Summary
Mutations in the mitochondrial kinase PINK1 and the cytosolic E3 ligase Parkin can cause Parkinson’s disease. Damaged mitochondria accumulate PINK1 on the outer membrane where, dependent on kinase activity, it recruits and activates Parkin to induce mitophagy, potentially maintaining organelle fidelity. How PINK1 recruits Parkin is unknown. We show that endogenous PINK1 forms a 700 kDa complex with the translocase of the outer membrane (TOM) selectively on depolarized mitochondria whereas PINK1 ectopically targeted to the outer membrane retains association with TOM on polarized mitochondria. Inducibly targeting PINK1 to peroxisomes or lysosomes, which lack a TOM complex, recruits Parkin and activates ubiquitin ligase activity on the respective organelles. Once there, Parkin induces organelle selective autophagy of peroxisomes but not lysosomes. We propose that the association of PINK1 with the TOM complex allows rapid re-import of PINK1 to rescue repolarized mitochondria from mitophagy, and discount mitochondrial-specific factors for Parkin translocation and activation.
Introduction
In humans, loss of function mutations in the genes encoding PINK1 and Parkin have been linked to autosomal recessive forms of Parkinson’s Disease (PD) (Kitada et al., 1998; Valente et al., 2004). In Drosophila, PINK1 and Parkin function in the same pathway to maintain mitochondrial fidelity (Clark et al., 2006; Greene et al., 2003; Park et al., 2006; Yang et al., 2006) supporting prior studies indicating that mitochondrial dysfunction may be a contributing factor in PD (Schapira, 2010). More recent studies have outlined a role for these proteins in the selective elimination of damaged mitochondria through a process termed mitophagy (Geisler et al., 2010; Lee et al., 2010; Matsuda et al., 2010; Narendra et al., 2008; Narendra et al., 2010b; Vives-Bauza et al., 2010). PINK1 is a serine/threonine kinase localized to mitochondria while Parkin is an E3 ubiquitin ligase found in the cytosol. Through its kinase activity, PINK1 can recruit Parkin to depolarized mitochondria where Parkin ubiquitinates mitochondrial substrates and, along with additional factors, drives mitophagy.
MEF cells lacking the mitochondrial fusion proteins Mfn1 and Mfn2 have a mixed population of both healthy and damaged/depolarized mitochondria (Chen et al., 2005). In these cells, Parkin specifically accumulates on the subpopulation of depolarized mitochondria (Narendra et al., 2008). The PINK1 requirement for Parkin translocation raises the question of how PINK1 is activated selectively on damaged mitochondria. The answer lies in the regulation of PINK1 protein levels and also its submitochondrial location. In healthy mitochondria, PINK1 levels are kept low through membrane potential dependent import and constitutive turn over (Jin et al., 2010; Narendra et al., 2010b). This process involves PINK1 import to the inner membrane where it is processed to a smaller 52 kDa form by the mitochondrial rhomboid protease PARL (Deas et al., 2011; Jin et al., 2010; Meissner et al., 2011; Whitworth et al., 2008). This form of PINK1 is subsequently degraded by an MG132 sensitive protease thereby keeping steady state levels of PINK1 almost undetectable on polarized mitochondria. When a mitochondrion sustains damage that leads to a loss in membrane potential, PINK1 proteolysis mediated by import is blocked in that organelle and PINK1 accumulates on the outer membrane. This appears to act as a sensing mechanism for damaged mitochondria and allows PINK1 to specifically recruit Parkin from the cytosol to depolarized organelles. How PINK1 recruits Parkin remains unclear.
In this study we assess the role of mitochondrial factors in PINK1 mediated Parkin translocation and activation. We find that PINK1 forms a 700 kDa complex with the translocase of the outer membrane (TOM) on depolarized organelles. PINK1 fused to the transmembrane anchor of OPA3 retains association with TOM in the absence of uncoupler, suggesting a functional association between PINK1/TOM. PINK1 targeted to peroxisomes or lysosomes, that lack a TOM complex, recruits and activates Parkin on the respective organelles. Furthermore, PINK1 requires membrane localization to stimulate its ability to recruit and activate Parkin. We also find that Parkin targeted to peroxisomes by PINK1 is sufficient to drive pexophagy but this was not recapitulated on lysosomes. Although we rule out mitochondrial-specific factors for Parkin translocation and activation, PINK1 binding to TOM may function to rapidly re-import PINK1 to down-regulate the PINK1/Parkin pathway
Results
PINK1 forms a large multimeric complex on the mitochondrial outer membrane
We have previously shown that PINK1 is imported into mitochondria and constitutively turned over in a PARL mediated process that requires mitochondrial membrane potential (Jin et al., 2010). In the presence of carbonyl cyanide m-chlorophenyl hydrazone (CCCP), PINK1 import and degradation are inhibited forcing its accumulation on the mitochondrial outer membrane where it functions in recruiting Parkin.
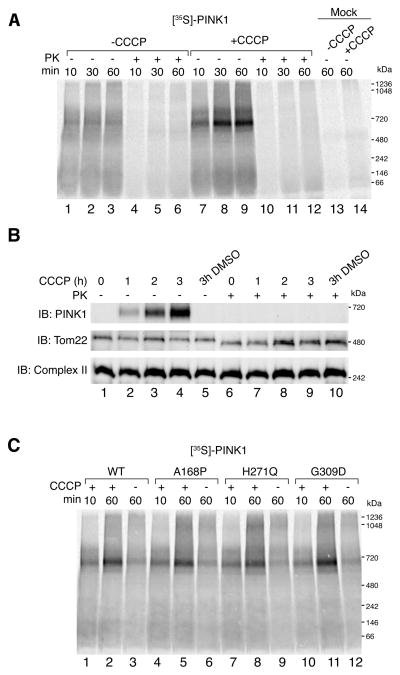
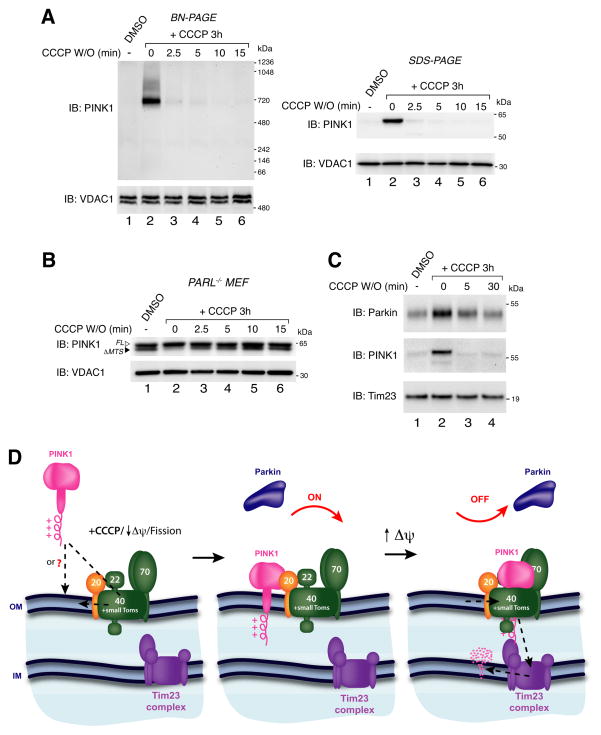
Using mitochondrial in vitro import assays coupled with BN-PAGE, we assessed the quaternary structure of PINK1 on the mitochondrial outer membrane. Given that in vitro translated PINK1 is imported into purified mitochondria, any assembly of PINK1 represents an interaction with preexisting proteins or complexes. As shown schematically (Fig. S1A), [35S]-labeled PINK1 was generated in vitro using rabbit reticulocyte lysates and incubated with freshly isolated HeLa mitochondria for different times with or without the mitochondrial uncoupler CCCP. External protease (Proteinase K) was added to half of the samples to degrade non-imported or outer membrane integrated PINK1. Samples were then solubilized in a 1% digitonin containing buffer and subjected to BN-PAGE followed by detection of radioactive protein using phosphorimaging (Fig. 1A). In polarized mitochondria, [35S]-PINK1 did not assemble into a prominent complex (lanes 1–3), however following the addition of CCCP, [35S]-PINK1 was found to assemble into a 700 kDa complex that accumulated over time (lanes 7–9). External protease (lanes 10–12) degraded the PINK1 containing complex suggesting that it forms on the mitochondrial outer membrane. Mock import of [35S]-PINK1 in the absence of mitochondria (lanes 13 and 14) as well as import of [35S]-PINK1 Δ110 lacking its N-terminal targeting sequences (Fig. S1B) confirmed that the complex formation was dependent on PINK1 import into mitochondria and not an artifact of aggregation. Furthermore, import of PINK1 into PARL−/− MEF mitochondria confirmed that in the absence of CCCP, the PINK1 complex does not form, nor does it resolve in its monomeric range on BN-PAGE (Fig. S1C).
