Abstract
Methanotrophic bacteria, organisms that oxidize methane, produce a small copper chelating molecule called methanobactin (Mb). Mb binds Cu(I) with high affinity and is hypothesized to mediate copper acquisition from the environment. Recent advances in Mb characterization include revision of the chemical structure of Mb from Methylosinus trichosporium OB3b and further investigation of its biophysical properties. In addition, Mb production by several other methanotroph strains has been investigated and preliminary characterization suggests diversity in chemical composition. Initial clues into Mb biosynthesis have been obtained by identification of a putative precursor gene in the M. trichosporium OB3b genome. Finally, direct uptake of intact Mb into the cytoplasm of M. trichosporium OB3b cells has been demonstrated and studies of the transport mechanism have been initiated. Taken together, these advances represent significant progress and set the stage for exciting new research directions.
Introduction
Copper plays an important role in many bacteria, serving as a cofactor in essential enzymes such as cytochrome oxidase, NADH dehydrogenase, and superoxide dismutase.1 An array of proteins, including metallochaperones, metalloregulatory proteins, P1B-type transport ATPases, and proton-substrate antiporters,2–6 ensures that copper is provided to these enzymes, but does not accumulate to toxic levels.7 Most of these homeostatic proteins are involved in removal of excess copper, and it remains largely unclear how bacteria import necessary copper.3, 4
Copper acquisition is particularly important for methanotrophs, gram-negative bacteria that utilize methane as their sole carbon source.8 The first step in methanotroph metabolism is the oxidation of methane to methanol by the copper-dependent enzyme particulate methane monooxygenase (pMMO).8–12 pMMO is membrane-bound13, 14 and accounts for up to 20% of the total cellular protein in methanotrophs.15 Not only is copper catalytically important for pMMO, but in some methanotroph strains, copper also controls expression of an alternate iron-dependent16, 17 methane oxidation system, soluble methane monooxygenase (sMMO). In these methanotrophs, copper represses transcription of the sMMO genes and leads to formation of intracytoplasmic membranes that house pMMO.11, 13, 18 This “copper switch” also affects expression of various other proteins.8 Although there has been some progress characterizing MmoR and MmoG, two proteins involved in copper-dependent transcriptional regulation of sMMO,19 the molecular mechanism of the copper switch and how it affects pMMO regulation, Mb biosynthesis, and intracellular membrane formation remain unknown.
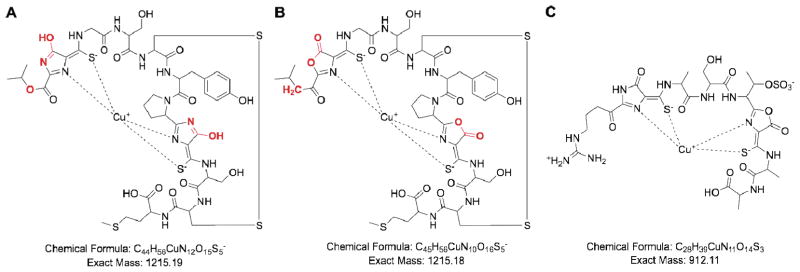
To meet their high requirement for copper, methanotrophs produce and secrete small copper chelating molecules. The existence of these copper binding compounds, originally called CBCs, was first postulated from studies of Methylosinus trichosporium OB3b mutant strains that do not switch from sMMO to pMMO expression under copper replete conditions.20, 21 Small molecules (382–1218 Da) that bind copper with high affinity were subsequently isolated from the spent media of several strains of methanotrophs under copper starvation conditions.22–24 A pure sample of one such CBC was obtained from M. trichosporium OB3b25, 26 and its crystal structure determined.27 This initial structure revealed a 1217 Da molecule with the sequence N-2-isopropyl ester-(4-thionyl-5-hydroxyimidazolate)-Gly1-Ser2-Cys3-Tyr4-pyrrolidine-(4-hydroxy-5-thionylimidazolate)-Ser5-Cys6-Met7 (Figure 1a). A single copper ion, determined to be Cu(I) from X-ray photoelectron (XPS),27 X-ray absorption (XAS),28 and electron paramagnetic resonance (EPR)28, 29 spectroscopic data, is coordinated by two nitrogen and two sulfur atoms.
Figure 1.
Methanobactin structures. (a) Initial structure of M. trichosporium OB3b Cu-Mb determined by X-ray crystallography. (b) Revised solution structure of M. trichosporium OB3b Cu-Mb. Differences between the two structures are highlighted in red. (c) Proposed structure of Methylocystis strain SB2 Cu-Mb.
This compound was named methanobactin (Mb), and is sometimes referred to as a “chalkophore” because its peptidic nature resembles many iron siderophores (chalko- is derived from the Greek word for copper; sidero- is from the Greek word for iron).27 The primary function of Mb is proposed to be copper uptake, particularly from insoluble mineral sources.30, 31 It is widely believed that Mb is secreted in the apo form (apo-Mb) to obtain copper from the environment and then internalized in its copper bound form (Cu-Mb). Many aspects of this model have not been tested in detail, however. Ancillary functions of Mb have also been proposed, including oxidative stress defense32 and enhancing pMMO activity,29 although strong evidence for the latter has not been obtained.33
Since the last comprehensive review of Mb in 2008,34 the molecular structure of M. trichosporium OB3b Mb has been revised and its biophysical properties have been investigated further. In addition, significant progress has been made toward addressing three key questions surrounding Mb structure and function. First, the question of whether other methanotroph strains produce Mb and, if so, whether it has the same structure and chemical composition, has been pursued vigorously. Second, some initial clues into Mb biosynthesis have been obtained. Third, the proposed copper acquisition function of Mb has been investigated experimentally, providing direct evidence for copper uptake via Mb as well as some insight into the nature of the uptake machinery. These recent studies, summarized here, represent major advances and suggest new research directions for the field.
