Abstract
Background & Aims
It is a challenge to develop direct-acting antiviral agents (DAAs) that target the NS3/4A protease of hepatitis C virus (HCV) because resistant variants develop. Ketoamide compounds, designed to mimic the natural protease substrate, have been developed as inhibitors. However, clinical trials have revealed rapid selection of resistant mutants, most of which are considered to be pre-existing variants.
Methods
We identified residues near the ketoamide-binding site in X-ray structures of the genotype 1a protease, co-crystallized with boceprevir or a telaprevir-like ligand, and then identified variants at these positions in 219 genotype 1 sequences from a public database. We used side-chain modeling to assess the potential effects of these variants on the interaction between ketoamide and the protease, and compared these results with the phenotypic effects on ketoamide resistance, RNA replication capacity, and infectious virus yields in a cell culture model of infection.
Results
Thirteen natural binding-site variants with potential for ketoamide resistance were identified at 10 residues in the protease, near the ketoamide binding site. Rotamer analysis of amino acid side-chain conformations indicated that 2 variants (R155K and D168G) could affect binding of telaprevir more than boceprevir. Measurements of antiviral susceptibility in cell culture studies were consistent with this observation. Four variants (Q41H, I132V, R155K, and D168G) caused low-to-moderate levels of ketoamide resistance; 3 of these were highly fit (Q41H, I132V, and R155K).
Conclusions
Using a comprehensive sequence and structure-based analysis, we showed how natural variation in the HCV protease NS3/4A sequences might affect susceptibility to first-generation DAAs. These findings increase our understanding of the molecular basis of ketoamide resistance among naturally existing viral variants.
Keywords: virology, genetic, drug resistance, treatment
Until recently, the standard of care (SOC) for patients with chronic hepatitis C virus (HCV) infection has consisted of a combination of pegylated interferon-α plus ribavirin (Peg-IFN/RBV), administered for 24- to 48-weeks depending on the HCV genotype1–3. The sustained virologic response (SVR) rate for this SOC has been only about 50% in patients infected with genotype 1 HCV, the most prevalent genotype in Europe and North America. The addition of a direct-acting antiviral agent (DAA) targeting the NS3/4A serine protease of HCV significantly improves the SVR rate, and two such drugs have recently been approved for clinical use in the United States. The ketoamide compounds boceprevir (SCH503034) and telaprevir (VX-950) were both designed to mimic the natural substrate of the protease4–6. Clinical trials in treatment-naïve genotype 1-infected patients have revealed significant improvements in the kinetic of the virologic response with the addition of a DAA to the prior SOC, leading to improved SVR rates of up to 74%7–9.
Unlike Peg-IFN/RBV, the selection of drug-resistant virus variants during treatment with protease inhibitors (PIs) is a major concern. According to recent calculations by Rong et al.10, most, if not all, potential drug-resistant viral variants pre-exist at low frequencies within the viral quasispecies population in untreated patients. The highly replicative nature of HCV infection, with approximately 1012 new virions produced each day in the typical infected individual11, coupled with the lack of proofreading activity in the RNA-dependent RNA polymerase, NS5B, results in the generation of every possible viral variant every day. Thus, each patient is infected with a viral quasispecies “cloud” comprised of genetically distinct but closely related viral genomes. In the absence of concomitant Peg-IFN/RBV therapy, drug-resistant viral variants are rapidly selected and may emerge at frequencies as high as 5–20% in the quasispecies of patients as early as the second day of treatment. Unless suppressed by concomitant PegIFN/RBV, these pre-existing resistant variants are likely to be selected with subsequent treatment failure10.
The NS3/4A protease plays an essential role in the HCV replication cycle by proteolytically processing non-structural proteins from the viral polyprotein downstream of the NS2–3 junction12. The protease domain of NS3, comprising the amino-terminal third of the protein contains a catalytic triad, H57, D81 and S139, and an “oxyanion hole” at G137. It acts in concert with its cofactor, NS4A, which intercalates into its structure and is required for full enzymatic activity and proper subcellular localization. The carboxy-terminal two-thirds of NS3 comprises a DExD-box RNA helicase domain that is essential for productive viral infection13. NS3 thus appears to be a critical component of the macromolecular viral RNA replicase that directs the synthesis of new viral RNAs. Genetic evidence indicates that NS3 has an additional distinct function in assembly of virus particles14, 15. Since viral RNA replication capacity and virus assembly are crucial determinants of viral fitness, mutations in NS3 that contribute to PI resistance can also profoundly influence virus fitness16. The probability of a resistant variant emerging from the quasispecies population during treatment with a DAA is determined not only by its degree of resistance, but also by its fitness. Many mutations associated with PI resistance negatively impact the replication of genotype 1a HCV RNA in cell culture, while some have additional effects on the production of infectious virus14. Compensatory second-site mutations may enhance the fitness of resistant viruses16, but current understanding of these is rudimentary.
Given the genetic diversity that exists among different HCV strains, it is possible that naturally occurring polymorphisms in the NS3/4A sequence could provide a priori resistance to DAAs, and thus negatively impact the success of future treatment regimens. Here, we have studied the variation in amino acid residues that neighbor ketoamide compounds in the ligand-binding site of the protease. We identified natural amino acid substitutions at these positions in NS3 among genotype 1a sequences deposited in a public database, and modeled their side-chain conformations to assess their potential impact on ketoamide binding. To corroborate these in silico predictions, we then introduced these amino acid substitutions into a cell culture-infectious genotype 1a virus (H77S.3)14 and determined their impact on both susceptibility to ketoamide PIs and replication fitness in a cell culture system.
MATERIALS AND METHODS
Details of the materials and methods can be found in the Supplementary Material.
In silico analysis
We used X-ray structures of the genotype 1a HCV NS3/4A protease from the Protein Databank RCSB PDB17 co-crystallized with boceprevir (PDB 2OC8) or a telaprevir-like ligand (TLL, PDB 2P59) to deduce sets of ketoamide-neighboring residues. We designated the P4 to P1 and P1’ groups for ligands and their corresponding specificity pockets within the ligand-binding site, S4 to S1 and S1’, according to the numbering scheme of Schechter and Berger18. We then analyzed 219 genotype 1a HCV NS3/4A sequences deposited in the European HCV database19, which contains sequences collected from around the world, to identify potential natural binding site variants (BSVs) at residues that neighbor the ketoamides within the structure of the protease. The side-chain conformations of these BSVs were modeled using IRECS20 (details in Supplementary Material).