Figure 1.
In vitro import and BN-PAGE analysis of PINK1. (A) [35S]-PINK1 was incubated with isolated HeLa mitochondria with or without 1 μM CCCP for increasing times as indicated. Samples were treated with or without Proteinase K (PK) and solubilized in 1% digitonin containing buffer. Mock import samples lacking mitochondria were treated as above as indicated. (B) Mitochondria were isolated from HeLa cells that were either untreated or treated with CCCP or vehicle control (DMSO). Isolated mitochondria were treated with or without external protease (PK) and subjected to BN-PAGE and immunoblotting using antibodies against PINK1 (outer membrane), Tom22 (outer membrane) and Complex II (inner membrane). (C) Radiolabeled WT PINK1 and PINK1 patient mutants A168P, H271Q and G309D were imported into isolated HeLa mitochondria as in (A). Radiolabeled proteins were detected by phosphorimage analysis. See also Figure S1.
We also analyzed endogenous PINK1 complex formation using mitochondrial extracts from living cells. HeLa cells were either untreated or treated with vehicle or CCCP for increasing times prior to mitochondrial isolation and BN-PAGE immunoblotting analysis (Fig. 1B). The 700 kDa PINK complex was observed following 1h CCCP treatment (Fig. 1B, lane 2, top row) and accumulated with increasing times (lanes 3 and 4). The PINK1 complex was not observed in mitochondria from untreated or vehicle treated cells (lanes 1 and 5, top row). External Proteinase K treatment led to the degradation of the PINK1 complex, and proteolytic processing of the exposed cytosolic facing domains of the TOM complex (Fig. 1B, lanes 6–10, middle row) but not the inner membrane complex II (bottom row). Additionally, a fraction of these samples was also subjected to SDS-PAGE and immunoblotted for various mitochondrial markers to confirm intactness of the organelle (Fig. S1D). Taken together, these results reveal that both in vitro imported and endogenous PINK1 accumulate into a 700 kDa complex on the outer membrane of depolarized mitochondria.
Next we assessed the complex assembly of PINK1 PD patient mutants A168P, H271Q and G309D using the in vitro import assay (Fig. 1C). The accumulation of PINK1 mutants into the 700 kDa complex was comparable to the WT PINK1 control suggesting that kinase activity may not be required for complex formation. Indeed, import of a PINK1 kinase dead mutant (Beilina et al., 2005), showed no defect in complex formation (Fig. S1E). Thus PINK1 complex formation occurs independently of its kinase activity.
Analysis of Parkin association with the PINK1 complex
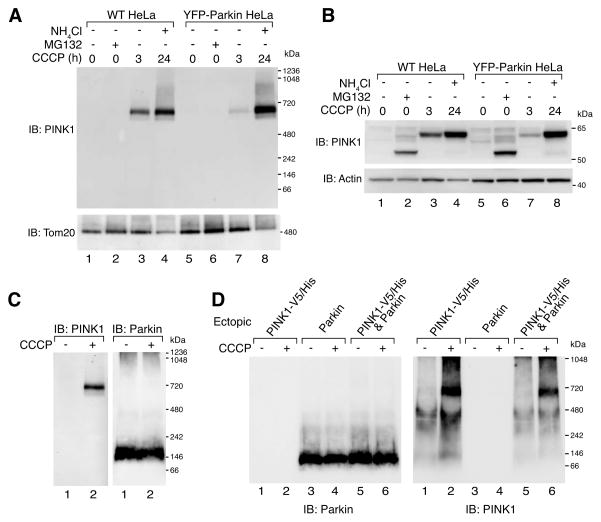
We asked whether Parkin expression impacts PINK1 complex assembly or shows stable Parkin association with the 700 kDa complex. To assess this, PINK1 complex assembly was monitored in stably transfected YFP-Parkin HeLa cells that lack endogenous Parkin. Once cells were treated with CCCP for 3h (Fig. 2A, lanes 3 and 7) or for 24h supplemented with ammonium chloride to block mitophagy (lanes 4 and 8) the PINK1 complex accumulated. However, no significant difference in the complex was observed in cells with and without expression of YFP-Parkin. Cells that were treated with MG132 did not show PINK1 complex formation (Fig. 2A, lanes 2 and 6) suggesting that the 52 kDa cleaved form of PINK1 that accumulates following MG132 treatment (Fig. 2B, lanes 2 and 6) does not integrate into the complex. The accumulation of both 52 kDa and full length forms of PINK1 was confirmed by SDS-PAGE and immunoblotting using antibodies against PINK1 (Fig. 2B and Fig. S1F). Additionally, whole cell lysates from YFP-Parkin expressing cells treated with or without CCCP were subjected to BN-PAGE and immunoblotted using anti-Parkin antibodies. YFP-Parkin was found in an ~150 kDa complex, but no signal was observed in the range of the 700 kDa PINK1 complex following CCCP treatment (Fig. 2C). Given that the ratio of endogenous PINK1 to ectopically expressed YFP-Parkin is likely to be low, we also co-expressed PINK1-V5/His. Even under these conditions, a PINK1/Parkin complex was not observed (Fig. 2D).
Figure 2.
PINK1 and Parkin complex analysis. (A-B) WT HeLa or YFP-Parkin HeLa cells were treated as indicated and subjected to BN-PAGE (A) or SDS-PAGE (B) and immunoblotted using α-PINK1 and either α-Tom20 (A) or α-actin (B) antibodies. (C) YFP-Parkin HeLa cells treated with DMSO or 20 μM CCCP for 3h were subjected to BN-PAGE and immunoblotting using α-PINK1 (left panel) and α-Parkin (right panel) antibodies. (D) HeLa cells transfected with either PINK1-V5/His or Parkin, or both were treated as in (C) before being subjected to BN-PAGE and immunoblotting using anti Parkin (left panel) or PINK1 (right panel) antibodies. Samples subjected to BN-PAGE were solubilized in 1% digitonin buffer. See also Figure S1.
In vitro import of [35S]-PINK1 into mitochondria from YFP-Parkin cells with or without prior CCCP treatment to accumulate Parkin on mitochondria (Fig. S1G and H), as well as import of [35S]-Parkin (data not shown), also did not resolve a PINK1/Parkin complex. Our findings indicate that PINK1 and Parkin do not form a stable complex on BN-PAGE and that Parkin does not affect PINK1 complex formation. Thus, either they do not stably associate or the PINK1/Parkin interaction is labile to digitonin or other components of the BN-PAGE system. We also found that in the neuronal cell line SH-SY5Y, that expresses endogenous Parkin and may be more relevant for PD, WT and PINK1 mutants displayed the same profile of mitochondrial PINK1 complex assembly as in HeLa cell mitochondria. (Fig. S1I).
