Abstract
Insects produce a group of antimicrobial peptides (AMPs) in response to microbial infections. Most AMPs are synthesized as inactive precursors/pro-proteins and require proteolytic processing to generate small active peptides. Here we report identification and functional analysis of two lebocin-related proteins (Leb-B and Leb-C) from the tobacco hornworm, Manduca sexta. The mRNA levels of Leb-B and Leb-C increased significantly in larval fat body and hemocytes after injection of Escherichia coli, Micrococcus luteus and Saccharomyces cerevisiae. Western blotting using rabbit polyclonal antibody to Leb-B showed accumulation of large protein(s) and small peptide(s) in larval hemolymph after microbial injection. This result and the presence of RXXR motifs in the deduced amino acid sequences led to our postulation that Leb-B/C may be inactive precursors that are processed in larval hemolymph to generate short active peptides. To test this hypothesis, we expressed and purified full-length and various fragments of Leb-B and Leb-C as thioredoxin (TRX) fusion proteins. We found that fusion proteins could be cleaved by induced larval plasma, and the cleavage sites were determined by protein sequencing. Antibacterial activity of peptide fragments was also verified using synthetic peptides, and active M. sexta lebocin peptides were located at the N-termini of Leb-B/C, which are different from Bombyx mori lebocins 1–4 that are located close to the C-termini. In addition, we found that synthetic Leb-B22–48 peptide not only had higher antibacterial activity but also caused agglutination of E. coli cells. Our results provide valuable information for studying processing of lebocin precursors in lepidopteran insects.
Keywords: Antimicrobial peptide, Lebocin, Insect immunity, Manduca sexta
1. Introduction
Antimicrobial peptides (AMPs) with antibacterial or antifungal activities are produced in invertebrates and vertebrates as a defense mechanism (Bulet et al., 2004). Most AMPs are cationic or amphipathic peptides that can destroy bacterial membrane or inhibit intracellular functions of bacteria (Brogden, 2005; Yeaman and Yount, 2003). AMPs can be classified into α-helix containing peptides, cysteine-containing cyclic peptides and specific residue-rich peptides (Bulet et al., 2004; Lemaitre and Hoffmann, 2007). Insect AMPs are produced by the fat body (an equivalent organ of human liver), hemocytes and epithelial cells (Lemaitre and Hoffmann, 2007). AMP expression is mainly regulated by the Rel family of transcription factors, Dorsal, Dif and Relish, in Drosophila melanogaster via activation of the Toll and IMD (immune deficiency) pathways (Hetru and Hoffmann, 2009; Lemaitre and Hoffmann, 2007). Bacterial peptidoglycan (PG), lipopolysaccharide (LPS) and lipoteichoic acid (LTA) are major bacteria-associated molecular patterns recognized by insects and activate expression of AMPs (Lemaitre and Hoffmann, 2007; Rao and Yu, 2010).
In the tobacco hornworm, Manduca sexta, six classes of AMPs have been identified: attacin (Kanost et al., 1990), cecropin (Dickinson et al., 1988), a lebocin-related precursor protein (Rayaprolu et al., 2010), moricin (Dai et al., 2008), gloverin (Zhu et al., 2003), and defensin (Genbank accession number: HQ400765). Lebocin was originally identified in Bombyx mori (Chowdhury et al., 1995; Furukawa et al., 1997; Hara and Yamakawa, 1995b), and it belongs to a family of lepidoptera-specific proline-rich AMPs (Otvos, 2002). Lebocin precursor proteins have also been identified in Samia cynthia (Bao et al., 2005), Trichoplusia ni (Liu et al., 2000), Pseudoplusia includens (Lavine et al., 2005), and M. sexta (Rayaprolu et al., 2010). Lebocin precursor proteins are secreted proteins that can be processed to produce active lebocin peptides and active B. mori lebocins are 32-residue peptides located closely to the C-termini of precursor proteins (Chowdhury et al., 1995; Furukawa et al., 1997). Glycosylation of the Thr15 in the 32-residue active B. mori lebocins is critical for antibacterial activity (Hara and Yamakawa, 1995b). B. mori lebocins 1–3 show lower antibacterial activities than cecropin B1, and synthetic lebocin peptides show even lower or no activities compared with natural products (Hara and Yamakawa, 1995b). B. mori lebocin-3 can increase permeability of E. coli-type liposomes and reduce minimal inhibitory concentration of cecropin D at low ionic-strength conditions (Hara and Yamakawa, 1995a).
Lebocin precursor proteins from other lepidopteran insects align well only with the N-terminal segments of B. mori lebocin precursor proteins, but other parts of lebocin sequences are quite different (Bao et al., 2005). It is unclear how active lebocin peptides are generated from precursors, and which peptides are active against microorganisms. Here we report an integrated study of molecular cloning, protein expression and functional analysis of two lebocin-related proteins (Leb-B and Leb-C) in M. sexta. We determined the cleavage sites in M. sexta lebocin precursors by protein sequencing of the fragments recovered from recombinant proteins treated with induced plasma. We also verified activity of synthetic peptides against bacteria and fungi, and found that active M. sexta Leb-B/C peptides were located at the N-termini, which are different from B. mori lebocins that are located closely to the C-termini. In addition, we showed that synthetic peptide Leb-B22–48, the active lebocin peptide of Leb-B, not only had higher antibacterial activity but also caused agglutination of E. coli cells, a property that has not been reported before.
2. Materials and methods
2.1 Insects, pathogenic bacteria, and fungi
M. sexta eggs were purchased from Carolina Biological Supply (Burlington, NC, USA), and larvae were reared on artificial diet at 25°C (Dunn and Drake, 1983). The fifth instar larvae (Day 2) were used for bacteria injection, plasma preparation, hemocytes and fat body collection. Dry yeast (Saccharomyces cerevisiae) and Gram-positive Micrococcus luteus were purchased from Sigma-Aldrich (MO, USA). Escherichia coli strain XL1-blue was purchased from Stratagene (CA, USA). Salmonella typhimurium was kindly provided by Professor Michael O’Connor, Staphylococcus aureus and Bacillus cereus were provided by Professor Brian Geisbrecht, S. cerevisiae (BY4741) and Cryptococcus neoformans (alpha) were provided by Professor Alexander Idnurm, School of Biological Sciences at University of Missouri-Kansas City. GFP-E. coli K12 was a gift from Professor Brenda Beerntsen at University of Missouri-Columbia.