Cell culture and reagents
Details of the cells and reagents used in this study are provided in Supplementary Material.
Plasmids
pH77S.3 and pH77S.3/GLuc2A are molecular clones of the genotype 1a H77 strain of HCV. Synthetic RNA transcribed from these plasmids replicates in transfected Huh7 cells and produces infectious virus14. pH77S.3/GLuc2A RNA also produces secreted Gaussia luciferase (GLuc) reporter protein. Amino acid substitutions in BSVs expected to impact ketoamide binding were created in these plasmids by site-directed mutagenesis14.
Virus fitness and antiviral resistance
Genome-length RNA was transcribed from the mutated pH77S.3 and pH77S.3/GLuc2A plasmids in vitro, and studies to assess antiviral resistance and viral fitness carried out as described previously14.
RESULTS
To identify amino acid residues in NS3 that are in close proximity to ketomides in the ligand-binding site of the protease, we analyzed PDB structure 2OC8 in which boceprevir is co-crystallized with the NS3/4A protease21. Since no similar co-crystallized structure is publically available for telaprevir, we used PDB structure 2P59 in which the protease is co-crystallized with a telaprevir-like ligand (TLL) that has two small differences from telaprevir22. Its P2 group and the P4 capping group are slightly modified, with the P4 providing a pyrrole NH for H-bond interactions with the protease (Supplementary Fig. S1)23. We identified 20 residues interacting with or neighboring boceprevir and TLL in these X-ray structures (Table 1 and Supplementary Materials and Methods), and analyzed genotype 1a HCV sequences in the European HCV database to identify differences from the consensus sequence at these residues. The residues were relatively well conserved in 219 genotype 1a sequences from diverse geographic regions. However, we identified 13 different binding site variants (BSVs) involving 10 ketoamide-neighboring residues (Supplementary Table S1 and Fig. S2), 8 of which have not been described previously as PI resistance-associated variants. None of the patients from which these sequences were derived appear to have been treated previously with a DAA (see Supplementary Table S3). Most of these naturally occurring BSVs were single amino acid substitutions without changes in other ketoamide-neighboring residues. However, T42A and K136R were both present within a single sequence (EU677251). Two different BSVs were identified as substitutions at T42 (A/S), V55 (A/I) and D168 (G/E), while single BSVs were identified at the other 7 residues (Supplementary Table S1 and Fig. S2). We examined the first-neighbor residues of these BSVs in an effort to identify potential second-site variants in the BSV strains (see Supplementary Material and Methods, Fig. S3 and Table S2). While there were no additional substitutions at these first-neighbor residues in most BSVs, T54S was found in both V55I strains (EF407443 and EU781818) and I170V was found in a R155K strain (EU781805).
Table 1.
Ketoamide-neighboring residues in NS31.
| Boceprevir | Telaprevir-like ligand (TLL) | ||
|---|---|---|---|
| RCSB PDB | UniProtKB | RCSB PDB | UniProtKB |
| 2OC8 (Chain B) | P27958 | 2P59 (Chain B) | P27958 |
| Q41 | Q41 | Q1067 | Q41 |
| T42 | T422 | T1068 | T42 |
| F43 | F43 | F1069 | F43 |
| V55 | V55 | - | - |
| H57 | H57 | H1083 | H57 |
| D81 | D81 | D1107 | D81 |
| R123 | R123 | R1149 | R123 |
| I132 | I132 | I1158 | I132 |
| L135 | L135 | L1161 | L135 |
| K136 | K136 | K1162 | K136 |
| G137 | G137 | G1163 | G137 |
| S138 | S138 | S1164 | S138 |
| S139 | S139 | S1165 | S139 |
| F154 | F154 | F1180 | F154 |
| R155 | R155 | R1181 | R155 |
| A156 | A156 | A1182 | A156 |
| A157 | A157 | A1183 | A157 |
| V158 | V158 | V1184 | V158 |
| C159 | C159 | C1185 | C159 |
| - | - | T1186 | T160 |
| D168 | D168 | D1194 | D168 |
Residues are numbered relative to the PDB residue sequence and H77c reference sequences (UniProtKB P27958).
Residues shown in italics do not directly interact with boceprevir or TLL, but are within 5.0 Å distance of the ligand (see Supplementary Materials and Methods).
Rotamer analysis of BSVs and expected impact on ketoamide binding
We modeled the energetically most favorable side-chain conformations for the genotype 1a consensus and BSV sequences (Fig. 1). Since V55 is buried in the protease domain24, it was not amenable to rotamer analysis. Molecular dynamics simulation of variants at this position will be reported separately (Welsch et al., in revision).
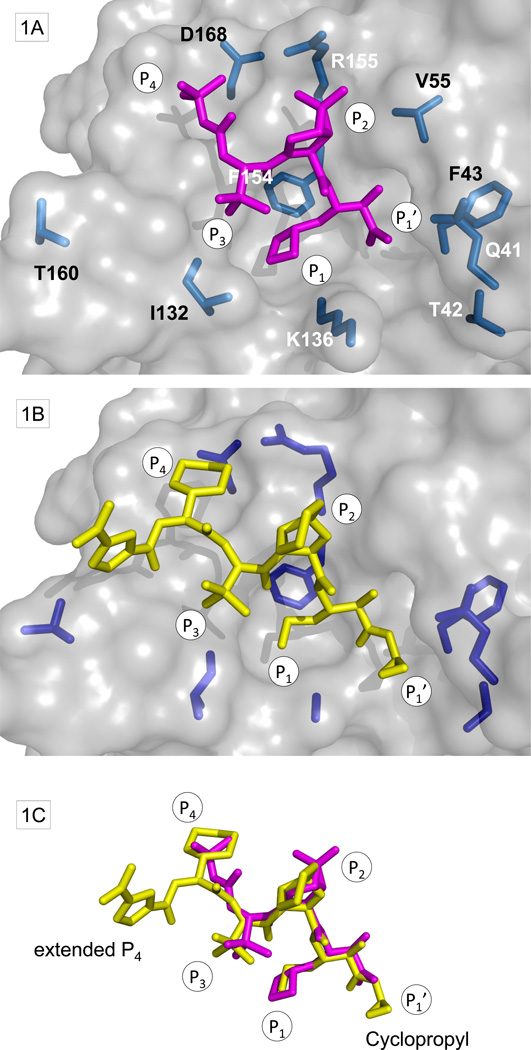
Figure 1. Ligand-binding site of NS3/4A with ketoamides and BSVs.