Identification of PINK1 complex components
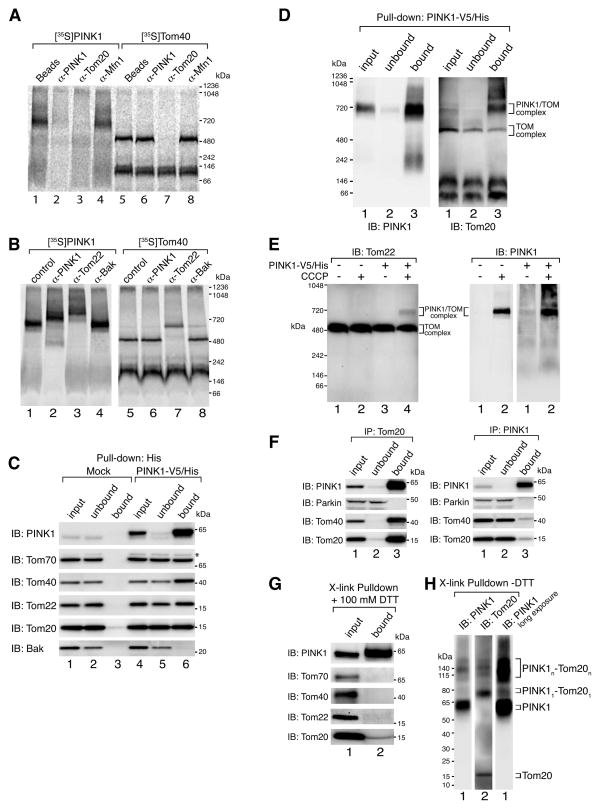
Mitochondrial precursors such as PINK1, that are destined for the mitochondrial inner membrane initially translocate through the outer membrane via the translocase of the outer membrane (TOM) complex (reviewed in Schmidt et al., (2010)). We reasoned that PINK1 could become sequestered within the TOM complex, retaining it on the mitochondrial surface when its import into the inner mitochondrial membrane is blocked by uncoupling with CCCP. To test this, we performed immunodepletion experiments after [35S]-PINK1 was imported into mitochondria in the presence of CCCP. Following import, mitochondria were lysed in 1% digitonin buffer and subjected to immunoprecipitation using antibodies against PINK1, Tom20 or Mfn1. PINK1 protein complexes not bound by these antibodies were analyzed by BN-PAGE (Fig. 3A). The PINK1 complex was specifically depleted following incubation with immobilized α-PINK1 antibodies but not with α-Mfn1 antibodies or beads alone. Interestingly, the PINK1 complex was depleted also by α-Tom20 antibodies (lane 3). As a control, [35S]-labeled Tom40 was imported into mitochondria in the absence of PINK1 and incubated with the same antibodies as above (Fig. 3A, lanes 5–8). Only the α-Tom20 antibody depleted the 500 kDa TOM complex (lane 7).
Figure 3.
PINK1 complex is associated with components of the TOM machinery. (A) Radiolabeled proteins were imported into isolated HeLa mitochondria in the presence ([35S]-PINK1) or absence ([35S]-Tom40) of 1 μM CCCP for 60 min. Samples were solubilized in 1% digitonin buffer and complexes were immunodepleted using indicated antibodies or beads alone as a control followed by BN-PAGE analysis. (B) Radiolabeled proteins were imported into isolated HeLa mitochondria in the presence ([35S]-PINK1) or absence ([35S]-Tom40) of 1 μM CCCP for 60 min. Samples were solubilized in 1% digitonin buffer followed by the addition of antibodies as indicated, and subjected to BN-PAGE. (C) Mock transfected and PINK1-V5/His stably transfected HeLa cells were treated with 20 μM CCCP for 3 h followed by mitochondrial isolation and immunocapture using α-His antibodies coupled to beads. Bound proteins were eluted with 6xHis peptides and various fractions as indicated were subjected to SDS-PAGE followed by immunoblotting using antibodies as indicated. *, non-specific band. (D) Samples treated as in (C) were analyzed using BN-PAGE and immunoblotting with α-PINK1 (left panel) and α-Tom20 (right panel) antibodies. (E) HeLa cells were either mock transfected or transfected with PINK1-V5/His and treated with or without 20 μM CCCP for 3 h before solubilization in 1% digitonin buffer followed by BN-PAGE and immunoblotting using α-Tom22 (left panel) or α-PINK1 (right panels) antibodies. (F) HEK293 cells were treated with 20 μM CCCP for 3h and then harvested and lysed in 1% digitonin containing buffer. Clarified lysates were used for immunopreciptation using α-Tom20 (left panel) or α-PINK1 (right panel) antibody coupled beads. Input, unbound and bound fractions were subjected to SDS-PAGE and immunoblotted using antibodies against PINK1, Parkin, Tom40 and Tom20. (G) Immunocaptured PINK1-V5/His complex as in (C) was incubated in 0.1 mM dithiobis[succinimidyl propionate] for 20 min on ice. After crosslinking, samples were incubated in 1% SDS containing buffer for 5 min at 95° and then subjected to immunoprecipitation using α-His beads. Crosslinker was cleaved with 100 mM DTT before SDS-PAGE and immunoblotting with antibodies as indicated. (H) Samples were treated as in (G) and subjected to SDS-PAGE in the absence of DTT followed by immunoblotting using α-PINK1 and α-Tom20 antibodies. Radiolabeled proteins were detected by phosphorimage analysis. See also Figure S2.
We performed antibody shift analysis directed against another component of the TOM complex, the import receptor Tom22, to further demonstrate the presence of TOM components within the PINK1 complex. As can be seen in Fig. 3B, antibodies against Tom22 shifted the TOM complex control (lane 7) as well as the PINK1 complex generated by WT PINK1 (lane 3) to a higher molecular weight. Shifts were not observed using antibodies against a control outer membrane protein Bak. As expected, antibodies against PINK1 shifted the PINK1 complex (lane 2) but not the TOM complex (lane 6) that is devoid of PINK1 under these conditions.
We set out to further characterize PINK1’s association with components of the TOM machinery by purifying the PINK1 complex under blue native gel extraction conditions. To achieve this we generated a stable HeLa cell line expressing PINK1 with a V5/His tag. In vitro import of [35S]-PINK1-V5/His showed that the tag did not affect complex formation as compared to WT PINK1 (Fig. S2A). HeLa cells stably expressing PINK1-V5/His were either untreated or treated with CCCP for 3h to accumulate the PINK1 complex followed by mitochondrial isolation and solubilization in 1% digitonin containing buffer. The PINK1 complex was pulled down using 6xHis antibody coupled beads and eluted under native conditions with 6xHis peptides. A fraction of the eluted protein subjected to either BN-PAGE (Fig. S2B top panel, lane 6) or SDS-PAGE (bottom panel, lane 6) and immunoblotted for PINK1 confirmed the successful isolation of the PINK1 complex from CCCP treated cells. We then tested for the presence of TOM components in this immuno-purified PINK1 complex and identified Tom20, Tom22, Tom40 and Tom70 as PINK1 complex interacting proteins but not the outer membrane control Bak (Fig. 3C, lanes 4–6). Mock transfected HeLa cells were used as controls to confirm specific immunocapture of TOM components (lanes 1–3).
To further test that TOM subunits associate in a complex with PINK1, we analyzed purified PINK1 complex samples using BN-PAGE by immunoblotting for Tom20 (Fig. 3D). As Tom20 was identified in a 700 kDa complex that co-migrates with PINK1, we conclude that PINK1 forms a complex with the TOM complex (Fig. 3D, right panel). However it should be noted that a portion of Tom20 was also found in the TOM complex lacking PINK1. This may result from partial dissociation of the TOM complex from PINK1 under the conditions used for BN-PAGE. This is supported by the presence of an ~200 kDa smear in the bound fraction when immunoblotting for PINK1 (left panel, lane 3). Furthermore, PINK1 appears to associate with only a fraction of the more highly expressed TOM complex given that complete depletion of TOM components was not observed in unbound fractions from Fig. 3C and D. This is further supported by the observation that endogenous levels of PINK1 have no effect on the Tom complex following CCCP treatment and BN-PAGE analysis (Fig. 3E). However a small fraction of the TOM complex was shifted to the 700 kDa PINK1/TOM complex in cells expressing ectopic PINK1-V5/His and treated with CCCP (Fig. 3E, lane 4).