2.2 Sequence analysis
Sequences of lebocin-related precursor proteins were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment was performed using Mega5 with a gap opening penalty of 10 and gap extension penalty of 0.2 (Tamura et al., 2011). Signal peptide sequences were predicted with SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al., 2004).
2.3 Genome walking and RACE
Genomic DNA was prepared from hemocytes of M. sexta larvae with PureLink Genomic DNA Mini Kit (Invitrogen). Genomic DNA was digested with Dra I, EcoR V, Pvu II and Stu I, respectively, and purified by phenol-chloroform extraction and ethanol precipitation. Purified fragments were ligated with a synthetic adaptor GenomeWalkerAP with T4 DNA ligase (New England BioLabs). The primary PCR was performed with AP1 and MsLebocinGSP1 primers (Table 1) using a two-step PCR: 7 cycles of 94°C for 25 sec, 72°C for 3 min, 32 cycles of 94°C for 25 sec, 55°C for 30 sec, and 72°C for 3 min. Nested PCR was performed with AP2 and MsLebocinGSP2 primers (Table 1) with 1μL of 1:100 diluted primary PCR product as the template. To determine cDNA sequences, 5′ and 3′ RACE were performed using smarter race kit (Clontech). PCR products were cloned into pGEM-T vector (Promega) and sequenced at DNA Core Facility, University of Missouri-Columbia.
Table 1.
Primers for genome walking, lebocin cloning and real-time PCR
| Primer | Sequence (5′→3′) |
|---|---|
| GenomeWalkerAP | GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT 3′-H2N-CCCGACCA-PO4-5′ |
| MsLebocinGSP1 | CGCAGATTATGAGTTACGAC |
| MsLebocinGSP2 | CAGATTATGAGTTACGACGA |
| AP1 | GTAATACGACTCACTATAGGGC |
| AP2 | ACTATAGGGCACGCGTGGT |
| msLebocin1-RT-N | ATCCATTTTCGCGCTTGGAGTTATC |
| msLebocin1-RT-C | GTTCGTGATGCTCCTGAGACGAG |
| msLebocin2-RT-N | AGTACTTGTACTTTGCGTTGTTGCG |
| msLebocin2-RT-C | CCGCTGGACGTAGAGTGG |
| rpS3-N | ACATCCCGGAACAGTCCGTGGA |
| rpS3-C | CGTCACCAGGATGTGGTCTGGCT |
2.4 Tissue distribution and microbial induction of lebocin mRNAs and time-course induction of lebocin proteins
Naïve or E. coli XL-1 blue immunized fifth instar larvae (24h after injection) were dissected, hemocytes, fat body, midgut, epidermis and malpighian tubules were collected and washed 3 times in anti-coagulant (AC) saline (4 mM NaCl, 40 mM KCl, 8 mM EDTA, 9.5 mM citric acid-monohydrate, 27 mM sodium citrate, 5% sucrose, 0.1% polyvinylpyrollidone, 1.7 mM PIPES). Total RNA was prepared with TRI reagent (Sigma-Aldrich). Reverse transcription was performed with Oligo-dT primer and ImProm-II reverse transcriptase (Promega) following the manufacturer’s instructions. For real-time PCR analysis, fifth instar larvae were injected with saline, heat-killed E. coli strain XL1-blue (108 cells/larva), dry M. luteus (120 μg/larva) or yeast (S. cerevisiae) (107 cells/larva). Twenty-four hours after injection, hemocytes and fat body were collected separately. Total RNA and cDNA were prepared as described above. Real-time PCR was performed with SYBR Premix (Takara) on a 7500 system (Applied Biosystems) with msLebocin1-RT-N and msLebocin1-RT-C (for Leb-B), msLebocin2-RT-N and msLebocin2-RT-C (for Leb-C), rpS3-N and rpS3-C (for ribosomal protein subunit 3 (rpS3), an internal control) primers (Table 1). Data from three replicates of each sample was analyzed with SDS software (ABI) using a comparative method (2−ΔΔCt) and these experiments were repeated with 3 different biological samples.
To determine time-course induction of lebocin proteins by Western blot analysis, the fifth instar larvae were injected with E. coli, M. luteus or yeast (S. cerevisiae) as described above (4 larvae in each group). Hemolymph (~50 μL) was collected from each larva every two hours after injection. Hemolymph samples were centrifuged at 3000g for 10 minutes and cell-free plasma samples were collected. Plasma samples (0.125 μL from each larva, 0.5 μL in total from four larvae) were analyzed by 10% Tris-Tricine gels. Western blot was performed using rabbit anti-Leb-B antiserum (see below) (1:2000 dilution) (Cocalico Biologicals, Inc) and Goat anti-Rabbit IgG, HRP-conjugate (1:10000 dilution) (Millipore). Images were developed using Amersham ECL Plus Western Blotting Detection Reagents on a typhoon phosphorimager (GE Healthcare).
2.5 Recombinant protein expression and in vitro cleavage assay
Full-length (Leb-B22–143 and Leb-C21–143) or various fragments (Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-C49–92 and Leb-B93–143) (Table 2) of Leb-B and Leb-C were expressed as soluble Thioredoxin (TRX) fusion proteins at 16°C after IPTG induction in Rosetta (DE3) cells using pET32a vector (Novagen). Bacterial pellets were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and sonicated for 10 min on ice. Cell lysate was centrifuged for 20 min at 18,000g. The clear supernatant was used for protein purification using Ni-NTA agarose. Leb-B (residues 22–143) was also expressed using the H6pQE60 vector (Lee et al., 1994) in E. coli XL1-blue cells as inclusion bodies, purified under denaturing conditions with Ni-NTA agarose (Qiagen), and used as an antigen to produce rabbit polyclonal antiserum against Leb-B at Cocalico Biologicals, Inc (Pennsylvania, USA).
Table 2.