(A) Surface representation from PDB structure 2OC8 with boceprevir shown in magenta. Side-chains at amino acid residues with BSVs are shown in stick format (light blue). (B) Similar representation of TLL within the PDB structure 2P59 with side-chains at amino acid residues with BSVs (dark blue). (C) Superposition of boceprevir and TLL.
Expected impact of Q41H and K136R at the ketoamide P1’ position
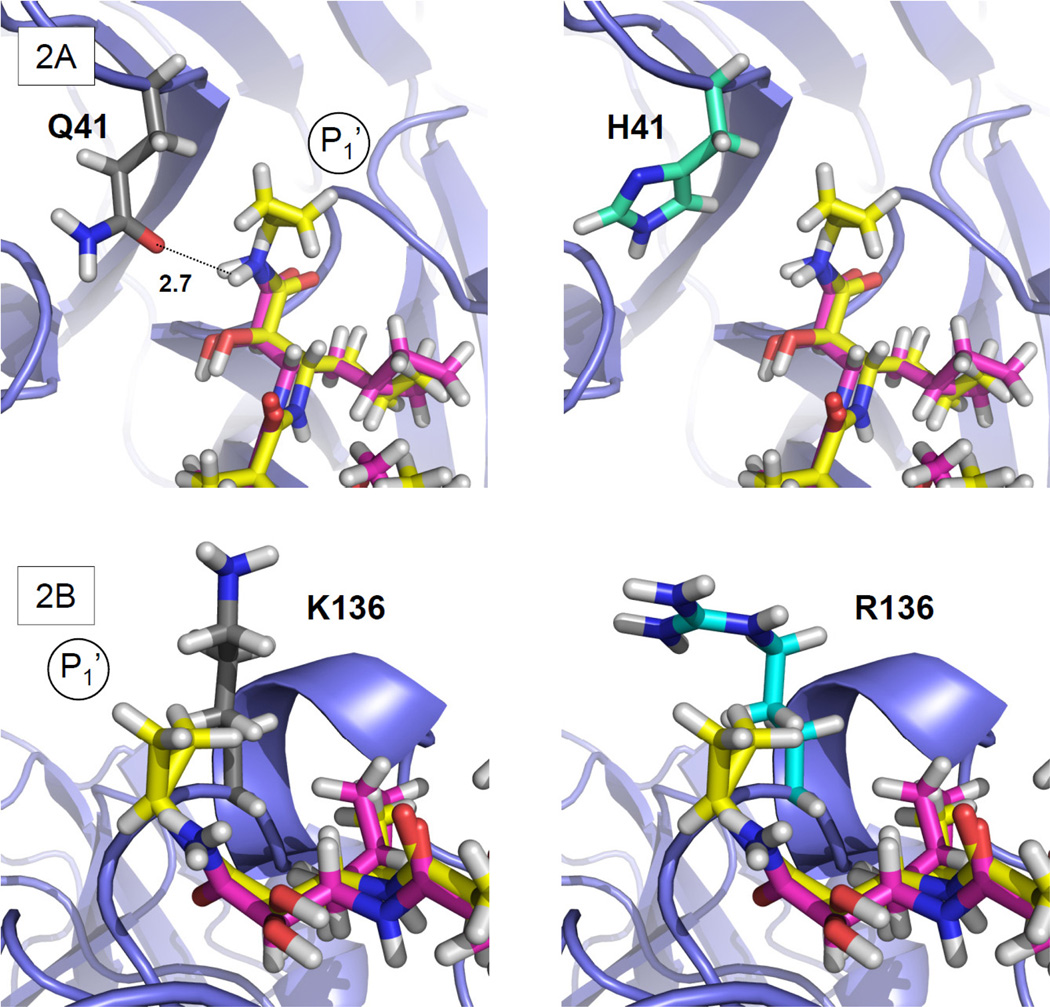
Q41 is located adjacent to S1’. The H41 variant represents a non-conservative change from the consensus sequence involving substitution of an uncharged polar side chain with a charged aromatic residue. Telaprevir possesses a cyclopropyl group at P1’ that is oriented away from the Q41 side chain without noncovalent interactions. The conformation of Q41 suggests a potential H-bond interaction of its carboxamide chain, including the H-bond donor NH2 and acceptor C=O, with the backbone of boceprevir and TLL (OE1—HN distance for Q41-ketoamide: 2.7 Å). However, the H41 BSV has no side-chain group allowing an H-bond interaction with the ligand (Fig. 2A). This suggests H41 will be associated with a minor decrease in binding affinity for boceprevir and telaprevir.
Figure 2. Rotamer analysis of Q41H and K136R.
Detail of PDB structure 2OC8 showing boceprevir (magenta) and the superposed TLL (yellow). The panels on the left show the side-chain conformation of the consensus residue, while those on the right show the predicted BSV conformation. (A) (left) The Q41 side-chain OH-group with predicted H-bond interaction (dotted line) with ketoamide backbone Cα, (right) this H-bond interaction is absent in H41. (B) Predicted R136 side-chain conformation and the TLL P1’ cyclopropyl showing closer van der Waals contacts in (right) R136 than in (left) the consensus K136.
In contrast, K136R is a conservative change between polar residues, both with long, flexible and charged side chains at S1’. The K136 side chain is not defined in the electron density map of PDB structure 2P59. As modeled, it has close contacts with the TLL P1’ cyclopropyl group (Fig. 2B). Boceprevir possesses no comparable P1’ group (Supplementary Fig. S1). We predict the R136 side-chain conformation is similar to the wild-type K136, but that it will wrap around the TLL cyclopropyl group providing an increasing number of van der Waals contacts (Fig. 2B). This suggests that the R136 variant may have stronger effects on telaprevir than on boceprevir binding. We expect hydrophilicity to play a role in ketoamide binding at S1’, and the R136 side chain possesses more polar contacts and potentially binds more water molecules than the wild-type K136. Thus, the release of water molecules bound to the R136 side chain could increase systemic entropy and potentially add to the free binding energy and affinity for telaprevir, although decreased enthalpy may compensate for the effects on entropy. Since the R136 side chain does not have a tight H-bond or salt bridge interaction at its end, electrostatic attraction between the nonpolar P1’ cyclopropyl group of telaprevir and the polar R136 side chain is unlikely. Overall, however, the impact of K136R on binding of telaprevir is not readily predicted by rotamer analysis alone.