Although only a small fraction of TOM is bound by PINK1 (Fig. 3C, D and E), co-immunoprecipitations of endogenous proteins using antibodies against Tom20 revealed that of all of the detectable PINK1 was associated with the TOM complex (Fig. 3F, left panel). Endogenous Parkin was not co-immunoprecipitated with α-Tom20 antibodies supporting our findings above (Fig. 2), that show Parkin does not stably associate with the PINK1/TOM complex. It should also be noted that immunoprecipitations using α-PINK1 antibodies also failed to pull down endogenous Parkin (Fig. 3F, right panel). This suggests that while the buffer conditions used were suitable to maintain a stable PINK1/TOM interaction, they may be labile for PINK1/Parkin. Otherwise the interaction between endogenous proteins may be transient.
To identify which TOM component directly associates with immunopurified PINK1 the complex was crosslinked using the thiol cleavable crosslinker dithiobis[succinimidyl propionate] (DSP). Proteins were dissociated using SDS followed by immunoprecipitation of PINK1. Immuno-captured crosslinked products were analyzed by reducing SDS-PAGE to identify individual components resulting from crosslinker cleavage. Of the TOM subunits analyzed, Tom20 was found to specifically crosslink with PINK1 (Fig. 3G). SDS-PAGE using non-reducing conditions was subsequently employed to determine the oligomeric state of PINK1-Tom20 crosslinks (Fig. 3H). Both PINK1 (lane 1, (right panel=long exposure)) and Tom20 (lane 2) immunostaining was observed at ~78 kDa and above 115 kDa. The combined molecular weight of PINK1 (62 kDa) and Tom20 (16 kDa) is predicted to be ~78 kDa in agreement with the 78 kDa band representing a single crosslink between the two. Taken together, our findings indicate that when mitochondria lose their membrane potential, PINK1 accumulates into a large multimeric assembly with the TOM complex and directly interacts with at least Tom20.
PINK1 was not found to crosslink with the import channel Tom40 suggesting that PINK1’s association with the TOM complex does not result from stalled import upon depolarization. Therefore, to address whether PINK1 has the potential to bind the TOM complex in the absence of CCCP treatment, when import is not impaired, we imported PINK1Δ110-YFP fused to the N-terminal mitochondrial anchor of OPA3 a.a. 1–30 (OPA3™-PINK1Δ110-YFP). In the absence of CCCP, OPA3™-PINK1Δ110-YFP has previously been shown to recruit Parkin and promote mitophagy in the same manner as WT PINK1 in the presence of CCCP (Narendra et al., 2010b). Import of OPA3™-PINK1Δ110-YFP circumvents the use of CCCP to localize PINK1 on the outer membrane and therefore would not allow PINK1 to become stuck in transit within the TOM translocase upon uncoupling. Indeed, [35S]-OPA3™-PINK1Δ110-YFP was found to assemble into the 700 kDa complex in the absence of CCCP treatment (Fig. S2C, lanes 1–3) and was comparable to WT PINK1 that assembled in the presence of CCCP (lane 7). Immunodepletion of PINK1 and OPA3™-PINK1Δ110-YFP complexes as well as antibody shift analysis as described in Fig. 3A and B, confirmed that the OPA3™-PINK1Δ110-YFP complex is associated with the TOM machinery (Fig. S2D and E).
Unusually, [35S]-OPA3™-PINK1Δ110-YFP did not assemble into the 700 kDa complex in the presence of CCCP (Fig. S2C, lanes 4–6), suggesting that the OPA3 anchor may require a membrane potential for its import. This is supported by the observation that CCCP treatment did not affect the mitochondrial location of previously imported OPA3™-PINK1Δ110-YFP in cells (Fig. S2F), although a slight increase in cytosolic fluorescence was observed suggesting that import of newly synthesized OPA3™-PINK1Δ110-YFP was blocked. Furthermore, CCCP treatment after complex formation of OPA3™-PINK1Δ110-YFP did not alter its binding to the TOM complex (Fig. S2G). Given this, we conclude that the OPA3 anchor may unconventionally require a membrane potential for its import. The PINK1/TOM association is likely to occur through the cytosolic domain of PINK1 since the association is not lost upon deletion of N-terminal residues 1–110 and replacement with the OPA3 anchor. Furthermore, given that OPA3™-PINK1Δ110-YFP associates with the TOM complex in the absence of CCCP it suggests that the PINK1/TOM complex does not represent a stalled import intermediate consistent with prior observations on single membrane spanning mitochondrial proteins such as PINK1 (Frazier et al., 2003).
Is the PINK1/TOM complex required for Parkin recruitment and/or mitophagy?
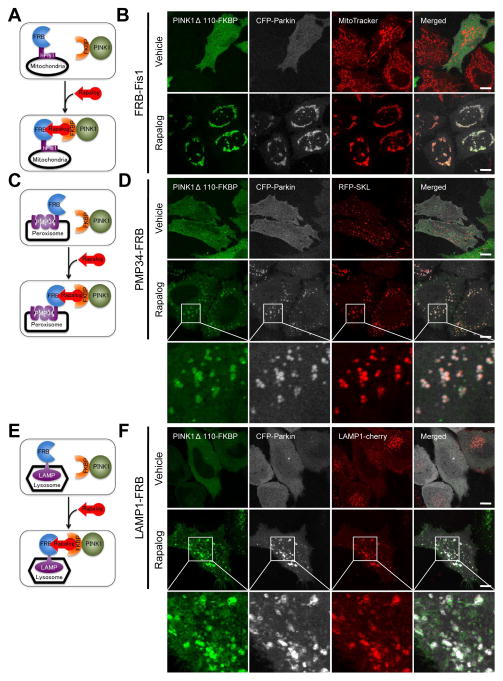
The TOM complex is exclusive to mitochondria. Targeting PINK1 to other organelles, such as peroxisomes and lysosomes allows us to investigate the potential requirement for a PINK1/TOM complex in Parkin recruitment and autophagy induction. To achieve this, we used a regulated heterodimerization system (Belshaw et al., 1996), in which the FRB domain, a 93 amino acid portion of human FK506 binding protein-12-rapamycin associated protein (also known as mTOR) was fused to different organelle membrane anchors. The FKBP domain (two tandem repeats of full-length human FK506 binding protein-12 (FKBP12)) was fused to the C-terminus of PINK1Δ110-YFP lacking the membrane spanning and mitochondrial targeting domains. Addition of a non-immunosuppressive analog of rapamycin (rapalog, also called AP21967) causes the FRB and FKBP domains of the fusion proteins to heterodimerize and thus co-localize. FRBs were fused to the following organelle-specific proteins, exposing FRB to the cytoplasm: 1) human Fis1 C-terminal tail (92–152) (FRB-Fis1) for mitochondria, 2) human 34 kDa peroxisomal integral membrane protein (PMP34-FRB) for peroxisomes, and 3) rat lysosomal-associated membrane protein 1 (LAMP1-FRB) for lysosomes (Fig. 4A, C and E). To confirm the functionality of these constructs, cells co-expressing EGFP fused to FKBP (EGFP-FKBP) and each of these organelle specific FRB fusion constructs were treated with or without rapalog and analyzed by confocal microscopy. FRB-Fis1, PMP34-FRB, and LAMP1-FRB successfully recruited cytosolic EGFP-FKBP to their corresponding organelles only in the presence of rapalog (Fig. S3A, B and C).
Figure 4.
Ectopically localized PINK1 can recruit Parkin to mitochondria, peroxisomes and lysosomes. (A, C, E) Schematic diagrams of organelle specific heterodimerization using FRB-Fis1 for mitochondria (A), PMP34-FRB for peroxisomes (C), and LAMP1-FRB for lysosomes (E) are illustrated. Treatment of cells with rapalog induces the heterodimerization of FRB and FKBP causing cytosolic PINK1Δ110-YFP-FKBP to attach to the organelle specified by the FRBs. (B, D, F) HeLa cells were transfected with PINK1Δ110-YFP-FKBP, CFP-Parkin, one of the organelle specific FRBs and an organelle targeted fluorescent marker protein: (RFP-SKL for peroxisomes (D) LAMP1-cherry for lysosomes (F)). For mitochondria, cells were stained with MitoTracker Red before rapalog treatment (B). After 48 h of transfection, cells were treated with or without rapalog for 2 h and imaged by confocal microscopy. (D, F) The third rows are enlarged images of the white boxes in the second rows. White bar, 10 μm. See also Figures S3 and S4.