Synthetic peptides used for antibacterial assay
| Leb-B22–48 | WRSDLPIILPTYKPPRTPSTVIIRTVR |
| Leb-C21–48 | GWNKNNGGIILPTFRPPPIWPGITRTVR |
| Leb-B49–92 | EAGDKPLWLYQGDDHPRAPSSGDHPVLPPIIDDVKLDPNRRYAR |
| Leb-B93–108 | SVNEPSSQEHHERFVR |
| Leb-B109–143 | SFDSRSSRHHGGSHSTSSGSRDTGATHPGYNRRNS |
| Leb-B117–143 | HHGGSHSTSSGSRDTGATHPGYNRRNS |
To determine cleavage of recombinant fusion proteins by proteinases in M. sexta larval plasma, induced plasma samples were collected from the fifth instar larvae at 12 hours after injection of E. coli XL-1 blue (107 cells/larva). Recombinant fusion proteins (1 μg each protein) were treated with 0.5 μL induced plasma in a total of 10μL in 10 mM Tris buffer at room temperature overnight. The cleavage products were separated on 15% SDS-PAGE and analyzed by Western blotting using mouse anti-His-Tag monoclonal antibody (Sigma-Aldrich). To determine the cleavage sites, TRX-Leb-B fusion protein (1 μg) was treated with induced plasma (0.5 μL) in a total of 10 μL at 16°C overnight. The cleavage products were separated on 10% Tris-Tricine gels and visualized by Coomassie Blue staining, or proteins were transferred to nitrocellulose membranes and analyzed by Western blotting using polyclonal rabbit anti-Leb-B antiserum or monoclonal mouse anti-His-Tag antibody (Sigma-Aldrich). For protein sequencing, the cleavage products were separated on Tris-Tricine gels and transferred to PVDF membrane (0.22 μm), the membrane was stained with 0.05% Coomassie brilliant blue R-250 prepared in water, and the small peptides were cut out and sent for protein sequencing at Biomolecular Resource Facility at the University of Texas Medical Branch.
2.6 Peptide synthesis
Six peptides (10 mg each) based on the sequences of Leb-B and Leb-C (Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-B93–108, Leb-B109–143, and Leb-B117–143) (Table 2) were synthesized by Shanghai Apeptide Co., LTD (Shanghai, China) and purified by HPLC to > 95%.
2.7 Antimicrobial activity assay
Antimicrobial activity of synthetic peptides was tested against four pathogenic bacterial strains: S. aureus, B. cereus, S. typhimurium and S. marcescens, and two fungal strains: S. cerevisiae (BY4741) and C. neoformans. A zone inhibition assay was performed for initial tests as described previously (Rao and Yu, 2010). A broth micro-dilution assay was used to generate growth curves (Rayaprolu et al., 2010). Briefly, overnight bacterial cultures were subcultured in tryptic soy broth (#091010717, MP Biochemicals) or fungal cultures were subcultured in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) and grown to mid-log phase. The bacterial and fungal cultures were centrifuged at 1,000g for 10 minutes at 4°C and washed once with 10 mM Tris, 0.1 mM EDTA. The bacterial cells were diluted to 5×105 cfu/ml in 5% tryptic soy broth and fungal cells were diluted to 1×105 cfu/ml in YPD medium, and the diluted bacterial cultures (190 μL) or fungal cultures (95 μL) were mixed with synthetic peptide (4 mg/mL, to a final concentration of 200 μg/mL) in 96-well plates. B. cereus and S. marcescens were cultured at 30°C with 220 rpm shaking, S. aureus and S. typhimurium were cultured at 37°C with 220 rpm shaking, and S. cerevisiae and C. neoformans cells were cultured at 30°C with 220 rpm shaking. OD600 was measured with Powerwave XS plate reader every hour (BioTek, VT, US). Bacterial growth curves were generated using the Graphpad Prism version 4.0 for Windows (GraphPad Software, La Jolla California USA, http://www.graphpad.com/).
2.8 Agglutination of GFP-E. coli by synthetic peptides
GFP-E. coli overnight culture (1 μL) was incubated with each synthetic peptide (5 μL, 4 mg/mL in H2O) or BSA (5 μL, 4 mg/mL in H2O) in a total of 10 μL in 10mM Tris buffer and incubated at room temperature for 30 minutes. Samples of bacterial cells were then applied to microscope slides and observed using an Olympus BX50 fluorescence microscope.
3. Results
3.1 Cloning and sequence analysis of M. sexta lebocin-related genes
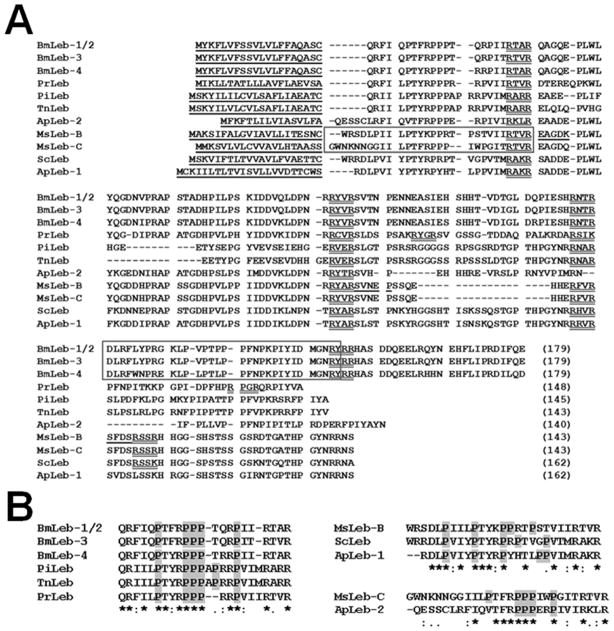
In a project to clone regulatory sequence of M. sexta lebocin gene by genome walking, we obtained two genomic fragments encoding two proteins that are homologous to lebocin-like proteins from other lepidopteran insects. To confirm the cDNA sequences, we performed 5′ and 3′ RACE to clone and sequence cDNA clones. We named the two clones as lebocin-related protein B (Leb-B, Genbank accession #: GU563900) and lebocin-related protein C (Leb-C, Genbank accession #: GU563901). Leb-B and Leb-C cDNAs were 90% identical, while protein sequences were 78% identical, but both proteins showed very low similarity (~20% identity) to a previously reported M. sexta lebocin-related protein (called Leb-A in this study) (Rayaprolu et al., 2010). Protein sequence alignment showed that the 143-residue Leb-B and Leb-C are similar to lebocin-related proteins from Samia cynthia (57 and 53% identity), Antheraea pernyi (56 and 51% to ApLeb-1, 40 and 42% to ApLeb-2), Pieris rapae (41 and 40%), Trichoplusia ni (30%), Pseudoplusia includens (33 and 32%), and to B. mori lebocins 1 to 4 (34–39%) (Fig. 1A). The 32-residue B. mori active lebocin peptides (Fig. 1A, boxed region) are not similar to the corresponding regions in other lepidopteran lebocin-related proteins. We also noticed presence of the RXXR motifs in all lebocin-related proteins (Fig. 1A). Thus M. sexta Leb-B/C may be precursor proteins that can be processed in the hemolymph by certain convertases (Devi, 1991; Veenstra, 2000).