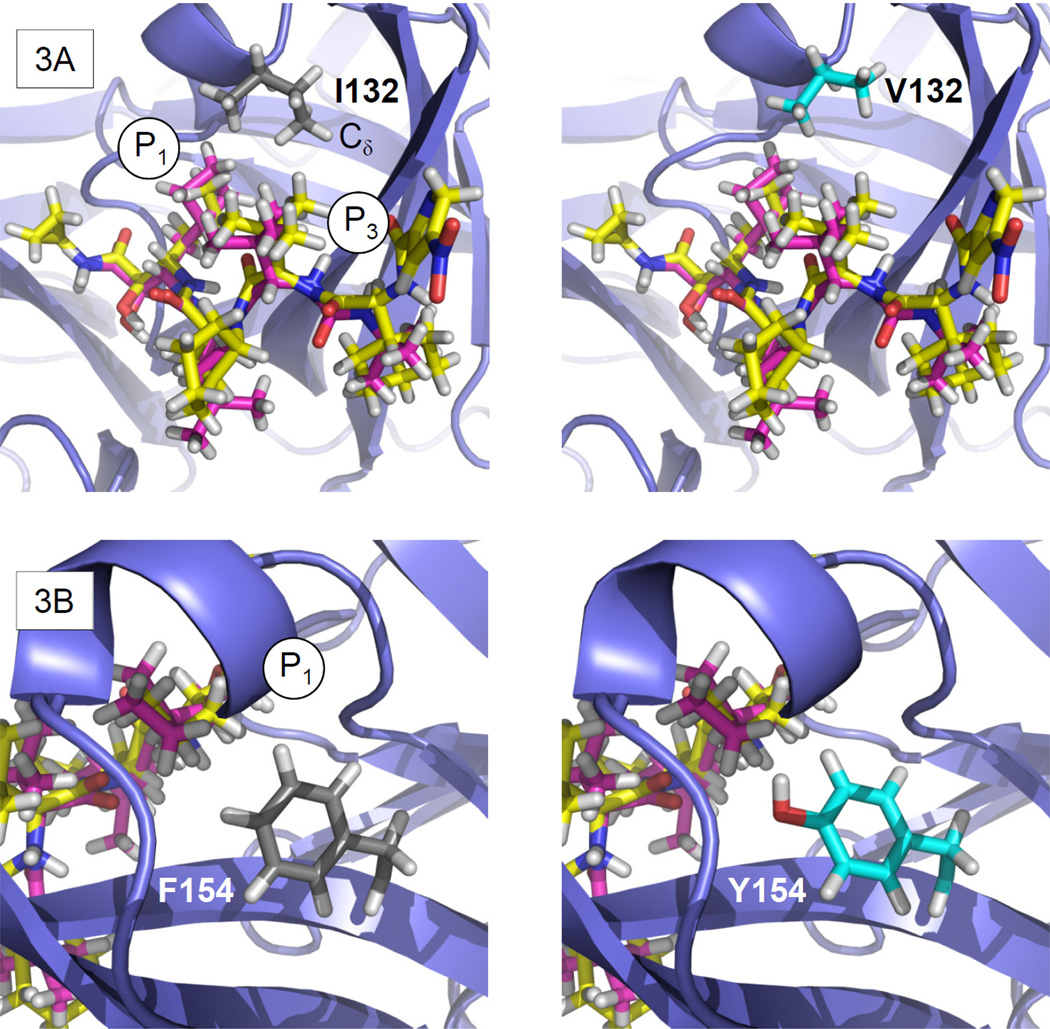
Expected impact of I132V and F154Y on ketoamide P1 and P3 interactions
I132 is located adjacent to S1 and S3, and its substitution with valine in V132 represents a conservative change between hydrophobic residues. The consensus I132 side-chain conformation forms a hydrophobic S1/3 interface with the ketoamide P1 and P3 groups. The I132 side-chain Cδ carbon points towards the ketoamide P3 group (Fig. 3A). The variant V132 shows a similar side-chain conformation but without a Cδ carbon. This suggests a loss of van der Waals contacts for V132, resulting in a minor reduction in binding affinity for boceprevir and telaprevir. F154Y is also a relatively conservative change, in this case from nonpolar towards polar among aromatic residues. This residue is close to the ketoamide P1 and P3 groups at the bottom of the S1 pocket. The wild-type F154 and the predicted variant Y154 side chains are oriented towards the ketoamide P1 group, potentially influencing boceprevir and telaprevir binding (Fig. 3B). The Y154 side-chain has an OH group that is not present in the wild-type F154. Ketoamides do not offer an opportunity for H-bond interactions at P1, but this OH group may provide for alternative binding of a water molecule. The polar, hydrophilic nature of the Y154 side chain reduces the hydrophobic properties of the S1 pocket, and this variant is expected to cause a significant reduction in binding affinity for boceprevir and telaprevir.
Figure 3. Rotamer analysis of I132V and F154T.
(A) (left) The I132 side chain provides a hydrophobic S1/3 interface for the ketoamide P1 and P3 moieties. The I132 Cδ carbon is oriented towards the ketoamide P3, while (right) V132 lacks a Cδ carbon. (B) (left) F154 neighbors S1 underneath the ketoamide, while (right) Y154 is predicted to assume a similar side-chain conformation but with an additional OH-group.