Isolation and characterization of Mb from M. trichosporium OB3b
Several detailed protocols are now available for isolation of M. trichosporium OB3b Mb.35, 36 In general, cells are grown either with no copper or with low levels of copper in the media and the spent media is harvested by centrifugation. The spent media is then concentrated using either reversed-phase tC18 Sep-Pak cartridges or for a larger scale, a Diaion HP20 column. Further purification is accomplished by reverse-phase HPLC. Typically, Cu(II) is added immediately after isolation of the spent media to stabilize Mb, although the less stable apo-Mb can also be isolated.35–37 The integrity of purified Cu-Mb can be assessed by optical spectroscopy and mass spectrometry. According to mass spectrometry analysis, the preparations typically contain some fraction of a variant that is missing the C-terminal methionine residue, Met7. This material, which structurally, spectroscopically, and functionally resembles intact Cu-Mb, is present in variable amounts and its presence has not been correlated with specific growth conditions.35, 38
Since 2008, two new structures of M. trichosporium OB3b Cu-Mb have been reported. The solution structure revealed two important differences from the X-ray structure (Figure 1b).39 First, the NMR data are consistent with an N-terminal isobutyl group rather than an isopropyl group. Second, analysis of the 15N spectrum indicates that the originally assigned hydroxyimidazolate rings are instead oxazolone rings. This revised structure is consistent with mass spectrometric data. The sequence of M. trichosporium OB3b Mb is therefore now believed to be 1-(N-[mercapto-{5-oxo-2-(3-methylbutanoyl)oxazol-(Z)-4-ylidene}methyl]-Gly1-Ser2-Cys3-Tyr4)-pyrrolidin-2-yl-(mercapto-[5-oxo-oxazol-(Z)-4-ylidene]methyl)-Ser5-Cys6-Met7 (Figure 1b). In addition, the crystal structures of intact Cu-Mb and Cu-Mb lacking the C-terminal methionine were determined to 0.92 and 1.00 Å resolutions, respectively.38 These structures are overall very similar to the original 1.15 Å resolution structure,27 and are consistent with the differences in chemical composition indicated by the NMR data. Loss of the C-terminal methionine does not affect the structure significantly, and the new C-terminal residue, Cys6, remains in a disulfide bond with Cys3.38
Several biophysical properties of M. trichosporium OB3b Cu-Mb have been further investigated as well. Molar extinction coefficients for apo-Mb (340 nm, 22.9 mM−1cm−1; 394 nm, 22.1 mM−1cm−1) and Cu-Mb (290 nm, 19.5 mM−1cm−1) have been measured and reported.38 The features at 340 and 394 nm are assigned to the oxazolone rings.39 The oxidation state of the two cysteine residues was also explored. Neither apo-Mb nor Cu-Mb reacts with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), indicating that the disulfide bond is present in both forms,38 consistent with earlier studies.22, 26 Thus, reduction of Cu(II) to Cu(I) by Mb is not coupled to disulfide bond formation, as suggested previously.28 The Cu(I) binding affinity of all forms of Mb, determined by competition experiments with bathocuproine disulfonate (BCS), is (6–7) × 1020 M−1.38 This value is several orders of magnitude higher than the affinity of the copper chaperones Atx1 and Atox1 for Cu(I)40 and raises the question of how Cu-Mb releases its Cu(I) ions. Finally, reduction potentials at pH 7.5 of 640 and 590 mV vs NHE were measured for Cu-Mb and Cu-Mb lacking the C-terminal methionine, respectively.38
Diversity of Mb
The production of Mb by methanotrophs other than M. trichosporium OB3b has been a major research focus over the past three years. One approach has been the development of an assay to detect Mb production in growing methanotroph cultures. The chrome azurol S (CAS) assay, often used to detect iron siderophores,41 was adapted to screen for Mb.42, 43 In this protocol, the disappearance of a blue Cu-CAS complex is monitored either in solution by optical spectroscopy or on agar plates by eye. If Mb is produced, it removes copper from CAS, leading to a color change from blue to orange. Experiments must be performed in parallel using Fe-CAS to avoid false positive results from Cu-siderophore complexes. According to this assay, Methylococcus capsulatus (Bath) and Methylomicrobium album BG8 produce Mb, but Methylocystis parvus OBBP does not.42 Neither M. album BG8 nor M. parvus OBBP have an sMMO system. Interestingly, siderophores are also secreted by Methylocystis species strain M, M. trichosporium OB3b and M. album BG8, but not by M. capsulatus (Bath) and M. parvus OBBP.42, 44
Consistent with the results of the Cu-CAS assay, candidate Mb molecules have been isolated from M. capsulatus (Bath) and M. album BG833, 45 with molecular masses of 1056.48 and 939.33 Da, respectively.45 These putative Mb molecules are less abundant than M. trichosporium OB3b Mb, and the amount present is apparently not correlated with initial copper concentrations in the media. The optical spectra of these Mb samples also differ significantly from that of M. trichosporium OB3b Mb. M. trichosporium OB3b Cu-Mb exhibits distinct features at 340 and 394 nm and M. trichosporium OB3b apo-Mb exhibits features in the 250–300 nm range.25, 26, 38 By contrast, the apo forms of Mb from the other two organisms display weak features at 260–270 and 400 nm, and an increase at 400 nm is observed upon copper addition. The fluorescence properties are similar to those of M. trichosporium OB3b Mb, although less