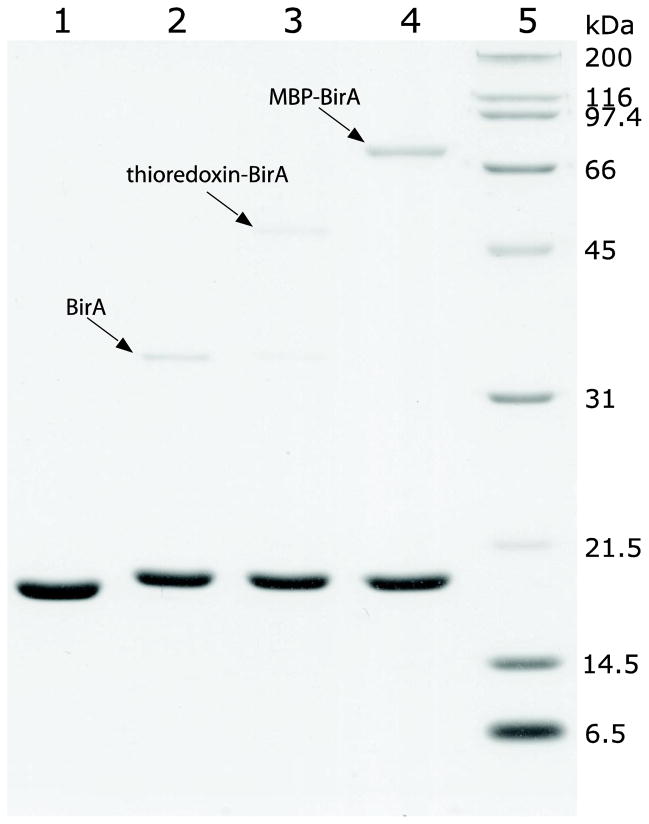
Fig. 10.
Activity assay for thioredoxin-BirA and MBP-BirA fusion proteins. Lane 1, negative control of the biotinylation reaction, which contains all the reagents except for the biotin ligase; Lane 2–4, biotinylation by released BirA (positive control), thioredoxin-BirA and MBP-BirA, respectively (enzyme to substrate molar ratio was 1:50 in all three cases and the reactions were proceeded at room temperature for 4 h); Lane 5, protein standards.
Discussion
BirA expressed by E. coli strain AVB101 was mainly found in the insoluble portion when protein expression was induced at 37 °C. The protein does not have an affinity tag attached to it and therefore is difficult to obtain in high purity. In this work, BirA was expressed as both thioredoxin and MBP fusions (Fig. 1). Fusion expression usually increases protein solubility and provides affinity handles for protein purification. Both fusion proteins were expressed at high levels. Although they remain largely insoluble when induced at 37 °C, most of the protein became soluble when the culture temperature was reduced to 18 °C after IPTG induction (Fig. 3). In both cases, the fusion proteins can be efficiently cleaved by TEV protease at engineered sites to release the ligase (Fig. 6). The liberated target protein can be effectively separated from the carriers using HisTrap HP column (Fig. 6). 24.7 and 27.6 mg BirA ligase per liter of culture were obtained from thioredoxin and MBP fusion constructs, respectively. This yield is significantly higher than that of previously reported for non-fusion expression, which typically yields 3 mg of protein per liter of culture [12].
The produced recombinant BirA ligase is highly active in catalyzing in vitro biotinylation. At an enzyme to substrate ratio of 1:100, the reaction reaches completion in 4 h at room temperature (Fig. 8B). Biotinylation of the Avi-tagged thioredoxin was unambiguously demonstrated by the substrate’s different migration rate on SDS-gel compared with the non-biotinylated form and its ability to interact with streptavidin (Fig. 9).
In conclusion, we developed a protocol for producing highly pure BirA ligase in relatively large amount. This recombinant ligase is enzymatically active and can be used for in vitro biotinylation of different Avi-tagged substrates for various purposes.
Highlights.
A novel protocol was developed for the production of BirA ligase in E. coli.
The target protein was expressed as both thioredoxin and MBP fusions and released by TEV protease cleavage.
Protein yield was significantly improved compared with that of previously reported non-fusion expression.
The produced recombinant BirA is highly active in catalyzing in vitro biotinylation.
Acknowledgments
This work was supported by departmental funding dedicated to the protein production core facility and NIH-GM52522 (to R.S.).
Footnotes
Abbreviations used: MBP, maltose-binding protein; TEV, tobacco etch virus; IPTG, isopropyl-β-D-thiogalactopyranoside; ATP, adenosine 5′-triphosphate
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weber PC, Ohlendorf DD, Wendoloski JJ, Salemme FR. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 2.Bayer EA, Wilchek M. Protein biotinylation. Methods Enzymol. 1990;184:138–160. doi: 10.1016/0076-6879(90)84268-l. [DOI] [PubMed] [Google Scholar]
- 3.Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981;146:451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- 4.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (NY) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 5.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- 7.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 8.Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 9.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 10.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tropea JE, Cherry S, Waugh DS. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol Biol. 2009;498:297–307. doi: 10.1007/978-1-59745-196-3_19. [DOI] [PubMed] [Google Scholar]
- 12.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]