Fig. 1. Multiple sequence alignment of lebocin-related proteins from some lepidopteran insects.
(A) M. sexta Leb-B and Leb-C were compared with B. mori lebocins 1–4 (AAB35218.1, BAA22883.1, and BAA22884.1), P. rapae (AEO21919.1), P. includens (AAS48093.1), T. ni (AAG44366.1), S. cynthia (BAD84189.1), and A. pernyi (EU557311, EU557312) lebocin-related proteins (sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) with the accession numbers). Predicted signal peptides were underlined and RXXR motifs were double underlined. The 32-residue active fragments of B. mori lebocins and the N-terminal active fragments of M. sexta Leb-B/C were shown in boxes. The five residues of three peptide fragments from M. sexta Leb-B determined by protein sequencing were also underlined. (B) Alignments of the N-terminal fragments of lebocin-related proteins based on their similarities.
3.2 Tissue distribution and transcriptional induction of Leb-B and Leb-C genes
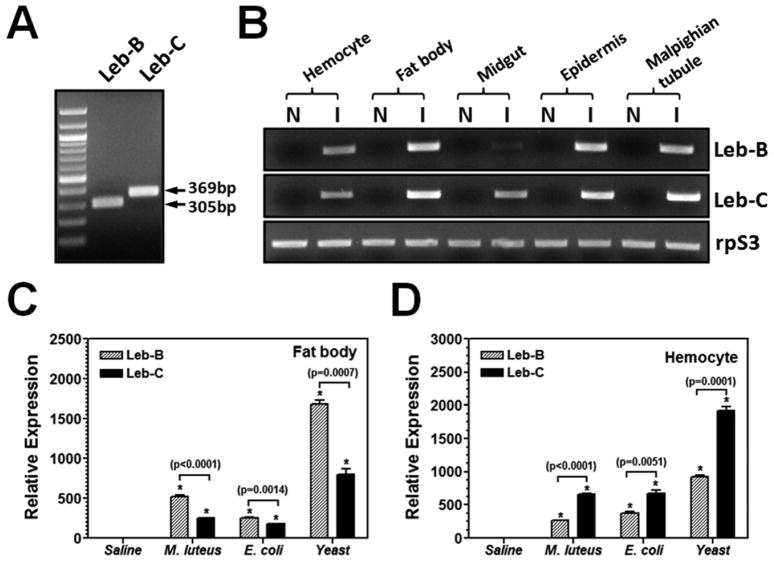
Antimicrobial peptide (AMP) genes are generally present in insects at very low basal levels. Upon detection of pathogens, AMP genes are transcriptionally up-regulated due to activation of the Rel-family of transcription factors (Ganesan et al., 2011). To study induction of M. sexta Leb-B and Leb-C genes, total RNA was prepared from different tissues of naïve and microorganism-induced larvae for cDNA synthesis. Two different pairs of primers were used to differentially amplify Leb-B and Leb-C genes (Fig. 2A). RT-PCR results showed that Leb-B and Leb-C mRNAs were not detected in the five tissues of naïve larvae, and Leb-B transcript was greatly induced in hemocytes, fat body, epidermis and malpighian tubule but weakly induced in midgut, while Leb-C transcript was greatly induced in all five tissues (Fig. 2B). Real-time PCR showed that both Leb-B and Leb-C mRNAs were induced in fat body and hemocytes after larvae were injected with E. coli, M. luteus and S. cerevisiae (Fig. 2C and D). In fat body, Leb-B transcript was induced to a higher level than Leb-C, but in hemocytes, Leb-C was induced to a higher level than Leb-B (Fig. 2C and D).
Fig. 2. Tissue distribution and induced expression of Leb-B/C in hemocytes and fat body.
(A) Differential amplification of Leb-B and Leb-C by two different pairs of primers. msLebocin1-RT-N/msLebocin1-RT-C (for Leb-B) and msLebocin2-RT-N/msLebocin2-RT-C (for Leb-C) primers (Table 1) were designed based on difference in cDNA sequences of Leb-B and Leb-C. (B) Up-regulation of Leb-B and Leb-C transcription in tissues by RT-PCR. Total RNAs from hemocytes, fat body, midgut, epidermis and malpighian tubule of naïve or E. coli induced larvae were reverse-transcribed to cDNA using Oligo-dT primer. The cDNA was used as template for PCR reactions using gene specific primers. PCR products were analyzed on 1% agarose gel and stained with ethidium bromide. (C) and (D) Induced expression of Leb-B and Leb-C mRNAs in fat body (C) or hemocytes (D) by real-time PCR. Larvae were injected with heat-killed E. coli (108 cells/larva), M. luteus (108 cells/larva), yeast (S. cerevisiae) (107 cells/larva) or saline (as a control), and hemocytes and fat body were collected at 12h post-injection. Total RNA was prepared immediately and reverse-transcribed to cDNA as described above for Real-time PCR. The bars represent the mean of three individual measurements ± S.E.M. Significant difference between Leb-B and Leb-C was determined by an unpaired t-test.
3.3 Induced expression of lebocin proteins in hemolymph
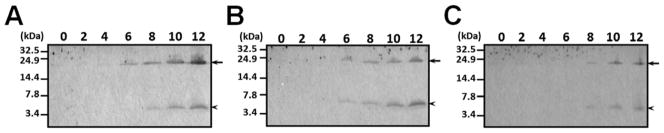
To confirm induced expression of M. sexta Leb-B and Leb-C proteins in hemolymph, we used a time-course Western blot to study accumulation of Leb-B and Leb-C proteins in the hemolymph after larvae were injected with microorganisms. Rabbit polyclonal anti-Leb-B antiserum was generated using recombinant Leb-B precursor as an antigen for the Western blot analysis. In the E. coli, M. luteus and yeast (S. cerevisiae) induced plasma samples, lebocin proteins (in between 14.4 and 24.9 kDa protein markers, arrows) became visible as early as 6h post-injection (Fig. 3A and 3B) and no later than 8 h post-injection (Fig. 3C). The protein concentration in hemolymph continued to increase until 12h post-injection. We also observed accumulation of small peptide(s) in between 3.4 and 7.8 kDa protein markers (Fig. 3, arrowheads). These results suggest that active M. sexta lebocins may be generated from precursor proteins upon immune stimulation.
Fig. 3. Induced expression of Leb-B/C proteins in hemolymph by Western blot.