quenching is observed upon copper addition. EPR analysis indicates that ~80% of the copper bound to these Mb molecules is Cu(I). Finally, these compounds bind copper with moderate affinities of 105–106 M−1, as determined by isothermal titration calorimetry.45 These values are not consistent with the high affinity of CAS for copper or the bindng constant of (6–7) × 1020 M−1 reported for M. trichosporium OB3b Mb,38 but comparing copper binding constants measured by different methods is generally fraught with technical difficulties.40 Given the lower abundance of these molecules, the uncertainty of the link between their production and copper availability, and the lack of structural information, more data will be required to identify them definitively as novel forms of Mb. It should also be noted that methanotrophs secrete other molecules in this molecular mass range, including flavins and siderophores.42, 44
The Mb from Methylocystis strain SB2, a recently isolated methanotroph that only produces pMMO and does not switch to sMMO upon copper depletion,46 has been characterized in more detail.47 The mass of Methylocystis strain SB2 Mb is 851.20 Da for the apo form and 912.11 Da for a sample with a single bound copper ion. The oxidation state of this copper ion was assigned as Cu(I) on the basis of XPS data. The optical spectrum reveals peaks at 338 and 387 nm, which are similar to the 340 and 394 nm features observed for M. trichosporium OB3b Mb. The chemical composition of Methylocystis strain SB2 Mb has been investigated by NMR spectroscopy and a partial sequence has been proposed. This sequence includes four intact amino acid residues, one oxazolone ring, one imidazolone ring, and modified arginine and threonine residues (Figure 1c). This structure has a calculated mass of 771.24 Da, which can be reconciled with the measured mass if a sulfonate group is present. It is suggested that this sulfonate group is appended to the threonine-like side chain (Figure 1c).47 Thus, Methylocystis strain SB2 Mb shares some structural features with M. trichosporium OB3b Mb, including the apparent coordination of Cu(I) by two five-membered heterocyclic rings and neighboring thioamide groups, but is smaller with a different amino acid composition.
Biosynthesis of Mb
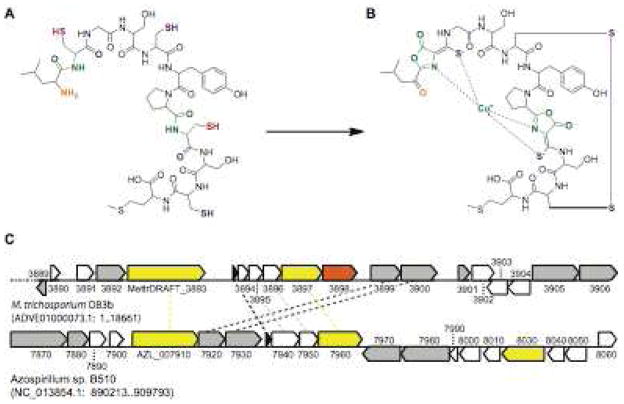
The availability of the M. trichosporium OB3b genome in 201048 combined with the revised M. trichosporium OB3b Mb structure39 has provided new insight into Mb biosynthesis. One proposed route for Mb biosynthesis involves a nonribosomal peptide synthetase (NRPS) system.25, 34 NRPS systems mediate siderophore biosynthesis49 and are present in both the M. trichosporium OB3b48 and, to a much lesser extent, M. capsulatus (Bath)50 genomes. A second possibility is that Mb is generated by modification of a ribosomally synthesized peptide.39 Preliminary evidence for this proposed route derives from identification in the M. trichosporium OB3b genome of a candidate precursor gene that encodes the peptide MTVKIAQKKVLPVIGRAAALCGSCYPCSCM47 (residues shown in bold are part of Mb; other residues may be a “leader sequence” as present in other natural products derived via posttranslational modification51) (Figure 2a). This peptide resembles a previously proposed starting peptide, LSGSCYPSSCM,39 with the substitution of cysteines for two of the serines. In that proposed mechanism, a set of posttranslational modification steps were suggested by analogy to biosynthesis of thiazolyl antibiotics such as microcin B17.39, 52 A similar mechanism can be envisioned for the peptide precursor LCGSCYPCSCM (Figure 2b), with the additional requirement that the two cysteines be converted to serines to allow formation of oxazolone rather than thiazolone rings.47 This conversion could be followed by a series of sulfuration, cyclization, and transamination steps to form the final compound (Figure 2b). There are precedents in other biosynthesis systems for cyclization of serines53 and for transamination reactions.54 The Mb from Methylocystis strain SB2 could be synthesized by a related series of modifications.47
Figure 2.
Possible pathway for biosynthesis of M. trichosporium OB3b Mb. (a) Putative precursor peptide. (b) M. trichosporium OB3b Cu-Mb. Necessary modifications would include replacement of the N-terminal amine group with a carbonyl group (orange), replacement of two cysteines with serines (red) prior to cyclization and formation of the oxazolone rings (green), and replacement of the oxygens in the carbonyl groups adjacent to the two initial cysteines with sulfur to form thioamide groups (blue). A disulfide bond must then be formed (purple) and the resultant apo-Mb loaded with copper (cyan). (c) Genomic neighborhood of the putative precursor gene in M. trichosporium OB3b and Azospirillium sp. B510 genomes. The M. trichosporium OB3b genome has not been fully assembled, and the location of the precursor peptide-containing 18.66 kb contig in relation to the other 172 contigs is not known. The Azospirillum sp. B510 genome is complete; an 18.66 kb region around the precursor peptide is displayed for purposes of comparison. Precursor peptide genes are shown in black, genes with no predicted annotated function are shown in white, genes with predicted annotated functions but unclear relevance to Mb are shown in gray, transporter genes are shown in yellow, and an aminotransferase gene is shown in orange. Potential homologs in the two genomes are connected by dashed lines.