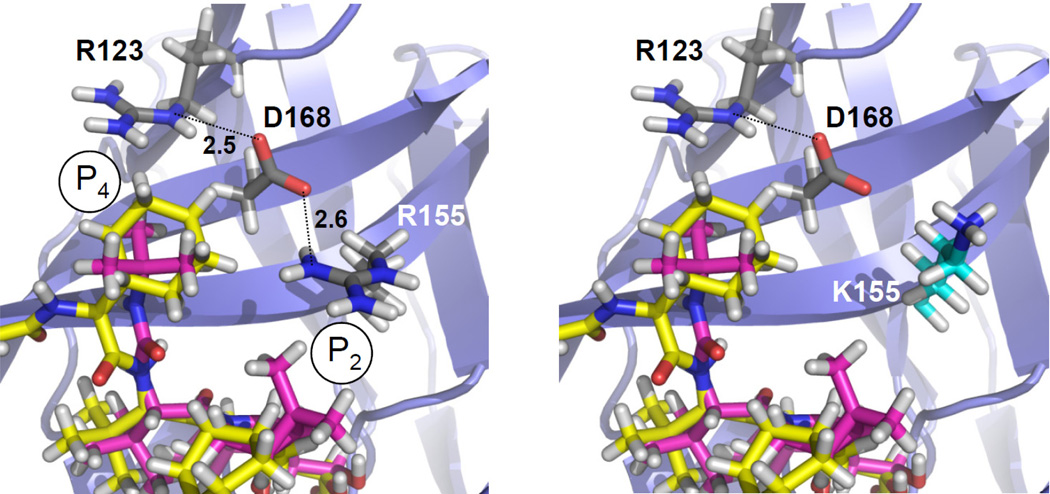
Expected impact of R155K, D168E and D168G on ketoamide P2 and P4 interactions
R155K is a conservative change between positively-charged residues within the S2 pocket The R155 side chain is predicted to participate in a pattern of noncovalent interactions involving its neighboring residues, R123 and D168, at S4 (the O-H distance for D168-R155 is 2.6 Å; for R123-D168, it is 2.5 Å) (Fig. 4). The combination of H-bond and electrostatic interactions is predicted to result in a particularly strong noncovalent salt ‘bridge’ interaction. Polar interactions between the D168, R123 and R155 side chains contribute to the strength of this noncovalent interaction network. The D168 side chain is predicted to be tightly fixed, allowing no other H-bond interaction and contributing to a nonpolar S2/4 interaction interface for the P2 and P4 groups in boceprevir and telaprevir. The variant K155 disrupts this nonpolar S2/4 interface by leaving the negatively-charged D168 unbound, and is expected to reduce binding affinity for boceprevir and telaprevir. A stronger effect is expected for telaprevir than for boceprevir, since the telaprevir P2 cyclopental-proline is larger than the respective boceprevir isopropyl-proline (Supplementary Fig. S1). Compared with R155, the K155 side-chain is also predicted to be shifted slightly away from the ketoamide P2 group. This shift is likely to cause a loss in van der Waals contacts with the ketoamide P2 group, and may reduce binding affinity for telaprevir. The effects on polarity and loss of van der Waals contacts suggest that there will be a significant decrease in binding affinity for both ketoamides, but a larger impact on telaprevir.
Figure 4. Rotamer analysis of R155K.
(left) The H-bond pattern (dotted lines) in the R155 structure with D168 serving as a nonpolar S4 contact interface for the ketoamide P4. (right) K155 disables this H-bond pattern, leaving D168 unbound with a polar OH-group at S4. The K155 side-chain conformation is also predicted to be shifted slightly away from the ketoamide P2.
Two variants were observed at the D168 position that contributes to the S4 pocket in the protease. D168G is a non-conservative change from aliphatic and polar to a smaller, nonpolar side chain. The wild-type D168 side-chain conformation is oriented towards the ketoamide P4 group, which is smaller in boceprevir than telaprevir. The extended P4 group in telaprevir (Fig. 1) points away from D168. Thus, the D168 variant is expected to have only minor effects on ketoamide binding. G168 is expected to have effects on the nonpolar S2/4 interface similar to K155, since it offers no polar side-chain interaction partner for R155. It is predicted to change the polarity of the S2/4 interface and expected to cause a minor decrease in binding affinity for both ketoamides. D168E is a conservative change as both side-chains are negatively charged and aliphatic. The side-chain conformation is predicted to be similar to the wild-type D168, and to preserve S2/4 interface polarity. D168E is not expected to impact ketoamide binding.
Additional BSVs without expected impact on ketoamide binding
T42A is located at the periphery of the S1’ pocket, and is a non-conservative change from aliphatic, polar and negative towards aliphatic and hydrophobic. A side-chain OH group is lost in the A42 variant, but preserved in the S42 variant at this position, allowing for a potential H-bond interaction. The wild-type T42 and variant A42 and S42 side-chain conformations are predicted to be similar. No ketoamide H-bond donor or acceptor group is found in close proximity to T42 or S42. Due to its relative distance from the ketoamide, the polarity change in T42A is predicted not to have a significant effect on ketoamide binding. Thus, the T42A/S variants are expected to have no impact on ketoamide binding. F43S is a non-conservative change from aromatic and nonpolar towards aliphatic and polar, and is found at the bottom of S1’. No change is predicted in the side-chain conformation. There is an additional OH group in the variant, but since there is no ketoamide H-bond donor or acceptor group in close proximity, this by itself is unlikely to have any impact on ketoamide binding. Nevertheless, F43S may cause ketoamide resistance since the F43 aromatic ring directly participates in the formation of the S1’ pocket, which impacts binding of the ketoamide P1’ group25. Substitutions at F43 have been shown previously to cause resistance to ketoamide compounds25. T160A is a non-conservative change from polar towards nonpolar and hydrophobic, located at a distance from S4 near the boundary of an extended S5 pocket. T160 interacts directly with the extended P4 group in TLL in the PDB structure 2P59, but has no noncovalent interactions with boceprevir in PDB structure 2OC8. The variant A160 side-chain is not expected to influence ketoamide binding.