Using the same approach, we assessed whether PINK1Δ110-YFP-FKBP when expressed at levels comparable to those of endogenous PINK1 (Fig. S4A) could be recruited to the different organelles and also whether it may recruit CFP-Parkin to the alternative compartments (Fig. 4B, D and F). In the absence of rapalog, both PINK1Δ110-YFP-FKBP and CFP-Parkin are localized to the cytosol in cells co-expressing Fis1-FRB (Fig. 4B, top panels). Upon rapalog treatment, PINK1Δ110-YFP-FKBP binds to mitochondria and also recruits CFP-Parkin to the same compartment (Fig. 4B, bottom panels). TMRE staining of cells and PINK1 immunoblotting confirmed that rapalog treatment does not depolarize mitochondria or induce stabilization of endogenous PINK1 (Fig. S4A and B). As a control, kinase dead PINK1-FKBP failed to recruit CFP-Parkin upon rapalog treatment (Fig. S4C). In contrast to endogenous PINK1 and OPA3™-PINK1Δ110-YFP, rapalog targeted PINK1Δ110-YFP-FKBP to mitochondria by FRB-Fis1 did not bind the TOM complex (Fig. S4D). The FRB and FKBP protein domains engineered into FRB-Fis1/PINK1Δ110-FKBP may sterically inhibit PINK1 binding to TOM in contrast to the more membrane proximal endogenous PINK1 and OPA3™-PINK1Δ110. Overall, these results show that PINK1 heterodimerized to the Fis1 mitochondrial anchor is functional in Parkin recruitment. Like EGFP, in the presence of rapalog, PINK1Δ110-YFP-FKBP was recruited to peroxisomes or lysosomes in cells co-expressing either PMP34-FRB or LAMP1-FRB, respectively (Fig 4D and F, bottom panels). Organelle recruitment was not observed in vehicle treated cells (Fig. 4D and F, top panels). Interestingly, CFP-Parkin was also recruited to peroxisomes or lysosomes specifically upon rapalog treatment (Fig. 4D and F). Thus, PINK1 can recruit Parkin to different subcellular organelles thereby excluding the putative requirement for other mitochondrial-specific factors as well as the PINK1/TOM complex in this process.
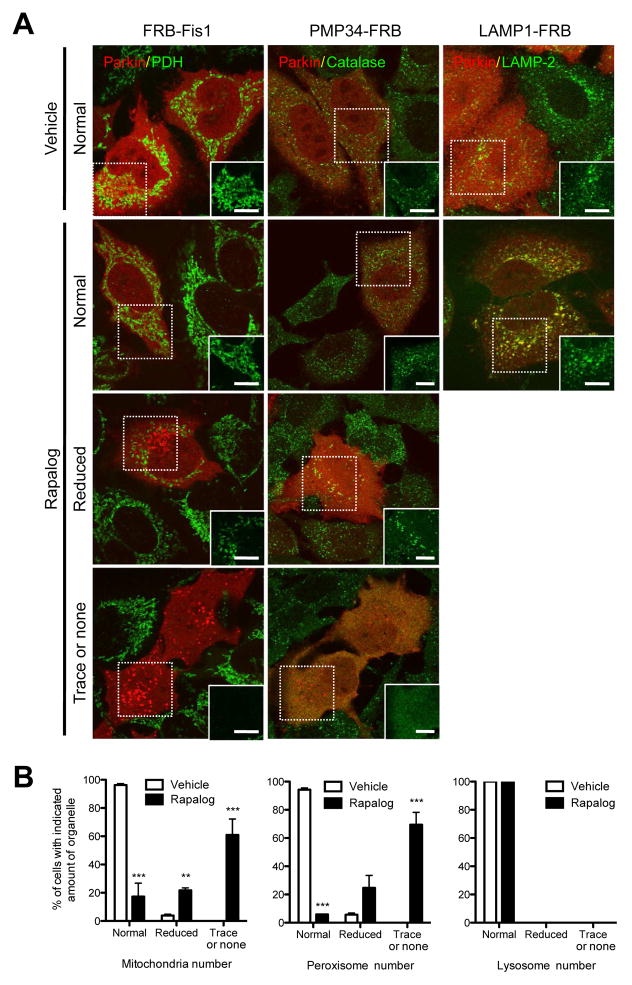
We also determined if Parkin recruited to alternative compartments by ectopic PINK1 can exert its downstream functions of ubiquitination and autophagy induction of the various organelles. Cells co-expressing PINK1Δ110-YFP-FKBP at levels comparable to those of endogenous PINK1 (Fig. S4A), the mitochondrial targeting construct (FRB-Fis1) and either CFP-Parkin or mCherry-Parkin did not display organelle ubiquitination or autophagic clearance in the absence of rapalog. (Fig. S5A and Fig. 5A top panels). However, upon treatment of cells with rapalog for 2 h, robust mitochondrial ubiquitination (Fig. S5A, middle panels) was observed. Extended incubation with rapalog for 48 h led to a large proportion of cells displaying reduced or no mitochondria (Fig. 5A and B, left panels), thereby confirming that ectopic targeting of PINK1 to mitochondria recapitulates CCCP induced PINK1/Parkin mediated mitophagy. When the same approach was applied with peroxisome targeting of PINK1, we found that after 2 hrs of rapalog incubation peroxisomes displayed ubiquitination (Fig. S5B), and 48 hr of rapalog incubation led to pexophagy (Fig. 5A and B, middle panels) in the presence of overexpressed mCherry-Parkin. To confirm that such organelle removal occurs through autophagy, we applied the same approach to Atg5−/− MEFs. We found a significant increase of cells showing no evidence of mitophagy or pexophagy and a significant decrease of cells showing reduction of the organelle number compared to Atg5+/+ MEFs (Fig. S5D and E). This indicates that the majority of the reduction in the number of mitochondria or peroxisomes in Figure 5B is caused by autophagy. Lysosomal targeting of PINK1 did not induce lysosome clearance after 48 hrs of rapalog incubation (Fig. 5A and B, right panels), even though an increased ubiquitination signal was observed in lysosomes after 2 hrs of rapalog incubation (Fig. S5C). As a control, the R42P Parkin PD mutant (Matsuda et al., 2010; Narendra et al., 2010b; Terreni et al., 2001) failed to ubiquitinate mitochondria, peroxisomes and lysosomes following rapalog targeting of PINK1 (bottom panels in Fig. S5A, B and C). Overall, these results exclude an essential role for the PINK1/TOM complex in Parkin recruitment and mitophagy. They also highlight that apart from PINK1, mitochondria-specific factors are not required for the PINK1/Parkin pathway to induce pexophagy. The ubiquitination of membrane proteins appears to be insufficient to induce clearance of lysosomes that may have specific restrictions to autophagic removal.
Figure 5.
Parkin recruited by ectopic PINK1 can induce autophagy of mitochondria and peroxisomes. (A) HeLa cells were transfected with PINK1Δ110-YFP-FKBP and cherry-Parkin together with one of the organelle specific FRBs (FRB-Fis1 (left column), PMP34-FRB (middle column) and LAMP1-FRB (right column)) for 24 h. After 48 h of treatment with (lower 3 rows) or without (top row) rapalog, cells were fixed, immunostained for organelle specific proteins (pyruvate dehydrogenase subunit E1α (PDH) for mitochondria (left column), catalase for peroxisomes (middle column) and LAMP-2 for lysosomes (right column)) and imaged with confocal microscopy. In rapalog treated cells, representative images of cells showing three different responses (normal, reduced and trace or none) of organelle mass are shown. White boxes on the right bottom corner show only the organelle markers from the dashed box to clearly show the change of organelle mass. White bar, 10 μm. (B) Cells having the indicated amount of each organelle in (A) were counted. To ensure the co-expression of the three constructs (PINK1-FKBP, Parkin and FRB) only the cells showing an abundant expression of mCherry-Parkin were counted. The graphs represent means ± SEM of counts in >100 cell per condition in three independent experiments and analyzed with 2 way ANOVA. ***; P<0.001, **; P<0.01. See also Figure S5.