The putative Mb precursor gene was not previously identified as a coding region and is found between genes MettrDRAFT_3893 and MettrDRAFT_3894 on Contig 00014 (NCBI reference sequence ADVE01000073.1) of the M. trichosporium OB3b draft genome (Figure 2c).47 Nearby genes encode a mutator type transposase (MettrDRAFT_3889) on the reverse strand and a neighboring hypothetical protein (MettrDRAFT_3890) on the forward strand, a sigma factor and FecR pair (MettrDRAFT_3891 and MettrDRAFT_3892), a TonB-dependent transporter (MettrDRAFT_3893), several unidentified and hypothetical proteins (MettrDRAFT_3894–3896), a member of the multidrug and toxic compound extrusion (MATE) family (MettrDRAFT_3897), an aminotransferase (MettrDRAFT_3898), a diheme enzyme (MettrDRAFT_3900) predicted to be a cytochrome c peroxidase and related to CorB from M. album BG8,55 and another gene product probably associated with the diheme enzyme (MettrDRAFT_3899) (Table 1). A sigma factor (MettrDRAFT_3901) and the unknown protein that follows it (MettrDRAFT_3902) are the last consecutive coding regions found on the forward strand. There are four other coding sequences on the contig, two unidentified putative proteins (MettrDRAFT_3903 and MettrDRAFT_3904) on the reverse strand followed by two genes related to type I restriction-modification systems (MettrDRAFT_3905 and MettrDRAFT_3906). It is not clear whether any of these proteins play a role in Mb biosynthesis or handling.
Table 1.
Annotated genes near the putative precursor peptides of M. trichosporium OB3b and Azospirillum sp. B510
| Gene | Annotation |
|---|---|
| MettrDRAFT_3889 | Transposase, mutator type |
| MettrDRAFT_3891 | RNA polymerase sigma 24 subunit, ECF subfamily |
| MettrDRAFT_3892 | FecR iron sensor protein |
| MettrDRAFT_3893 | TonB-dependent transporter |
| MettrDRAFT_3897 | Multidrug antimicrobial extrusion (MATE) protein |
| MettrDRAFT_3898 | Aminotransferase class I and II |
| MettrDRAFT_3899 | Similar to CorA (M. album BG8); part of two-gene unit with diheme enzyme |
| MettrDRAFT_3900 | Diheme cytochrome c peroxidase, similar to CorB (M. album BG8) |
| MettrDRAFT_3901 | RNA polymerase sigma 24 subunit, ECF subfamily |
| MettrDRAFT_3905 | N-6 DNA methylase (type I restriction-modification system, M subunit) |
| MettrDRAFT_3906 | Type I restriction-modification system, DNA specificity domain (S subunit) |
| AZL_007870 | Putative uncharacterized protein (contains domains similar to CopC and CopD) |
| AZL_007880 | SCO2 protein |
| AZL_007910 | TonB-dependent transporter |
| AZL_007920 | Similar to CorA (M. album BG8); part of two-gene unit with diheme enzyme |
| AZL_007930 | Diheme cytochrome c peroxidase, similar to CorB (M. album BG8) |
| AZL_007960 | Multidrug antimicrobial extrusion (MATE) protein |
| AZL_007970 | CydB (cytochrome bd-I oxidase subunit II) |
| AZL_007980 | CydA (cytochrome bd-I oxidase subunit I) |
| AZL_008030 | TonB-dependent transporter |
A similar precursor peptide is found in only one other genome, that of the rice endophyte Azospirillum sp. B510.47, 56 The significance of the absence of homologs in other sequenced methanotroph genomes is unclear, but may reflect the structural diversity of Mb molecules or the ability of methanotrophs to scavenge Mbs from other species. In the case of the Azospirillum sp. B510 genome, a 31-residue coding region encodes the peptide MTIKIAKKQTLSVAGRAGACCGSCCAPVGVN.47, 56 Several of the neighboring genes observed in the M. trichosporium OB3b genome are also conserved in Azospirillum sp. B510,47 including the TonB-dependent transporter (AZL_007910), the MATE family member (AZL_007960), the diheme enzyme and its neighbor (AZL_007930 and AZL_007920), and the unknown proteins MettrDRAFT_3894–3896 (AZL_007940, which has separate regions homologous to MettrDRAFT_3894 and MettrDRAFT_3895, and AZL_007950) (Figure 2c, Table 1). The significance of this similarity remains unclear, however. It is potentially relevant that Azospirillum sp. B510 produces siderophores.56