Replication capacity and infectious virus yield from RNAs containing BSVs
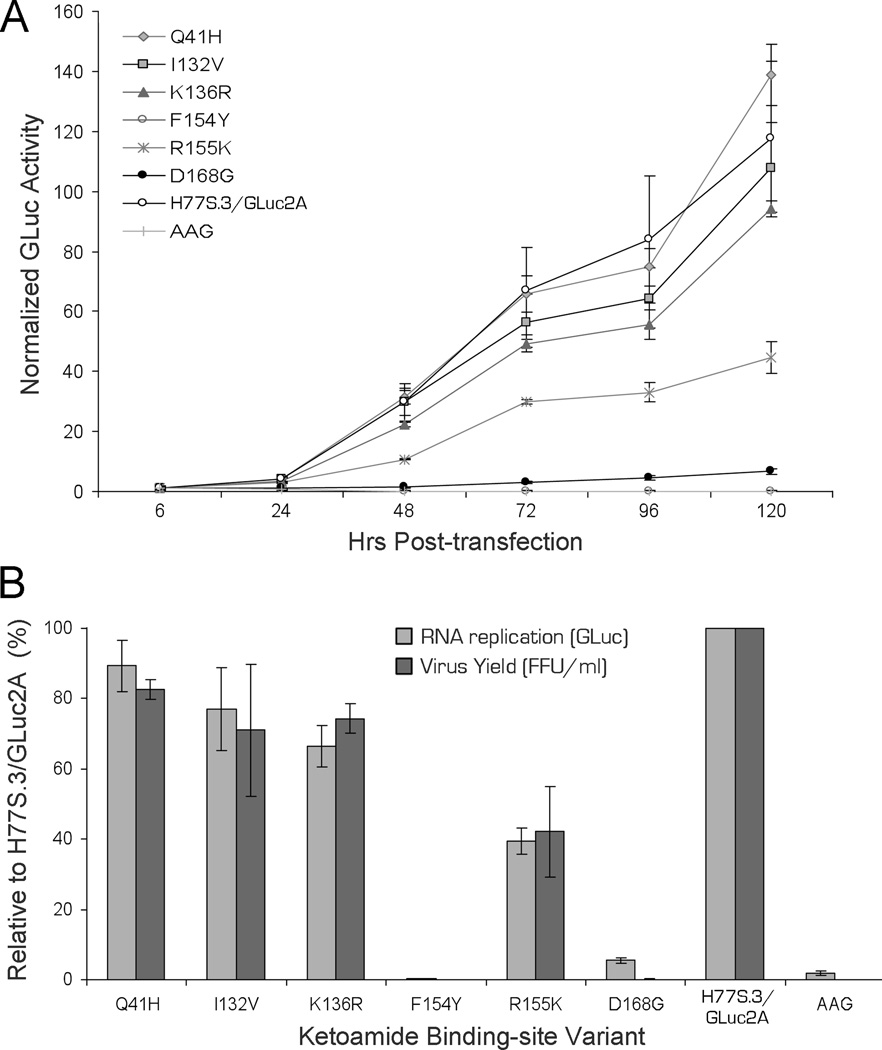
Those BSVs for which the rotamer analysis suggested a possible impact on ketoamide binding (Q41H, I132V, F154Y, R155K and D168G) were selected for phenotypic characterization. We similarly tested K136R, for which the rotamer analysis provided no clear predictions. The amino acid substitutions were created within the background of the genotype 1a H77S.3 genome, and their impact on replication of the viral RNA and production of infectious virus determined in RNA-transfected cells.
(1) RNA replication capacity
The replication capacity of H77S.3/GLuc2A RNA mutants with BSV substitutions in NS3 was assessed by measuring GLuc activities in supernatant media collected at 24h intervals following transfection of synthetic RNA, as described previously14. Results were normalized to the activity present at 6h after transfection, as this represents GLuc expressed directly by the transfected input RNA. The RNA replication capacity of the Q41H variant was similar to the parental H77S.3/GLuc2A RNA, while I132V and K136R were minimally impaired (Fig. 5A). In contrast, as shown previously14, the replication of R155K was moderately impaired while D168G was severely handicapped for RNA replication. The maximum RNA replication capacity observed was with Q41H (89% of parental RNA) while the lowest was with D168G (5.5%). The F154Y substitution had a lethal effect on RNA replication in the H77S.3 background, suggesting that viruses containing this sequence variant (and possibly also G168) possess one or more compensating changes at other amino acid positions (see Discussion). None of the BSVs demonstrated enhanced RNA replication capacity compared to the parental H77S.3/GLuc2A RNA (Fig. 5B).
Figure 5. Replication capacity and infectious virus yield of H77S.3 RNAs with BSV substitutions in NS3.
(A) RNA replication capacity reflected of H77S.3/GLuc2A BSV into Huh7.5 cells. Results are normalized to the 6h GLuc activity, and represent the mean of triplicate samples. (B) Comparison of RNA replication capacity (lightly shaded bars) and infectious virus yields (dark shaded bars). Both are normalized to those obtained with the relevant parental RNA (100%). Data represent the mean ± SD from at least 3 independent experiments
(2) Infectious virus yield
We also assessed the impact of these BSVs when placed within the background of H77S.3 RNA, which lacks the GLuc2A-coding sequence and produces infectious virus as described previously14. Cell culture supernatant fluids were collected 72h after transfection of the RNA, and subsequently inoculated onto naïve cells, with foci of infected cells detected by immunofluorescence 72h later. Each of the BSVs tested produced infectious virus yields in the range of that expected from their RNA replication capacity, although the low replication capacity of the D168G variant precluded a careful analysis on production of infectious virus (Fig. 5). Thus, none of these amino acid substitutions were documented to directly impair infectious virus assembly or release, as described previously for some resistance-associated NS3 variants14. Reductions in the fitness of these particular BSVs are confined primarily to defects in viral RNA replication.
Ketoamide resistance of BSVs
We measured the antiviral activities (EC50) of boceprevir and telaprevir against each of the H77S.3/GLuc2A mutants by determining the concentration of each compound required to cause a 50% reduction in RNA replication (GLuc expression by RNA-transfected cells). Resistance testing could not be performed for the F154Y variant because it was not competent for replication.
Boceprevir demonstrated antiviral activity against each of the BSVs (Table 2). The EC50 value against the parental H77S.3/GLuc2A was 870 ± 48 nM. The maximum fold-change in the EC50 for any of the BSVs was 2.1 for R155K14. Only R155K showed significant, albeit low-level resistance against boceprevir. Telaprevir showed greater molar activity than boceprevir, with an EC50 against the H77S.3/GLuc2A of 120 ± 10 nM. As with boceprevir, telaprevir was active against each of the BSVs, but low- to medium-level resistance was evident with Q41H, I132V, R155K and D168G (Table 2). The maximum fold-change in the EC50 was 8.8 for R155K. As expected from the in silico analysis, the range of fold-changes in EC50 was broader for telaprevir than boceprevir. In general, these changes were in good agreement with the impact of these BSVs on ketoamide binding predicted from the rotomer analysis, except for K136R which was difficult to predict and showed greater antiviral activity than anticipated against both ketoamide compounds (Table 2).
Table 2.