The fifth instar M. sexta larvae were injected with M. luteus (A), yeast (S. cerevisiae) (B) or heat-killed E. coli (C) as described in the Materials and Methods. Hemolymph samples (50 μL) were collected at 0, 2, 4, 6, 8, 10, 12h post-injection, and hemocytes were removed by centrifugation. Cell-free plasma samples (0.5 μL mixed plasma samples from 4 larvae) were loaded on 10% Tris-Tricine gel for Western blot analysis using rabbit anti-Leb-B serum. The arrows indicated Leb-B/C precursor proteins and the arrowheads indicated accumulation of processed small peptide(s).
3.4 Expression and purification of recombinant fusion proteins and in vitro cleavage assay
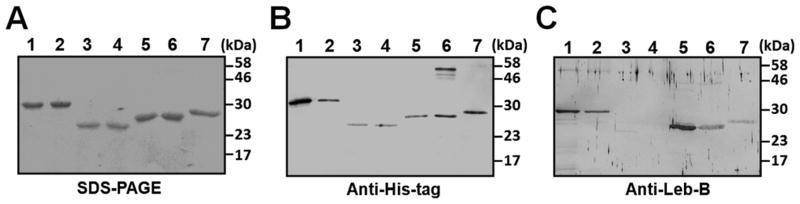
To study cleavage of lebocin precursor proteins, we expressed full-length and various fragments of Leb-B and Leb-C as fusion proteins to Thioredoxin (TRX)-tag at the N-termini (Table 2) (Fig. 4). The amino acid sequences of the fragments were selected based on the RXXR motifs as potential cleavage sites. All seven recombinant fusion proteins were purified to homogeneity as analyzed by SDS-PAGE (Fig. 4A), and they were all recognized by monoclonal anti-His-tag antibody (Fig. 4B). Five (lanes 1, 2, 5–7), but Leb-B22–48 and Leb-C21–48 fusion proteins (lanes 3 and 4), were also recognized by rabbit polyclonal antibody to Leb-B (Fig. 4C).
Fig. 4. Expression and purification of recombinant fusion proteins from Rosetta (DE3) cells.
Full-length or various fragments of Leb-B/C were expressed using the pET32a vector as fusion proteins to a thioredoxin (TRX) tag at the N-termini. Purified fusion proteins were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue (A), or by Western blot using mouse anti-His-tag monoclonal antibody (B) or rabbit anti-Leb-B polyclonal antibody (C). One microgram (1 μg) of each purified protein was used for the staining gel, and 0.5 μg of each protein for Western blot. Lanes 1–7 were Leb-B22–143, Leb-C21–143, Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-C49–92, and Leb-B93–143 fusion proteins, respectively.
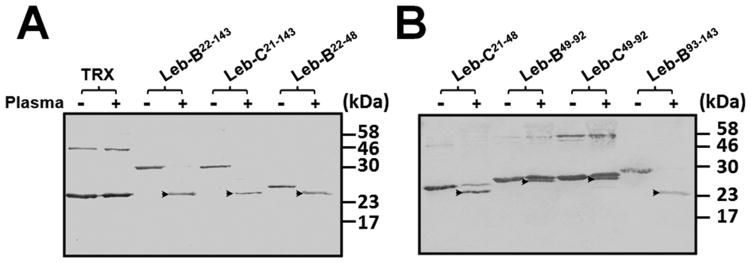
To determine cleavage of fusion proteins by proteinases in the hemolymph, purified fusion proteins and the control TRX-tag were treated with induced plasma from bacteria-injected M. sexta larvae. Western blot analysis showed that all seven fusion proteins but the control TRX-tag were cleaved by induced larval plasma, as cleavage products were detected after incubation of fusion proteins with induced plasma (Fig. 5, arrowheads), indicating that there are cleavage sites in Leb-B and Leb-C as well as in the five fragments (Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-C49–92 and Leb-B93–143) (Table 2). Leb-B22–143, Leb-C21–143, Leb-B22–48, Leb-C21–48 and Leb-B93–143 were cleaved more completely than Leb-B49–92 and Leb-C49–92. There may be more than one cleavage sites in Leb-B/C because naïve plasma sample could also cleave fusion proteins incompletely and generated more than one cleavage products (data not shown).
Fig. 5. In vitro cleavage of recombinant fusion proteins by induced plasma.
Purified fusion proteins and the TXR control (0.5 μg each) were incubated with 0.5 μL of E. coli induced plasma at 25°C overnight. Cleavage products were then separated on 15% SDS-PAGE gels and detected by Western blotting using Mouse anti-His-Tag monoclonal antibody. Arrowheads indicate the cleavage products of fusion proteins.
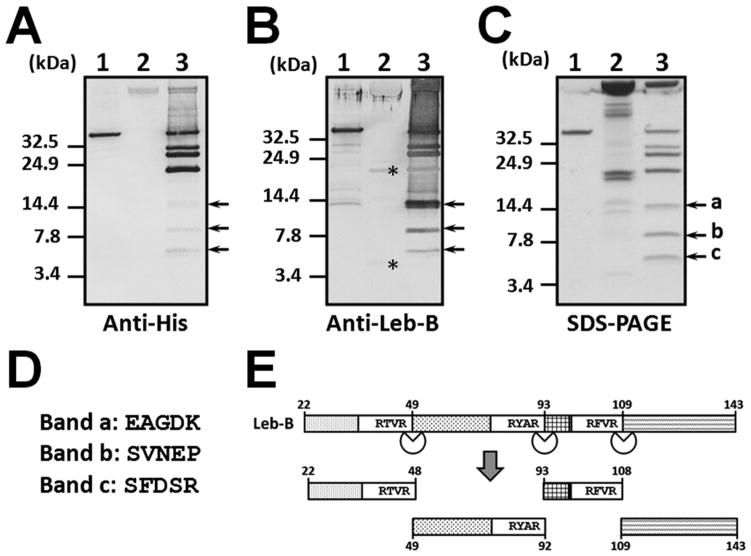
To further determine the cleavage sites, we recovered cleavage products from Leb-B fusion protein with Ni-NTA beads and separated them on Tris-Tricine gel (Fig. 6). Multiple cleavage products were confirmed by Western blot using anti-His-Tag and anti-Leb-B antibodies (Fig. 6A and B, lane 3). We observed three smaller cleavage products (Fig. 6C, Bands a, b and c in lane 3), suggesting that there may be at least three cleavage sites in Leb-B. Bands a, b and c were excised separately and sent for sequencing, and the first five amino acids were determined as EAGDK, SVNEP and SFDSR for Bands a, b and c, respectively (Fig. 6D), which perfectly matched the amino acid sequences right after the three RXXR motifs in Leb-B (Figs. 1A and 6E). The three bands were C-terminal fragments of Leb-B after cleavage at the three RXXR sites, and the calculated molecular masses of Bands a, b and c are 12.1, 7.2 and 5.3 kDa, respectively, which also matched the apparent masses of the three fragments in Tris-Tricine gel (Fig. 6C, arrows). In the induced plasma sample alone, anti-Leb-B antibody also recognized a protein and a peptide (Fig. 6B, lane 2, asterisks), which may represent Leb-B precursor protein and processed lebocin peptide(s), respectively.