PINK1 requires membrane localization to recruit Parkin
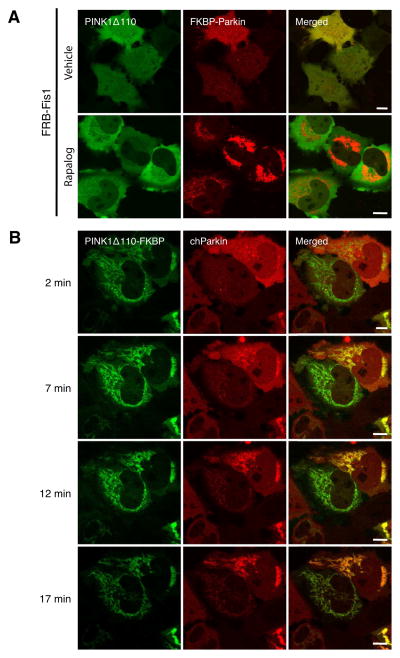
The ectopic recruitment of Parkin to PINK1 expressed in alternate locations is consistent with direct binding of PINK1 to Parkin, as previously reported (Sha et al., 2010; Shiba et al., 2009; Xiong et al., 2009), but not seen in BN gels (Fig. 2) or immunoprecipitations of endogenous proteins (Fig. 3F). A constitutively stable interaction between the PINK1 and Parkin suggests that Parkin targeted to the mitochondria would recruit a cytosolic form of PINK1 (Δ110), lacking its mitochondrial targeting information. To investigate this we fused the FKBP domain to mCherry-Parkin at the N-terminus of Parkin (mCherry-FKBP-Parkin). To confirm that Parkin in this recombinant protein is fully functional, we assessed mitophagy by CCCP treatment in cells expressing either mCherry-FKBP-Parkin, or YFP-Parkin as a control (Fig. S6A and B). Mitochondrial clearance of mCherry-FKBP-Parkin after 24 h CCCP treatment was similar to that of YFP-Parkin. We therefore used FRB-Fis1 to direct mCherry-FKBP-Parkin to mitochondria with rapalog to see if it induced mitophagy or recruited PINK1. In vehicle treated cells, both mCherry-FKBP-Parkin and PINK1Δ110-YFP were localized in the cytosol (Fig. 6A, top panels). Upon rapalog treatment for 2 h, mCherry-FKBP-Parkin was recruited to mitochondria but PINK1Δ110-YFP remained in the cytosol (Fig. 6A, bottom panels), arguing against a constitutive PINK1/Parkin interaction. Previous reports have shown that overexpressed PINK1 and Parkin interact by co-immunoprecipitation after lysis (Sha et al., 2010; Xiong et al., 2009), however our results in intact cells are not in agreement. Moreover, a time course rapalog treatment of cells co-expressing PINK1Δ110-YFP-FKBP along with FRB-Fis1 and mCherry-Parkin, revealed that PINK1Δ110-YFP-FKBP targeted to mitochondria in two minutes (Fig. 6B, top left panel). However mCherry-Parkin remained in the cytosol (Fig. 6B, top middle panel), and only accumulated on mitochondria after a >5 minute delay (lower panels). The differential kinetics of mitochondrial recruitment between the two raises the interesting possibility that upon membrane localization, latent PINK1Δ110-YFP-FKBP becomes active in Parkin recruitment.
Figure 6.
PINK1 not localized to membranes does not interact with Parkin. (A) Mitochondrial Parkin does not recruit cytosolic PINK1 to mitochondria. HeLa cells were transfected with PINK1Δ110-YFP, mCherry-FKBP-Parkin and FRB-Fis1 and treated with or without rapalog for 2 h. The live cells were imaged by confocal microscopy. (B) Delayed Parkin translocation following the translocation of cytosolic PINK1-FKBP induced by heterodimerization. HeLa cells were transfected with PINK1Δ110-YFP-FKBP, mCherry-Parkin and FRB-Fis1 for 48 h, treated with rapalog and applied for confocal live cell imaging over 17 min. The first image was taken after 2 min of rapalog treatment, and subsequent images were taken after every 5 min. White bar, 10 μm. See also Figure S6.
It is possible that the primary role of PINK1 is to recruit Parkin to mitochondria and once there, Parkin is able to ubiquitinate substrates and stimulate mitophagy. However mCherry-FKBP-Parkin targeted to mitochondria by FRB-Fis1 did not ubiquitinate mitochondria (Fig. S6C, upper panels), nor did it induce mitophagy (Fig. S6D, upper panels). Similar results were observed when mCherry-FKBP-Parkin was placed on peroxisomes (Fig. S6C and D, lower panels) by heterodimerization. Mitophagy was observed in rapalog treated cells co-expressing mCherry-FKBP-Parkin, PINK1Δ110-YFP-FKBP and FRB-Fis1 (Fig. S6E), indicating that mitochondrial Parkin localized on mitochondria through the heterodimerization can be activated by mitochondrial PINK1 to induce mitophagy. Taken together these results show that although FKBP-Parkin is functional, its localization on mitochondria is not sufficient for activity. PINK1 (Δ110) located in the cytosol is insufficient for both Parkin translocation and subsequent activation of E3 ligase activity but regains these activities upon ectopic targeting to alternate membranes.
PINK1 is rapidly re-imported following CCCP washout
Through ectopic expression of PINK1 on peroxisomes we have shown that, apart from PINK1, mitochondrial-specific factors are not required for Parkin mediated autophagy of organelles. This raises the question of the physiological function of PINK1 binding to the mitochondrial-specific TOM complex. It is possible that PINK1’s tight association with the TOM complex may facilitate its rapid re-import into mitochondria that regain their membrane potential. Indeed, we found that in HeLa cells endogenous PINK1 was degraded within two and a half minutes of CCCP washout using both BN-PAGE and SDS-PAGE analysis (Fig. 7A). TMRE staining of cells confirmed that mitochondria could regain their membrane potential within the two and a half minute washout period (Fig. S7). Furthermore, rapid re-import of PINK1 was confirmed in PARL−/− MEFs transfected with PINK1-V5/His (Fig. 7B). In these cells, imported PINK1 has its mitochondrial targeting sequence cleaved by the matrix processing peptidase (MPP), and appears as a slightly faster migrating species (described in Jin et al., (2010)). This imported form of PINK1 (ΔMTS) was absent after CCCP treatment (Fig. 7B, lane 2) and re-appeared within two and half minutes after washout where import mediated MPP processing of PINK1 resumed (lanes 3–6).
Figure 7.
PINK1 and Parkin after CCCP washout. (A) HeLa cells were treated with either DMSO or CCCP for 3 h before CCCP washout for increasing times as indicated. Cells were lysed in 1% digitonin buffer (BN-PAGE; left panel) or SDS sample buffer (SDS-PAGE; right panel) and immunoblotted using α-PINK1 and α-VDAC1 antibodies. (B) PARL−/− MEFs transfected with PINK1-V5/His were treated as in (A). Cells were lysed in SDS sample buffer and immunoblotted using α-PINK1 and α-VDAC1 antibodies. (C) HeLa cells were treated as in (A) and mitochondrial fractions were subjected to SDS-PAGE and immunoblotting using α-Parkin, α-PINK1 and α-Tim23 antibodies. W/O: washout, FL: full-length, ΔMTS: mitochondrial targeting sequence cleaved. (D) Model for PINK1 regulation. See discussion for description. See also Figure S7.
Since PINK re-import could act as a regulatory mechanism in the Parkin pathway, we analyzed whether Parkin is released from mitochondria upon PINK1 re-import. Cells transfected with untagged Parkin were treated with CCCP followed by washout for different times. Mitochondria were isolated from cells and immunoblotted for PINK1, Parkin and Tim23 as a loading control. As can be seen (Fig. 7C), the levels of Parkin in the mitochondrial fraction were reduced in conjunction with PINK1 following CCCP washout. This indicates that PINK1 on the outer membrane is required to maintain Parkin on depolarized mitochondria and supports a role for the PINK1/TOM complex in re-import downregulation of the pathway.