Predicted and measured impact of BSVs on antiviral activity of ketoamides1.
| Genotype 1a HCV |
IRECS | EC50 [nM] | Fold-change in EC50 | |||
|---|---|---|---|---|---|---|
| Boceprevir | TLL | Boceprevir | Telaprevir | Boceprevir | Telaprevir | |
| H77S.3/GLuc2A | - | - | 870 (±48) | 120 (±10) | 1.0 | 1.0 |
| Q41H | MiN2 | MiN | 1040 (±60) | 410 (±10) | 1.2 | 3.5 |
| T42A | NoI | NoI | - | - | - | - |
| T42S | NoI | NoI | - | - | - | - |
| F43S | NoI | NoI | - | - | - | - |
| I132V | MiN | MiN | 980 (±60) | 280 (±40) | 1.1 | 2.4 |
| K136R | NoI | NP | 440 (±19) | 100 (±10) | 0.5 | 0.9 |
| F154Y | SN | SN | ND3 | ND | ND | ND |
| R155K | MeN | SN | 1830 (±240) | 1010 (±240) | 2.1 | 8.8 |
| T160A | NoI | NoI | - | - | - | - |
| D168G | MiN | MeN | 490 (±50) | 260 (±30) | 0.6 | 2.2 |
| D168E | NoI | NoI | - | - | - | - |
The table compares the in silico predictions of the impact of BSVs on ketoamide binding from the IRECS20 analysis, and corresponding EC50 values determined from H77S.3/GLuc2A-transfected cell cultures. Results shown represent the mean ± SD and fold changes (FC) compared to wild-type.
Predicted impact: NoI, no impact; MiN, minor negative impact, change only in a single ketoamide-BSV interaction (H-bond, van der Waals) or in polarity; MeN, moderate negative impact, combined changes in ketoamide-BSV interaction and polarity; SN, strong negative impact, same as for MeN but at particularly close ketoamide-BSV interaction sites; NP, not predictable;
ND, not determined.
DISCUSSION
Mathematical arguments suggest that every possible drug-resistant viral variant is likely to pre-exist at a low frequency in the replicating viral quasispecies population of the typical HCV-infected patient10. Whether this is actually the case, and at what frequency such variants actually exist, may never be formally demonstrated due to technical difficulties. In this study, we analyzed the natural variation present among ketoamide-neighboring residues in 219 genotype 1a HCV sequences collected from geographically diverse sites and deposited in a public database. We cannot exclude the possibility that some of the BSVs we identified in this set of sequences may represent variants that were present at low frequency in their source patient, or even unrecognized sequencing errors. However, it is likely that they represent true variants present within the dominant quasispecies of the patients from which these sequences were derived, since multiple BSVs were identified at some residues (T42, V55, and D168) (Supplementary Fig. S2), while others (H41, A42, A55, I44, and K155) were present in more than one sequence. We identified BSVs at one or more ketoamide-neighboring residues in 17 of 219 (7.8%) genotype 1a sequences. Importantly, 8 of these variants (Q41H, T42A, T42S, V55I, I132V, K136R, F154Y, and T160A) have not been identified, to our knowledge, in previous in vivo or in vitro studies of ketoamide resistance.
Although they are both linear ketoamide compounds, boceprevir and telaprevir have distinct structural features (Supplementary Fig. S1). Telaprevir possesses an extended P4 capping group and a P1’ cyclopropyl group. The P2 group is different from the isopropyl-proline in boceprevir, which is smaller than the telaprevir P2 cyclopental-proline23. These structural differences likely contribute to the lower EC50 of telaprevir against genotype 1a HCV in cell culture (Table 2), but could also pose a higher risk for telaprevir resistance among BSVs. Rotamer analysis predicted that two BSVs (R155K and D168G) would exert a greater negative effect on the binding of telaprevir than boceprevir. The impact of the K136R substitution proved difficult to predict on the basis of rotamer analysis alone, however, and subsequent tests in cell culture demonstrated that it imposes no resistance against either ketoamide (Table 2).
On the other hand, the R155K and D168G substitutions led to a 2- to 4-fold greater increase in the EC50 of teleprevir compared with boceprevir, and almost a 9-fold increase in the teleprevir EC50. This was consistent with predictions from the rotamer analysis, which also agreed with previous crystallographic studies26. Whether such changes in the EC50 result in clinical resistance would be dependent upon the potency of a drug and the drug exposure achieved in a typical patient. A 2-fold change might not be clinically relevant, but a 9-fold increase such as that found with R155K is likely to be significant. Importantly, R155K has been associated previously with resistance to both ketoamide and macrocyclic compounds23, 26.
Three BSVs (Q41H, I132V and F154Y) were predicted to interact with ketoamide structural features common to both telaprevir and boceprevir, and thus to influence the binding of these compounds equally. Although Q41H was expected to cause only a minor decrease in affinity for both ketoamides, in vitro assays revealed a 3.5-fold increase in the EC50 for teleprevir versus 1.2 for boceprevir. Its greater impact on telaprevir potency is likely related to its P1’ cyclopropyl group and induced structural changes in the corresponding S1’ pocket. Such changes were not detectable using rotamer analysis, but nonetheless probably influenced the binding of telaprevir. A similar difference in the magnitude of the change in EC50 was observed for the I132V variant (1.1 fold-change for boceprevir versus 2.4 for teleprevir). This could result from a loss of van der Waals contacts with the ketoamide P3 group, which differs slightly in its orientation in the co-crystallized boceprevir and TLLstructures (see Fig. 1). The F154Y substitution was lethal for RNA replication when placed in the background of the H77S.3 virus, and thus we could not measure antiviral activity against it.
Viral fitness coupled with the degree of resistance conferred by a BSV are likely to be the major determinants driving selection of a variant from within the viral quasispecies during therapy. RNA replication capacity is one measure of the fitness of the virus, and this is dependent on proper processing of the polyprotein by NS3/4A. Ketoamides mimic the natural substrate of the protease at the site of NS3-NS4A scission, and it is likely that the negative influence of BSVs on RNA replication (Fig. 5A) reflects altered recognition of the polyprotein substrate related to structural changes similar to those leading to drug resistance. Despite this, there is no strong correlation between the degree of PI resistance and the impact on RNA replication14. This is reflected in the marked deficit in replication demonstrated by the D168G substitution (Fig. 1A), which confers only a minimal increase in the teleprevir EC50 (Table 2). There is no obvious structural or molecular explanation for this difference, and it is not possible to exclude the possibility that G168 might provide for more robust RNA replication when placed in the context of a different virus sequence.