Fig. 6. Determination of proteinase cleavage sites in Leb-B.
Purified Leb-B fusion protein (1 μg) was incubated at 16°C overnight with or without 0.5 μL of induced plasma. The cleavage products were then separated on 10% Tris-Tricine gels and subject to Western blotting analysis using mouse anti-His-Tag monoclonal antibody (A) or rabbit anti-Leb-B polyclonal antibody (B). The gel was also stained with Coomassie Brilliant Blue (C). Short cleavage products in (C) (Bands a, b and c) corresponding to the processed products of Leb-B were excised separately and sequenced, and the first five residues were determined by protein sequencing (D). Cleavage of Leb-B at the three RXXR sites was also shown in a schematic diagram (E). Lane 1: Leb-B fusion protein alone; lane 2: induced plasma alone; lane 3: Leb-B fusion protein treated with induced plasma. Asterisks in (B) indicate endogenous lebocin proteins and processed peptides in the induced plasma recognized by rabbit anti-Leb-B antibody.
3.5 Antimicrobial activities of synthetic peptides
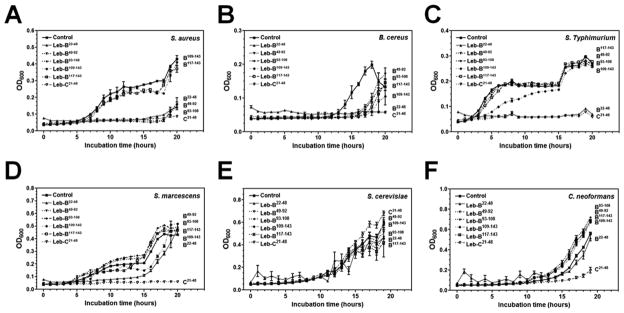
Since we determined the cleavage sites in Leb-B, six peptides (Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-B93–108, Leb-B109–143, and Leb-B117–143) (Table 2) were synthesized to verify activity of M. sexta lebocin peptides. We initially measured antibacterial activities of synthetic peptides using zone inhibition assay (Rao and Yu, 2010), but did not see inhibition zones (data not shown), maybe because these synthetic peptides have low activity as B. mori active lebocins (Hara and Yamakawa, 1995b). We then performed the micro-dilution assay (Rayaprolu et al., 2010) using four pathogenic bacterial strains and two fungal strains. We found that Leb-B22–48 and Leb-C21–48 had high activity against Gram-positive S. aureus, B. cereus, and Gram-negative S. typhimurium (Fig. 7A–C), Leb-C21–48 also had high activity against Gram-negative S. marcescens (Fig. 7D) and the fungus C. neoformans (Fig. 7F). Leb-B49–92 and Leb-B93–108 had high activity against S. aureus (Fig. 7A), Leb-B109–143 had low activity against S. typhimurium (Fig. 7C), Leb-B22–48 and Leb-B109–143 had low activity against S. marcescens (Fig. 7D), and Leb-B49–92, Leb-B93–108, Leb-B109–143 and Leb-B117–143 had low activity against B. cereus (Fig. 7B). These results suggest that Leb-B22–48 and Leb-C21–48, which were located at the N-termini of Leb-B and Leb-C, may be the major active M. sexta lebocin peptides.
Fig. 7. Antimicrobial activities of synthetic peptides.
Diluted bacterial (S. aureus, B. cereus, S. marcescens and S. typhimurium) and fungal (S. cerevisiae and C. neoformans) cultures were incubated with synthetic peptides (200 μg/mL final concentration) or water (Control) in 96-well plates with 220 rpm shaking at 30°C or 37°C. OD600 was recorded every hour up to 20 hours after incubation. The points represent the mean of three individual measurements ± SEM. Growth curve of each peptide was labeled on the right to show its activity.
3.6 Agglutination of GFP-E. coli by Leb-B22–48 peptide
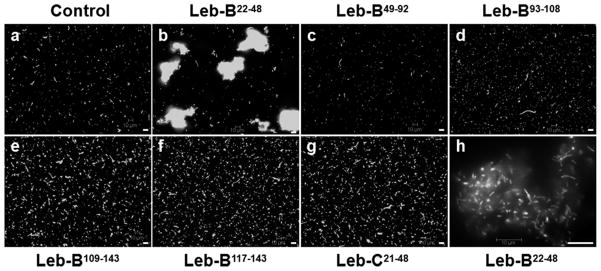
When testing antimicrobial activities of synthetic peptides, we also incubated an E. coli strain that expresses GFP with synthetic peptides and observed bacterial cells under fluorescence microscope. Surprisingly, we found that synthetic Leb-B22–48 peptide could agglutinate E. coli cells (Fig. 8b and h); while other five synthetic peptides, including Leb-C21–48 that had high antibacterial activity as Leb-B22–48, didn’t have this agglutination activity (Fig. 8c–g). These E. coli cells formed large clumps in the presence of Leb-B22–48, but lysis of bacterial cells was not observed (Fig. 8h).
Fig. 8. Agglutination of GFP-E. coli cells by synthetic Leb-B22–48.
Overnight culture of GFP-E. coli (1 μL) was incubated with 5 μL synthetic peptides (4 mg/mL) or BSA (Control) in a total of 10 μL at room temperature for 30 minutes and subject to fluorescence microscope imaging. Bars: 10 μm.