Copper Uptake Function
The proposed copper uptake function of Mb derives from early observations linking the presence of Mb in the spent media with copper limitation during growth.22–24 In support of a role in copper uptake, Mb can promote the switch from sMMO expression to pMMO expression, as demonstrated by monitoring protein levels, by monitoring transcript levels of pmoA (encodes pMMO pmoA subunit) and mmoX (encodes sMMO α subunit), or by detecting sMMO activity.30, 36, 38 Whereas pMMO has a limited substrate specificity, sMMO oxidizes a number of larger substrates,57 including naphthalene, which is amenable to a colorimetric assay.58 A copper-mediated switch from sMMO to pMMO can be observed in M. trichosporium OB3b upon addition of copper as either CuCl2 or Cu-Mb, but only Cu-Mb stimulates the switch in the presence of Cu-doped Fe oxide or Cu-doped borosilicate glass,30 and Cu-Mb can actually increase the dissolution rate of Cu-substituted glass.31 These findings provide insight into Mb function in nature and have been extended to correlate copper content of glass with methane oxidation rates of M. trichosporium OB3b.59 In these experiments, methane depletion was measured in the presence of manufactured borosilicate glasses with 0–800 ppm Cu, with maximum rates observed at 80 and 200 ppm, and lowest rates at 0 and 800 ppm. The methane oxidation rates are comparable to those measured using soluble copper, suggesting that insoluble copper can support methanotroph growth in nature. The current direction for this work is to assess the effect of Mb on growth using copper mineral sources other than Cu-doped silicates, including malachite, tenorite, cuprite, and chalcocite.60
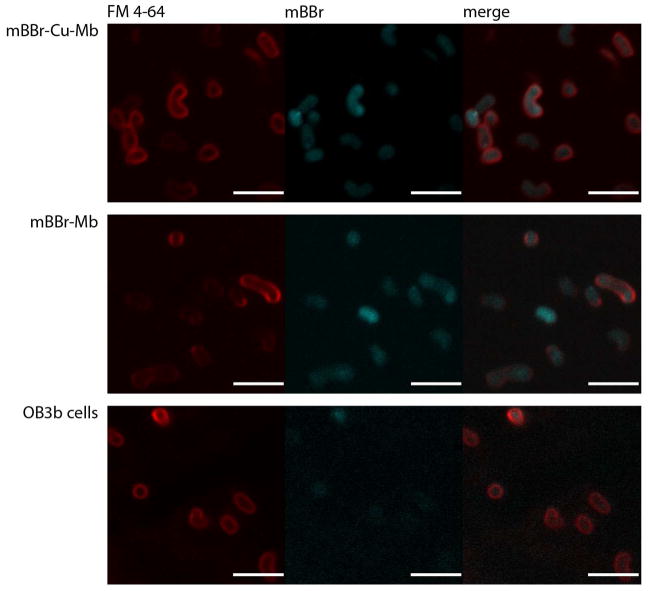
Although these data strongly support the copper uptake function, direct evidence for uptake of the Mb molecule has been lacking, and the specific mechanisms by which copper from Cu-Mb is internalized have not been established. These issues have been addressed recently by tracking copper and Mb uptake by isotopic and fluorescent labeling.61 An increase in intracellular copper content of M. trichosporium OB3b cells treated with 65Cu-Mb was correlated with an altered 63Cu/65Cu ratio, indicating that Cu bound to Mb is internalized. The high affinity of Mb for copper suggests that intact Cu-Mb is taken up, and the labeling of Cu-Mb with the fluorescent probe monobromobimane (mBBr-Cu-Mb) permitted verification of mBBr-Cu-Mb uptake into the cytoplasm by confocal microscopy (Figure 3). Interestingly, the apo form of labeled Mb (mBBr-Mb) is also taken up by M. trichosporium OB3b cells (Figure 3). Other methanotroph strains, including Methylocystis sp. strain M, M. capsulatus (Bath), and Methylomicrobium alcaliphilum 20Z also internalize Cu-Mb from M. trichosporium OB3b, but E. coli does not, suggesting that the phenomenon is specific to methanotrophs.61
Figure 3.
Imaging Mb uptake by confocal microscopy. Fluorescence images were taken at 405 nm (cyan, mBBr-Mb) and 543 nm (red, the lipophilic membrane dye FM 4–64). The merged images show the distribution of both signals. The scale bar corresponds to 4 μm. M. trichosporium OB3b cells incubated with mBBr-Cu-Mb and FM 4–64 (top) or mBBr-Mb and FM 4–64 (middle) exhibit primarily cytoplasmic localization of the compound. The background fluorescence of M. trichosporium OB3b cells, which presumably includes the intrinsic fluorescence of internally produced Mb (bottom, also in the presence of FM 4–64) is also detectable, but is of a much lower magnitude.