Discussion
It is currently under debate whether PINK1-dependent phosphorylation of Parkin or another substrate drives mitochondrial translocation and activity of Parkin (Kim et al., 2008b; Narendra et al., 2010b; Sha et al., 2010; Vives-Bauza et al., 2010). PINK1 lacking its mitochondrial targeting sequences resides in the cytosol and does not activate Parkin translocation to mitochondria (Matsuda et al., 2010; Narendra et al., 2010b). This raises the question whether mitochondrial factors could function with PINK1 to recruit Parkin.
Mitochondrial in vitro import of PINK1 coupled with BN-PAGE revealed that PINK1 efficiently assembles into a 700 kDa complex on the outer membrane of uncoupled mitochondria. In the absence of uncoupler, PINK1 has been identified in complexes ranging from 130–460 kDa using sucrose gradients (Liu et al., 2009). That 700 kDa PINK1 complex formation was observed when mitochondria were depolarized and import to the inner membrane was inhibited raised the possibility that PINK1 was trapped in the TOM complex. The TOM complex mediates the import of most mitochondrial proteins and is an essential molecular machine for mitochondrial function (Kunkele et al., 1998; Model et al., 2002). Using different approaches, we found that the 700 kDa PINK1 complex contained the import receptors Tom70, Tom22 and Tom20, as well as the translocation pore Tom40. Specifically, we found that PINK1 directly interacts with Tom20, the main receptor for N-terminal presequences (Abe et al., 2000; Brix et al., 1997). Although Parkin was not found to stably interact with the PINK1/TOM complex, PINK1 mediated recruitment of Parkin to mitochondria was found to induce degradation of TOM complex components (Chan et al., 2011; Yoshii et al., 2011).
In yeast, Oxa1 and Cox18 were found to form a 500 kDa import intermediate within the TOM complex following mitochondrial depolarization (Frazier et al., 2003). Like PINK1, both Oxa1 and Cox18 contain a cleavable MTS signal but differ from PINK1 owing to their multispanning transmembrane domains. In the same study, the 500 kDa TOM complex associated intermediate was not identified with soluble matrix proteins or proteins like PINK1 that contain a cleavable MTS and a single spanning transmembrane domain. To stably trap precursors within the TOM complex researchers typically tag precursors with DHFR, an enzyme that adopts a tight fold in the presence of methotrexate and blocks precursor transit within TOM (Dekker et al., 1997; Ryan et al., 1999). Also, precursors can be crosslinked with Tom40 and other TOM components within the TOM complex (Kanamori et al., 1999; Wiedemann et al., 2001). These approaches are often combined with mitochondrial depolarization.
Even though precursors such as PINK1 do not stall in the TOM complex (Frazier et al., 2003), and PINK1 did not crosslink with the translocation pore Tom40, it is still a possibility that PINK1 becomes trapped within the TOM en route to being imported upon CCCP treatment. We used the N-terminus of OPA3 to target PINK1 to the outer mitochondrial membrane independently of import inhibition with CCCP and found that PINK1 still bound the TOM complex on polarized mitochondria, arguing against the ‘stalled import’ hypothesis. This analysis also revealed that PINK1 can associate with Tom20 through its cytosolic domain and not its N-terminal targeting sequences, further arguing against PINK1 residing within the TOM complex import channel. However as the structural basis of the PINK1-TOM complex is not resolved thus one cannot rule out that PINK1 occupies the TOM channel.
In the presence of CCCP, OPA3™-PINK1Δ110-YFP import and complex assembly was unexpectedly blocked. Single spanning signal-anchored proteins like OPA3 do not typically require a membrane potential for their import nor do they require any of the known import components for their insertion and to date, little is known about their import (reviewed in Dukanovic and Rapaport, (2011)). Thus it is not clear why CCCP should impact import of the OPA3 anchor, however it is interesting to note that currently there is discrepancy regarding membrane potential mediated processing of the outer mitochondrial membrane protein Mcl-1 (Warr et al., 2011; Yang-Yen, 2011).
A fully assembled TOM complex has a reported molecular mass of ~440–600 kDa (Ahting et al., 2001; Frazier et al., 2003; Kunkele et al., 1998), and in this report TOM lacking PINK1 was observed in the 500 kDa range on BN-PAGE. Considering the size of monomeric PINK1 (62 kDa) and the change in size from the 500 kDa TOM to the 700 kDa PINK1/TOM complex, it appears likely that more than one PINK1 or additional factors may be associated in the complex. This is also supported by the comparably modest change in TOM complex size from ~440 kDa to 500 kDa observed for Oxa1 (45 kDa) and Cox18 (36 kDa) intermediate complexes (Frazier et al., 2003). A PINK1 dimer may be associated with TOM as PINK1 can interact with itself (Liu et al., 2009), and kinases often function as dimers (Huse and Kuriyan, 2002). The presence of additional factors in the PINK1/TOM complex, such as PGAM5 (Imai et al., 2010), Miro and Milton (Weihofen et al., 2009) is also possible.
We also addressed the role of the PINK1/TOM complex in Parkin recruitment by targeting PINK1 to organelles that lack a TOM complex using a regulated heterodimerization system. Interestingly, PINK1 localization to peroxisomes or lysosomes was sufficient to recruit Parkin and stimulate its E3 ligase activity, as evidenced by organelle ubiquitination. In the absence of PINK1, Parkin ectopically targeted to mitochondria, peroxisomes or lysosomes had no detectable activity, confirming PINK1’s essential role (Matsuda et al., 2010; Narendra et al., 2010b). These findings eliminate the requirement for PINK1 binding to TOM for Parkin translocation and importantly, suggest that other mitochondrial factors are not required. We also found that Parkin targeted to peroxisomes by PINK1 was sufficient to signal pexophagy. Ubiquitin on the surface of peroxisomes can signal their degradation (Kim et al., 2008a), although ubiquitin on mitochondria does not signal mitophagy (Narendra et al., 2010a). Furthermore, autophagic removal of lysosomes was not observed, supporting the notion that ubiquitination is not sufficient for all organellar autophagy. However it is also possible that lysosomes may have inherent restrictions for autophagic removal.
PINK1’s ability to direct Parkin to different subcellular compartments is consistent with the model that they constitutively interact. However, mitochondrial targeted Parkin did not recruit PINK1Δ110 to mitochondria suggesting a stable interaction does not exist when PINK1 is not membrane bound. This indicates that PINK1 requires membrane localization for Parkin activation. PINK1 autophosphorylation has been reported in an in vitro system although its physiological function is not clear (Beilina et al., 2005; Silvestri et al., 2005). PINK1 activation may occur through a concentration dependent process (Zhang et al., 2006) that rises considerably once localized on a membrane potentially allowing it to dimerize. Alternatively, PINK1’s proximity to lipid may increase its activity by inducing a conformational change, as has been reported for the PTEN phosphatase PIP2 (Leslie et al., 2008).
PINK1 is rapidly re-imported and degraded in HeLa cells when mitochondria regain their membrane potential. This is supported by the rapid displacement of mitochondrial Parkin that occurs in conjunction with PINK1 re-import. A very recent report has shown that proteins can be laterally released from the TOM complex to the outer membrane (Harner et al., 2011). This supports a model whereby PINK1 can re-enter the import pathway through lateral opening of the TOM pore. In healthy cells, mitochondria continually undergo cycles of fission and fusion. Although mitochondrial fission events often generate depolarized units that are less likely to re-fuse and are removed by autophagy, occasionally depolarized organelles regain their membrane potential and re-fuse with the network (Twig et al., 2008). Thus, PINK1’s association with the TOM complex would allow for tight regulation of PINK1 levels (Fig. 7D). If mitochondria re-establish membrane potential, PINK1 accumulated on the outer mitochondrial membrane is readily re-imported and degraded to deactivate Parkin, terminate mitophagy and maintain healthy organelles in the network.
Materials and Methods
Cloning Procedures
The construction of plasmids is described in the Extended Experimental Procedures.