4. Discussion
Induced production of antimicrobial peptides (AMPs) is an important humoral response in insects (Lemaitre and Hoffmann, 2007). Fat body and hemocytes are major tissues to produce AMPs and secrete them into hemolymph. Epidermal cells also can synthesize AMPs and initiate localized immune responses (Lemaitre and Hoffmann, 2007). AMP gene expression is mainly regulated by the Rel-family of transcription factors, which bind to cis-regulatory elements in the promoters of AMP genes and initiate transcription (Ganesan et al., 2011). We found that M. sexta Leb-B and Leb-C mRNAs were up-regulated in several tissues that may directly contact pathogens (Fig. 2B). Gram-negative E. coli, Gram-positive M. luteus and yeast (S. cerevisiae) all were able to stimulate expression of Leb-B and Leb-C in hemocytes and fat body, and Leb-B was induced to a higher level in fat body than Leb-C, while Leb-C was induced to a higher level in hemocytes than Leb-B (Fig. 2C and D). Induced expression of Leb-B and Leb-C proteins and accumulation of processed peptide(s) in the hemolymph were also confirmed by Western blotting analysis (Fig. 3).
Most AMPs are synthesized as precursor proteins or pro-proteins, and they require proteolytic processing to produce active AMPs. Proteinases in insect hemolymph are important for cleavage of signaling molecules and activation of the prophenoloxidase system and immune signaling pathways (Gál et al., 2007; Jiang and Kanost, 2000; Jiang et al., 1999; Kim et al., 2008; Volz et al., 2005). Analysis of lebocin protein sequences showed that lebocin precursor proteins contain several RXXR motifs (Fig. 1A), which can be recognized by processing enzymes (Devi, 1991; Veenstra., 2000), suggesting that active lebocins are generated from precursor proteins by processing enzymes in vivo. Two RXXR motifs are highly conserved in all lebocin precursors, while M. sexta Leb-B/C and B. mori lebocins 1–4 precursors contain two more RXXR motifs (Fig. 1A). In order to study processing of M. sexta Leb-B/C in vitro, recombinant Leb-B and Leb-C precursors and different fragments of Leb-B and Leb-C containing RXXR motif(s) were expressed as fusion proteins to a thioredoxin (TRX)-tag. These fusion proteins were soluble and purified under native conditions, but they did not have antimicrobial activity (data not shown). All seven recombinant fusion proteins (Leb-B and Leb-C (4 RXXR motifs), Leb-B22–48, Leb-C21–48, Leb-B49–92, Leb-C49–92 (each contains one RXXR motif), and Leb-B93–143 (two RXXR motifs)), but not the TRX-tag alone, could be processed by induced plasma from M. sexta larvae (Fig. 5), indicating that these RXXR motifs indeed were recognized by processing enzymes in the hemolymph. Using Leb-B fusion protein and protein sequencing of cleavage products, three cleavage sites were determined (Fig. 6). Leb-B precursor was cleaved mainly at the first three RXXR motifs, RTVR45–48, RYAR89–92, and RFVR105–108 (Figs. 1A and 6E). Naïve plasma could also process the fusion proteins, but the cleavage was incomplete compared to the induced plasma (data not shown), suggesting that some proteinases also require activation.
Leb-B and Leb-C are both 143-residue long with a 21-residue signal peptide for Leb-B and 20-residue signal peptide for Leb-C. Leb-B and Leb-C shared 78% identity, but they showed low similarity (~20% identify) to a previously reported lebocin-related protein (Leb-A) in M. sexta (Rayaprolu et al., 2010). Leb-A contains several repeats of a 27-residue fragment, and it also contains multiple RXXR motifs. To test antimicrobial activity of processed peptides from M. sexta Leb-B and Leb-C, several peptides based on the cleavage of recombinant Leb-B by induced plasma were synthesized (Table 2). We initially used a zone inhibition assay to test activity of synthetic peptides, but didn’t observe formation of inhibition zones (data not shown), suggesting that antibacterial activity of these synthetic peptides may be low. This may be because synthetic peptides lack necessary glycosylations that are important for antibacterial activities as in the case of B. mori lebocins (Hara and Yamakawa, 1995b). High salt concentration (171 mM NaCl) in the normal LB agar plate may also inhibit the activity of synthetic peptides (Hara and Yamakawa, 1995b). Therefore, we used a modified micro-dilution assay (Rayaprolu et al., 2010) to test antimicrobial activities of these peptides in 5% tryptic soy broth (4.2 mM NaCl) or in YPD medium (1% yeast extract, 2% peptone, 2% dextrose).
Leb-B22–48 and Leb-C21–48 (N-terminal fragments) showed very low similarity, but both fragments contain 5 proline residues, and they may be proline-rich active peptides. Antimicrobial activity assay showed that both synthetic Leb-B22–48 and Leb-C21–48 peptides had higher activity against Gram-positive S. aureus and B. cereus, as well as Gram-negative S. typhimurium and S. marcescens than other four peptides (Fig. 7). Leb-B49–92 and Leb-C49–92 are almost identical and differ only in 4 residues, while Leb-B93–143 and Leb-C93–143 are completely identical. Synthetic Leb-B49–92, Leb-B93–108, Leb-B109–143, and Leb-B117–143 peptides all showed some activity against B. cereus (Fig. 7B), Leb-B49–92 and Leb-B93–108 had high activity against S. aureus (Fig. 7A), whereas Leb-B109–143 had low activity against S. typhimurium and S. marcescens (Fig. 7C and D). These results suggest that lebocin precursors may be processed by proteinases to produce different peptides, and these processed peptides may function together to exert maximal activity against different microorganisms. Among the six peptides, only Leb-C21–48 had activity against the fungus C. neoformans (Fig. 7F), and Leb-C21–48 also had higher activity against Gram-negative S. marcescens than Leb-B22–48 (Fig. 7D), suggesting that Leb-C21–48 may have higher activity and broader spectrum against microorganisms than Leb-B22–48. We also observed that Leb-B precursor could be expressed and purified from E. coli, but re-folded Leb-B precursor did not have activity against bacteria (data not shown). However, Leb-C precursor could not be expressed in E. coli since bacterial culture stopped growing and became clear after addition of IPTG (data not shown), suggesting that some products of Leb-C may be toxic to E. coli.