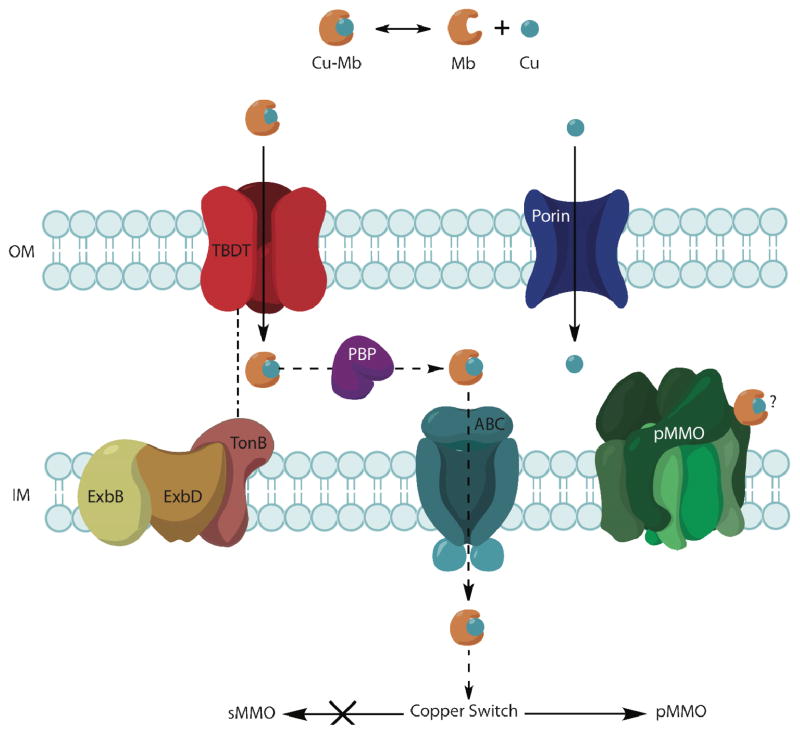
Initial insight into the mechanism of Cu-Mb uptake has also been obtained. In gram-negative bacteria, nutrients are transported across the outer membrane either by passive diffusion using porins or by active transport using TonB-dependent transporters (TBDTs).62 TBDTs interact with the inner membrane TonB-ExbB-ExbD protein complex and transduce energy from the inner membrane proton motive force to translocate substrates across the outer membrane. These systems are involved in siderophore uptake63 and have been suggested as potential Cu-Mb transporters.34 Porins and TBDTs can be inhibited selectively using spermine and uncouplers such as cyanide m-chlorophenyl hydrazone (CCCP) or methylamine, respectively. Treatment of M. trichosporium OB3b cells with spermine inhibits uptake of unchelated copper (provided as CuSO4), but not Cu-Mb, and treatment with CCCP or methylamine inhibits uptake of Cu-Mb, but not unchelated copper.61 Therefore, Cu-Mb is not transported by porins, but in an energy-dependent process that might involve TBDTs (Figure 4). The existence of specific outer membrane receptors for Mb is supported by the observation that apo-Mb can compete with Cu-Mb for copper uptake.61
Figure 4.
Hypothesized uptake mechanisms for methanobactin and copper. Copper (Cu) is transported across the outer membrane (OM) by a passive transport process likely involving porins whereas Cu-Mb is transported via an active process potentially mediated by TonB-dependent transporters (TBDTs). The mechanism of translocation across the inner membrane (IM) is unknown, but by analogy to siderophores, could involve an ATP-binding cassette (ABC) transporter and a periplasmic binding protein (PBP). It remains unclear whether intact Cu-Mb interacts directly with pMMO or with proteins involved in the “copper switch.”
Finally, Mb may have practical applications beyond its copper uptake function in methanotrophs. For example, M. trichosporium OB3b Mb was recently tested as a therapeutic in a rat model of Wilson disease, a human genetic disorder characterized by toxic copper overload in the liver due to reduced biliary excretion.64 In this study, Mb was demonstrated to remove copper from metallothionein and to promote excretion of copper in bile.65 Thus, Mb may represent a new route to treatments for Wilson disease. Such efforts may be enhanced by development of synthetic Mb molecules.66
Summary
The structure and biophysical properties of M. trichosporium OB3b Mb are well understood,38, 39 and internalization of Mb by M. trichosporium OB3b and other methanotrophs is now established. Uptake occurs through an active transport process, perhaps using TBDTs.61 Identifying which, if any, of the approximately 45 TBDTs in the M. trichosporium OB3b genome is involved represents a significant future challenge. Moreover, transport of Mb across the inner membrane has not been explored at all, but by analogy to siderophores, could conceivably involve ABC transporters and periplasmic binding proteins (Figure 4). How copper is released from Cu-Mb and whether Cu-Mb directly provides copper to the pMMO active site or directly triggers the copper switch also remain unclear. Methanotrophs besides M. trichosporium OB3b produce Mb-like molecules that can chelate copper,42 and in the case of Methylocystis strain SB2, preliminary characterization suggests a chemical composition that recapitulates some, but not all, features of M. trichosporium OB3b Cu-Mb.47 Further detailed structural and chemical analyses will be critical to determining the prevalence and diversity of Mb in methanotrophs. A first clue into Mb biosynthesis has been obtained with the identification of a potential precursor peptide in the M. trichosporium OB3b genome.47 A major goal for the future is to determine whether this peptide is indeed involved and if so, to unravel the complete biosynthetic mechanism. Also completely unexplored is the means by which methanotrophs secrete Mb. Addressing these main issues, identification of the transport machinery, structural characterization of new Mb molecules, and elucidating the mechanism of Mb biosynthesis, will be the central focus of Mb research in the next few years.
Acknowledgments
Work in the Rosenzweig laboratory on methanobactin is supported by the National Science Foundation (MCB0842366). G. E. K. is supported in part by National Institutes of Health Training Grant GM08061. We thank R. Balasubramanian for valuable discussions.