Cell culture, transfection and mitochondrial isolation
HeLa cells were cultured in Dulbecco’s modified Eagles medium (DMEM; GIBCO-BRL) containing 10% (vol/vol) fetal calf serum (FCS) at 37°C under an atmosphere of 5% CO2. To transfect cells for confocal microscopy, cells were plated in 2 well coverglass chambers. Constructs as indicated in figure legends, were mixed with Fugene HD at 1:3 ratio in Opti-MEM (GIBCO-BRL). After 15 min, the mixture was added to the culture and incubated for 24 h or 48 h before 250 nM rapalog treatment for 48 h to assess autophagy or 2 h rapalog treatment to assess Parkin translocation and ubiquitination. For mitochondrial isolation, cells were homogenized in 20 mM HEPES (pH7.6), 220 mM mannitol, 70 mM sucrose, 1 mM EDTA and 0.5 mM phenylmethylsulfonyl fluoride. Cell homogenates were centrifuged at 800 x g at 4°C for 10 min to obtain a post-nuclear supernatant and then mitochondria were pelleted by centrifugation at 10,000 x g at 4°C for 20min.
Immunocytochemistry and confocal imaging
For immunostaining, cells were fixed with 4% PFA in PBS, permeabilized with 0.5% Triton X-100 in PBS, and blocked with 10% BSA in PBS supplemented with 0.5% Triton X-100. Indicated primary antibodies and corresponding secondary antibodies were serially added. To image mitochondria or lysosomes, cells were incubated for 5 min with MitoTracker® Red CMXROS or LysoTracker® Red DND-99 at 100 ng/ml. The fixed, immuno-labeled cells or live cells were imaged using an inverted confocal microscope (LSM510 Meta; Carl Zeiss, Inc.) with a 63x 1.4 NA oil differential interference contrast Plan Apo objective. Image contrast and brightness were adjusted with Volocity (PerkinElmer).
CCCP Washout Cell Imaging
HeLa cells were transiently transfected with mito-YFP (YFP targeted to the mitochondrial matrix) for 24h prior to imaging. Cells in CO2-independent media (Invitrogen), were incubated with TMRE (Tetramethylrhodamine ethyl ester perchlorate, Invitrogen) or CCCP (Carbonyl cyanide m-chlorophenyl hydrazone, Sigma) for the indicated times at concentrations of 10nM and 10μM, respectively. For washout, cells were washed in 1mL of CO2-independent media. Images were acquired on an UltraView LCI confocal microscope (PerkinElmer) at 37°C with a 63x/1.4-numerical aperture Apochrome objective.
Mitochondrial in vitro import and BN-PAGE
Mitochondrial imports were essentially carried out as previously described (Stojanovski et al., 2007). A master import tube containing freshly isolated mitochondria in import buffer (20 mM HEPES (pH 7.4), 250 mM sucrose, 80 mM potassium chloride, 5 mM magnesium acetate) was split in half and one half was supplemented with 5 mM ATP, 10 mM Na succinate and vehicle control (DMSO) to a final concentration of 0.1% (v/v). The other half was supplemented with 1 μM CCCP. PINK1 translation products were incubated with mitochondria at 24°C for various times as indicated in the figure legends. At each time point the equivalent of 50 μg mitochondria was removed from each import tube and placed on ice. For protease treatment samples were incubated on ice for 10 min with 50 μg/ml Proteinase K (Sigma), followed by the addition of 1 mM PMSF for a further 10 min on ice. For BN-PAGE, all mitochondrial pellets (50 μg protein) were resuspended in 50 μl 1% (w/v) digitonin (Wako), 50 mM NaCl, 10% (v/v) glycerol, 20 mM Bis-Tris pH 7.0). BN-PAGE antibody shift assays were performed as previously described (Johnston et al., 2002). Radiolabeled proteins were detected by phosphorimaging (STORM 840, Amersham Biosciences).
PINK1-V5/His complex immunocapture and crosslinking
HeLa cells stably expressing PINK1-V5/His were generated by retroviral infection using the pBMN-IRES-EGFP vector system (Allele Biotech). Details for the approach used to immunocapture PINK1-V5/His as well as crosslinking experiments are described in the Extended Experimental Procedures.
Supplementary Material
Highlights.
PINK1 forms a 700 kDa complex with TOM for rapid re-import & PINK1 pathway regulation
PINK1 targeted to peroxisomes or lysosomes can recruit Parkin to ectopic locations
Peroxisomal PINK1 activates Parkin mediated pexophagy
PINK1 requires membrane localization to stimulate its interaction with Parkin
Acknowledgments
We thank Drs. Koji Yamano and Derek Narendra for valuable suggestions, Dr. Mike Ryan for kindly providing the Tom22 antibody, Dr. Bart De Strooper for kindly sharing the PARL−/− MEFs and Dr. Chunxin Wang for the PINK1-V5/His HeLa cells. This work is supported by the National Institute of Neurological Disorders and Stroke intramural program.
Footnotes
Supplemental information includes Extended Experimental Procedures, seven figures and supplemental references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Martin F, Maarse AC, Bomer U, Muller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukanovic J, Rapaport D. Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim Biophys Acta. 2011;1808:971–980. doi: 10.1016/j.bbamem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Frazier AE, Chacinska A, Truscott KN, Guiard B, Pfanner N, Rehling P. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol Cell Biol. 2003;23:7818–7828. doi: 10.1128/MCB.23.21.7818-7828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Neupert W, Deponte M. Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J. 2011;30:3232–3241. doi: 10.1038/emboj.2011.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Hoogenraad J, Dougan DA, Truscott KN, Yano M, Mori M, Hoogenraad NJ, Ryan MT. Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J Biol Chem. 2002;277:42197–42204. doi: 10.1074/jbc.M205613200. [DOI] [PubMed] [Google Scholar]
- Kanamori T, Nishikawa S, Nakai M, Shin I, Schultz PG, Endo T. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc Natl Acad Sci USA. 1999;96:3634–3639. doi: 10.1073/pnas.96.7.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008a;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008b;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kunkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27:5464–5476. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- Liu W, Vives-Bauza C, Acin-Perez R, Yamamoto A, Tan Y, Li Y, Magrane J, Stavarache MA, Shaffer S, Chang S, et al. PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson’s disease. PLoS One. 2009;4:e4597. doi: 10.1371/journal.pone.0004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- Model K, Prinz T, Ruiz T, Radermacher M, Krimmer T, Kuhlbrandt W, Pfanner N, Meisinger C. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J Mol Biol. 2002;316:657–666. doi: 10.1006/jmbi.2001.5365. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Muller H, Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J Biol Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Complex I: inhibitors, inhibition and neurodegeneration. Exp Neurol. 2010;224:331–335. doi: 10.1016/j.expneurol.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K, Arai T, Sato S, Kubo S, Ohba Y, Mizuno Y, Hattori N. Parkin stabilizes PINK1 through direct interaction. Biochem Biophys Res Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Terreni L, Calabrese E, Calella AM, Forloni G, Mariani C. New mutation (R42P) of the parkin gene in the ubiquitinlike domain associated with parkinsonism. Neurology. 2001;56:463–466. doi: 10.1212/wnl.56.4.463. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Mills JR, Nguyen M, Lemaire-Ewing S, Baardsnes J, Sun KL, Malina A, Young JC, Jeyaraju DV, O’Connor-McCourt M, et al. Mitochondrion-dependent N-terminal processing of outer membrane Mcl-1 protein removes an essential Mule/Lasu1 protein-binding site. J Biol Chem. 2011;286:25098–25107. doi: 10.1074/jbc.M111.218321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. discussion 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Wang D, Chen L, Choo YS, Ma H, Tang C, Xia K, Jiang W, Ronai Z, Zhuang X, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Yen HF. Does N-terminal processing of Mcl-1 occur at mitochondrial outer membrane or matrix? J Biol Chem. 2011;286:le15. doi: 10.1074/jbc.L111.218321. author reply le16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin Mediates Proteasome-dependent Protein Degradation and Rupture of the Outer Mitochondrial Membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.