Sequence alignment showed that the N-terminal fragments of B. mori lebocins 1–4, P. rapae, P. includens, and T. ni lebocin precursors share high similarity, the N-terminal fragments of M. sexta Leb-B, S. cynthia and A. pernyi lebocin-1 precursors share high similarity, while the N-terminal fragments of M. sexta Leb-C and A. pernyi lebocin-2 have high similarity, which are more similar to those of B. mori lebocins 1–4 than to M. sexta Leb-B, S. cynthia and A. pernyi lebocin-1 precursors (Fig. 1B). All these N-terminal fragments are proline-rich (4–6 proline residues). The active peptides of B. mori lebocins 1–4 did not align with the corresponding regions in other lebocin precursors (Fig. 1A). Since the N-terminal fragments (Leb-B22–48 and Leb-C21–48) of M. sexta Leb-B and Leb-C were active peptides (Fig. 7), we think that the N-terminal proline-rich fragments of lebocins are likely active peptides after processing by proteinases in lepidopteran insects. We also found an interesting result that Leb-B22–48 not only had higher antibacterial activity than other peptides, but also caused agglutination of E. coli cells (Fig. 8b and h), a property that has not been reported for lebocins so far. Leb-C21–48 though had high antibacterial activity as Leb-B22–48 (Fig. 7), couldn’t agglutinate E. coli cells (Fig. 8g). But lysis of the agglutinated E. coli cells by Leb-B22–48 was not observed (Fig. 8h). These results suggest that even though lebocin peptides may have lower direct antibacterial activity compared to other AMPs, agglutinating activity of some lebocin peptides can help other AMPs to kill bacterial cells more effectively, as it has been reported that B. mori lebocins can decrease minimum inhibitory concentration of cecropin D (Hara and Yamakawa, 1995a). Future work is to investigate how lebocin peptides act together with other AMPs to kill different bacteria.
Two lebocin-related proteins (Leb-B and Leb-C) are identified in Manduca sexta and both are immune inducible.
Leb-B and Leb-C are precursors that could be cleaved at the RxxR motifs and the cleavage sites are determined.
Active lebocin peptides (Leb-B22–48 and Leb-C21–48) are located at the N-termini of precursors.
Synthetic Leb-B22–48 and Leb-C21–48 can inhibit the growth of some pathogenic bacteria and fungi.
Leb-B22–48, but not Leb-C21–48, can cause agglutination of E. coli cells.
Acknowledgments
This work was supported by National Institutes of Health Grant GM066356. The nucleotide sequences reported in this paper have been submitted to the Genbank/EBI Data Bank with accession numbers GU563900 and GU563901.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao Y, Yamano Y, Morishima I. A novel lebocin-like gene from eri-silkworm, Samia cynthia ricini, that does not encode the antibacterial peptide lebocin. Comp Biochem Physiol B: Biochem Mol Biol. 2005;140:127–131. doi: 10.1016/j.cbpc.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Taniai K, Hara S, Kadono-Okuda K, Kato Y, Yamamoto M, Xu J, Choi SK, Debnath NC, Choi HK, et al. cDNA cloning and gene expression of lebocin, a novel member of antibacterial peptides from the silkworm, Bombyx mori. Biochem Biophys Res Commun. 1995;214:271–278. doi: 10.1006/bbrc.1995.2284. [DOI] [PubMed] [Google Scholar]
- Dai H, Rayaprolu S, Gong Y, Huang R, Prakash O, Jiang H. Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin. J Pept Sci. 2008;14:855–863. doi: 10.1002/psc.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L. Consensus sequence for processing of peptide precursors at monobasic sites. FEBS Lett. 1991;280:189–194. doi: 10.1016/0014-5793(91)80290-j. [DOI] [PubMed] [Google Scholar]
- Dickinson L, Russell V, Dunn PE. A family of bacteria-regulated, cecropin D-like peptides from Manduca sexta. J Biol Chem. 1988;263:19424–19429. [PubMed] [Google Scholar]
- Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J Invertebr Pathol. 1983;41:77–85. [Google Scholar]
- Furukawa S, Taniai K, Ishibashi J, Hara S, Shono T, Yamakawa M. A novel member of lebocin gene family from the silkworm, Bombyx mori. Biochem Biophys Res Commun. 1997;238:769–774. doi: 10.1006/bbrc.1997.7386. [DOI] [PubMed] [Google Scholar]
- Gál P, Barna L, Kocsis A, Závodszky P. Serine proteases of the classical and lectin pathways: similarities and differences. Immunobiology. 2007;212:267–277. doi: 10.1016/j.imbio.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-κB/Rel Proteins and the Humoral Immune Responses of Drosophila melanogaster. Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S, Yamakawa M. Cooperative antibacterial relationship between lebocin and cecropin D, antibacterial peptides isolated from the silkworm, Bombyx mori. Appl Entomol Zool. 1995a;30:606–608. [Google Scholar]
- Hara S, Yamakawa M. A novel antibacterial peptide family isolated from the silkworm, Bombyx mori. Biochem J. 1995b;310 (Pt 2):651–656. doi: 10.1042/bj3100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetru C, Hoffmann JA. NF-kappaB in the immune response of Drosophila. Cold Spring Harb Perspect Biol. 2009;1:a000232. doi: 10.1101/cshperspect.a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Four serine proteinases expressed in Manduca sexta haemocytes. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Kawooya JK, Law JH, Ryan RO, Van Heusden MC, Ziegler R. Insect Haemolymph Proteins. In: Evans PD, Wigglesworth VB, editors. Adv Insect Physiol. Academic Press; 1990. pp. 299–396. [Google Scholar]
- Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, Yang Y, Park JW, Lee HH, Ha NC, Kang HJ, Nonaka M, Soderhall K, Lee BL. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283:7599–7607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Chen G, Strand MR. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochem Mol Biol. 2005;35:1335–1346. doi: 10.1016/j.ibmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The Host Defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Liu G, Kang D, Steiner H. Trichoplusia ni lebocin, an inducible immune gene with a downstream insertion element. Biochem Biophys Res Commun. 2000;269:803–807. doi: 10.1006/bbrc.2000.2366. [DOI] [PubMed] [Google Scholar]
- Otvos L., Jr The short proline-rich antibacterial peptide family. Cell Mol Life Sci. 2002;59:1138–1150. doi: 10.1007/s00018-002-8493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao XJ, Yu XQ. Lipoteichoic acid and lipopolysaccharide can activate antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Dev Comp Immunol. 2010;34:1119–1128. doi: 10.1016/j.dci.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu S, Wang Y, Kanost MR, Hartson S, Jiang H. Functional analysis of four processing products from multiple precursors encoded by a lebocin-related gene from Manduca sexta. Dev Comp Immunol. 2010;34:638–647. doi: 10.1016/j.dci.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra JA. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch Insect Biochem Physiol. 2000;43:49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Volz J, Osta MA, Kafatos FC, Müller HM. The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. J Biol Chem. 2005;280:40161–40168. doi: 10.1074/jbc.M506191200. [DOI] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]