Keywords
- Methanotroph
a type of bacteria that lives on methane as its sole source of carbon and energy
- Methane monooxygenase
a metalloenzyme that oxidizes methane to methanol in methanotrophs
- Particulate methane monooxygenase
integral membrane methane monooxygenase that contains copper
- Soluble methane monooxygenase
cytoplasmic methane monooxygenase that contains a diiron active site
- Methanobactin
a small high affinity copper chelating molecule isolated from methanotrophic bacteria
- Chalkophore
high affinity copper binding molecule analogous to an iron siderophore
- TonB-dependent transporter
outer membrane protein in gram-negative bacteria that facilitates uptake of nutrients by transducing energy from the inner membrane protonmotive force
- Wilson disease
a genetic disorder of copper metabolism characterized by toxic copper accumulation in the liver
Abbreviations
- apo-Mb
metal free form of methanobactin
- CAS
chrome azurol S
- CBC
copper binding compound
- Cu-Mb
copper bound form of methanobactin
- mBBr
monobromobimane
- Mb
methanobactin
- mBBr-Cu-Mb
Cu-Mb labeled with monobromobimane
- pMMO
particulate methane monooxygenase
- sMMO
soluble methane monooxygenase
- TBDT
TonB-dependent transporter
References
- 1.Ridge PG, Zhang Y, Gladyshev VN. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS ONE. 2008;3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osman D, Cavet JS. Copper homeostasis in bacteria. Adv Appl Microbiol. 2008;65:217–247. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- 5.Kim EH, Nies DH, McEvoy MM, Rensing C. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011;193:2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Lett. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanian R, Smith SM, Rawat S, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman RL, Rosenzweig AC. The quest for the particulate methane monooxygenase active site. Dalton Trans. 2005;21:3390–3396. doi: 10.1039/b506651d. [DOI] [PubMed] [Google Scholar]
- 11.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 12.Chan SI, Yu SSF. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc Chem Res. 2008;41:969–979. doi: 10.1021/ar700277n. [DOI] [PubMed] [Google Scholar]
- 13.Prior SD, Dalton H. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath) J Gen Microbiol. 1985;131:155–163. [Google Scholar]
- 14.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 15.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J Am Chem Soc. 2007;129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 17.Tinberg CE, Lippard SJ. Dioxygen activation in soluble methane monooxygenase. Acc Chem Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murrell JC, McDonald IR, Gilbert B. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 2000;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 19.Scanlan J, Dumont MG, Murrell JC. Involvement of MmoR and MmoG in the transcriptional activation of soluble methane monooxygenase genes in Methylosinus trichosporium OB3b. FEMS Microbiol Lett. 2009;301:181–187. doi: 10.1111/j.1574-6968.2009.01816.x. [DOI] [PubMed] [Google Scholar]
- 20.Fitch MW, Graham DW, Arnold RG, Agarwal SK, Phelps P, Speitel GE, Jr, Georgiou G. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1993;59:2771–2776. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelps PA, Agarwal SK, Speitel GE, Jr, Georgiou G. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl Environ Microbiol. 1992;58:3701–3708. doi: 10.1128/aem.58.11.3701-3708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahn JA, DiSpirito AA. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD, Taylor AB. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol. 1998;180:3606–3613. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Téllez CM, Gaus KP, Graham DW, Arnold RG, Guzman RZ. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl Environ Microbiol. 1998;64:1115–1122. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Galeva N, Larive CK, Alterman M, Graham DW. Purification and physical-chemical properties of methanobactin: a chalkophore from Methylosinus trichosporium OB3b. Biochemistry. 2005;44:5140–5148. doi: 10.1021/bi047367r. [DOI] [PubMed] [Google Scholar]
- 26.Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, Hargrove MS, Pohl NL, Boyd ES, Geesey GG, Hartsel SC, Shafe PH, McEllistrem MT, Kisting CJ, Campbell D, Rao V, de la Mora AM, DiSpirito AA. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45:1442–1453. doi: 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 27.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PMA. Methanobactin, a copper-aquisition compound from methane oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 28.Hakemian AS, Tinberg CE, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds Cu(I) J Am Chem Soc. 2005;127:17142–17143. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi DW, Antholine WE, Do YS, Semrau JD, Kisting CJ, Kunz RC, Campbell D, Rao V, Hartsel SC, DiSpirito AA. Effect of methanobactin on the activity and electron paramagnetic resonance spectra of the membrane-associated methane monooxygenase in Methylococcus capsulatus Bath. Microbiol. 2005;151:3417–3426. doi: 10.1099/mic.0.28169-0. [DOI] [PubMed] [Google Scholar]
- 30.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci USA. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulczycki E, Fowle DA, Knapp C, Graham DW, Roberts JA. Methanobactin-promoted dissolution of Cu-substituted borosilicate glass. Geobiology. 2007;5:251–263. [Google Scholar]
- 32.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM, DiSpirito AA. The membrane-associated methane monooxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi DW, Semrau JD, Antholine WE, Hartsel SC, Anderson RC, Carey JN, Dreis AM, Kenseth EM, Renstrom JM, Scardino LL, Van Gorden GS, Volkert AA, Wingad AD, Yanzer PJ, McEllistrem MT, de la Mora AM, DiSpirito AA. Oxidase, superoxide dismutase, and hydrogen peroxide reductase activities of methanobactin from types I and II methanotrophs. J Inorg Biochem. 2008;102:1571–1580. doi: 10.1016/j.jinorgbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Balasubramanian R, Rosenzweig AC. Copper methanobactin: a molecule whose time has come. Curr Opin Chem Biol. 2008;12:245–249. doi: 10.1016/j.cbpa.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, Gilles VS, DiSpirito AA. Isolation of methanobactin from the spent media of methane oxidizing bacteria. Methods in Enzymology: Methods in Methane Metabolism. 2011;495(Pt B):259–269. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 36.Graham DW, Kim HJ. Production, isolation, purification, and functional characterization of methanobactins. Methods in Enzymology: Methods in Methane Metabolism. 2011;495(Pt B):227–245. doi: 10.1016/B978-0-12-386905-0.00015-2. [DOI] [PubMed] [Google Scholar]