In contrast F154Y was lethal for replication in the context of the H77S.3 virus. This substitution occurs at a central position within the ligand-binding site at the bottom of the S1 pocket. A recent crystallographic study found the F154 aromatic ring to directly contact the substrate P1 side-chain27. Thus, the complete loss of RNA replication observed with F154Y could be due to either intrusion of the Y154 side-chain into the space normally occupied by the polyprotein substrate, or a polarity shift within S1 due to the Y154 OH group. Nonetheless, the presence of Y154 in the database suggests that it is capable of functioning in an alternative sequence context. This highlights a limitation of the technical approach we have taken here to study the phenotypic effect of BSVs, as second-site substitutions in the same strain might compensate for defects fitness. In fact, the sequence of the F154Y BSV (EU677253) contains two additional substitutions in NS3 that differ from the genotype 1a consensus: R11G and H110R (Supplementary Fig. S2). H110R is in close structural proximity to F154Y and the natural substrate of the protease (Supplementary Fig. S3). While it might be preferable in investigating BSVs to engineer swaps of the complete NS3 sequence into the background of a replication-competent clone, it is not clear how often this approach would fail due to sequence incompatibilities between the “donor” and “recipient” viruses. NS3 appears to interact with several other viral proteins28.
Our findings provide a molecular basis for ketoamide resistance among BSVs that exist naturally as dominant quasispecies in some patients prior to treatment with DAAs (Supplementary Table S3). Such natural variants may be of limited clinical significance at present as they are likely to be suppressed by Peg-IFN/RBV in current SOC regimens, but they can be expected to be of substantial importance to the outcome of future interferon-sparing, all-DAA combination therapies. These variants might also affect future generations of inhibitors depending upon their chemical structures. Knowledge on the natural variability in structures targeted by antivirals, as presented in this study, may help guide the development of future generation PIs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ann D. Kwong and Govinda Rao for their critical reading of the manuscript.
Grant Support: This study was supported by a DFG grant to CW, TL, and SZ (Clinical Research Unit, KFO129, TP3 and TP6, LE 491/16-2 and LE 491/17-2) and a DFG Research Fellowship (WE 4388/3-1) to CW. SML was supported by the National Institutes of Health (NO1-AI25488, RO1-AI095690).
Abbreviations
- BSV
binding-site variant
- DAA
direct-acting antiviral agent
- EC
effective concentration
- FFU
single infectious focus-forming unit
- GLuc
Gaussia Luciferase
- HCV
hepatitis C virus
- NS
non structural protein
- PDB
Protein Databank
- PI
protease inhibitor
- RNA
ribonucleic acid
- SOC
standard-of-care
- SVR
sustained virological response
- TLL
telaprevir-like ligand
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: SZ has served as a consultant for Abbott, Achillion, Anadys, BMS, Boehringer, Gilead, iTherX, Janssen, Merck, Novartis, Pfizer, Pharmasset, Roche, Santaris, Tibotec, and Vertex. SML has served as a consultant for Abbott, Hoffmann-LaRoche, Juvaris Biotherapeutics, Merck, Novartis, and Pfizer; research in his laboratory is supported by Merck and Tibotec. The remaining authors disclose no conflicts.
Author contributions:
CW: obtain funding, study concept and design, acquisition, analysis and interpretation of data, writing and drafting of manuscript
TS, CH, YY: acquisition, analysis and interpretation of data
FSD: technical support, critical review of the manuscript
TL: obtain funding, critical review of manuscript
SZ, SML: obtain funding, study concept and design, interpretation of data, critical review of manuscript
References
- 1.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 2.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 3.Shiffman ML, Suter F, Bacon BR, et al. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124–134. doi: 10.1056/NEJMoa066403. [DOI] [PubMed] [Google Scholar]
- 4.Malcolm BA, Liu R, Lahser F, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006;50:1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prongay AJ, Guo Z, Yao N, et al. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]-- 6,6-dimethyl-3- azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization. J Med Chem. 2007;50:2310–2318. doi: 10.1021/jm060173k. [DOI] [PubMed] [Google Scholar]
- 6.Perni RB, Almquist SJ, Byrn RA, et al. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2006;50:899–909. doi: 10.1128/AAC.50.3.899-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hezode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360:1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 8.Kwo PY, Lawitz EJ, McCone J, et al. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010 doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- 9.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 10.Rong L, Dahari H, Ribeiro RM, et al. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2:30ra32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 12.Raney KD, Sharma SD, Moustafa IM, et al. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolykhalov AA, Mihalik K, Feinstone SM, et al. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3' nontranslated region are essential for virus replication in vivo. Journal of Virology. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimakami T, Welsch C, Yamane D, et al. Protease Inhibitor-Resistant Hepatitis C Virus Mutants With Reduced Fitness From Impaired Production of Infectious Virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Yates J, Liang Y, et al. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi M, Tong X, Skelton A, et al. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor: Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol.Chem. 2006;281:8205–8215. doi: 10.1074/jbc.M510246200. [DOI] [PubMed] [Google Scholar]
- 17.Kouranov A, Xie L, de la Cruz J, et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 19.Combet C, Garnier N, Charavay C, et al. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 2007;35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmann C, Antes I, Lengauer T. IRECS: a new algorithm for the selection of most probable ensembles of side-chain conformations in protein models. Protein Sci. 2007;16:1294–1307. doi: 10.1110/ps.062658307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prongay AJ, Guo Z, Yao N, et al. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1- dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3- azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization. J Med Chem. 2007;50:2310–2318. doi: 10.1021/jm060173k. [DOI] [PubMed] [Google Scholar]
- 22.Perni RB, Chandorkar G, Cottrell KM, et al. Inhibitors of hepatitis C virus NS3.4A protease. Effect of P4 capping groups on inhibitory potency and pharmacokinetics. Bioorg Med Chem Lett. 2007;17:3406–3411. doi: 10.1016/j.bmcl.2007.03.090. [DOI] [PubMed] [Google Scholar]
- 23.Tong X, Bogen S, Chase R, et al. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 2008;77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Susser S, Welsch C, Wang Y, et al. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 25.Welsch C, Domingues FS, Susser S, et al. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of the hepatitis C virus. Genome Biol. 2008;9:R16. doi: 10.1186/gb-2008-9-1-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Muh U, Hanzelka BL, et al. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J Biol Chem. 2007;282:22619–22628. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 27.Romano KP, Ali A, Royer WE, et al. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc Natl Acad Sci U S A. 2010;107:20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Anantpadma M, Timpe JM, et al. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J Virol. 2011;85:86–97. doi: 10.1128/JVI.01070-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.