- 37.Pesch ML, Christl I, Barmettler K, Kraemer SM, Kretzschmar R. Isolation and purification of Cu-free methanobactin from Methylosinus trichosporium OB3b. Geochem Trans. 2011;12:2. doi: 10.1186/1467-4866-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Ghazouani A, Basle A, Firbank SJ, Knapp CW, Gray J, Graham DW, Dennison C. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg Chem. 2011;50:1378–1391. doi: 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 39.Behling LA, Hartsel SC, Lewis DE, DiSpirito AA, Choi DW, Masterson LR, Veglia G, Gallagher WH. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J Am Chem Soc. 2008;130:12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao ZG, Brose J, Schimo S, Ackland SM, La Fontaine S, Wedd AG. Unification of the copper(I) biinding affinities of the metallochaperones Atx1, Atox1, and related proteins Detection probes and affinity standards. J Biol Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 42.Yoon S, Kraemer SM, DiSpirito AA, Semrau JD. An assay for screening microbial cultures for chalkophore production. Environ Microbiol Rep. 2010;2:295–303. doi: 10.1111/j.1758-2229.2009.00125.x. [DOI] [PubMed] [Google Scholar]
- 43.Yoon S, DiSpirito AA, Kraemer SM, Semrau JD. A simple assay for screening microorganisms for chalkophore production. Methods in Enzymology: Methods in Methane Metabolism. 2011;495(Pt B):247–258. doi: 10.1016/B978-0-12-386905-0.00016-4. [DOI] [PubMed] [Google Scholar]
- 44.Balasubramanian R, Levinson BT, Rosenzweig AC. Secretion of flavins by three species of methanotrophic bacteria. Appl Environ Microbiol. 2010;76:7356–7358. doi: 10.1128/AEM.00935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi DW, Bandow NL, McEllistrem MT, Semrau JD, Antholine WE, Hartsel SC, Gallagher W, Zea CJ, Pohl NL, Zahn JA, DiSpirito AA. Spectral and thermodynamic properties of methanobactin from gamma-proteobacterial methane oxidizing bacteria: A case for copper competition on a molecular level. J Inorg Biochem. 2010;104:1240–1247. doi: 10.1016/j.jinorgbio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Im J, Lee SW, Yoon S, DiSpirito AA, Semrau JD. Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ Microbiol Rep. 2011;3:174–181. doi: 10.1111/j.1758-2229.2010.00204.x. [DOI] [PubMed] [Google Scholar]
- 47.Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, Gallagher WH. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry. 2010;49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein LY, Yoon S, Semrau JD, DiSpirito AA, Crombie A, Murrell JC, Vuilleumier S, Kalyuzhnaya MG, Op den Camp HJ, Bringel F, Bruce D, Cheng JF, Copeland A, Goodwin L, Han S, Hauser L, Jetten MS, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T, Zeytun A, Klotz MG. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J Bacteriol. 2010;192:6497–6498. doi: 10.1128/JB.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Molec Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Metheacute B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonason I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nature Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scholz R, Molohon KJ, Nachtigall J, Vater J, Markley AL, Sussmuth RD, Mitchell DA, Borriss R. Plantazolicin, a novel microcin B17/streptolysin S-Like natural product from Bacillus amyloliquefaciens FZB42. J Bacteriol. 2011;193:215–224. doi: 10.1128/JB.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy RS, Gehring AM, Milne JC, Belshaw PJ, Walsh CT. Thiazole and oxazole peptides: biosynthesis and molecular machinery. Nat Prod Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 54.Li CX, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 55.Karlsen OA, Larsen O, Jensen HB. Identification of a bacterial di-haem cytochrome c peroxidase from Methylomicrobium album BG8. Microbiol. 2010;156:2682–2690. doi: 10.1099/mic.0.037119-0. [DOI] [PubMed] [Google Scholar]
- 56.Kaneko T, Minamisawa K, Isawa T, Nakatsukasa H, Mitsui H, Kawaharada Y, Nakamura Y, Watanabe A, Kawashima K, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Sato S. Complete genomic structure of the cultivated rice endophyte Azospirillum sp B510. DNA Res. 2010;17:37–50. doi: 10.1093/dnares/dsp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burrows KJ, Cornish A, Scott D, Higgins IJ. Substrate specificities of the soluble and particulate methane monooxygenases of Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:3327–3333. [Google Scholar]
- 58.Brusseau GA, Tsien H-C, Hanson RS, Wackett LP. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- 59.Kulczycki E, Fowle DA, Kenward PA, Leslie K, Graham DW, Roberts JA. Stimulation of methanotroph activity by Cu-substituted borosilicate glass. Geomicrobiol J. 2011;28:1–10. [Google Scholar]
- 60.Fru EC, Gray ND, McCann C, Baptista JdC, Christgen B, Talbot HM, El Ghazouani A, Dennison C, Graham DW. Effects of copper mineralogy and methanobactin on cell growth and sMMO activity in M. trichosporium OB3b. Biogeosciences Discuss. 2011;8:2851–2874. [Google Scholar]
- 61.Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. J Biol Chem. 2011;286:37313–37319. doi: 10.1074/jbc.M111.284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Molec Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Ann Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts EA, Schilsky ML. Diagnosis and treatment of Wilson disease: An update. Hepatol. 2008;47:2089–2111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- 65.Summer KH, Lichtmannegger J, Bandow N, Choi DW, DiSpirito AA, Michalke B. The biogenic methanobactin is an effective chelator for copper in a rat model for Wilson disease. J Trace El Med Biol. 2011;25:36–41. doi: 10.1016/j.jtemb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Pinder TA, Graham LA, Rabinovich D. Copper(I) bis(thione) complexes as synthetic analogs of methanobactin. Abstr Pap Am Chem Soc. 2009;237:307-INOR. [Google Scholar]