Abstract
Purpose
The aim of this research was to further investigate the contribution of CD20 antigen expression to rituximab activity and define the mechanisms responsible for CD20 downregulation in rituximab-resistant cell lines (RRCL).
Experimental Design
Rituximab-sensitive, rituximab-resistant cell lines, and primary neoplastic B-cells were evaluated by Chromium51-release assays, ImageStream image analysis, immunohistochemical staining, flow cytometric analysis, CD20 knockdown, promoter activity, chromatin immunoprecipitation (ChIP) analysis of CD20 promoter, and CD20 plasmid transfection experiments in order to identify mechanisms associated with CD20 regulation in rituximab-resistant cells.
Results
RRCL exhibited a gradual loss ofCD20 surface expressionwith repeated exposure to rituximab. We identified a CD20 antigen surface threshold level required for effective rituximab-associated complement-mediated cytotoxicity (CMC). However, a direct correlation between CD20 surface expression and rituximab-CMC was observed only in rituximab-sensitive cell lines (RSCL). CD20 promoter activity was decreased in RRCL. Detailed analysis of various CD20 promoter fragments suggested a lack of positive regulatory factors in RRCL. ChIP analysis showed reduced binding of several key positive regulatory proteins on CD20 promoter in RRCL. Interluekin-4(IL-4)induced higher CD20 promoter activityand CD20 expression, but modestly improving rituximab activity in RRCL and in primary B-cell lymphoma cells. Forced CD20 expression restored cytoplasmic but not surface CD20, suggesting the existence of a defect in CD20 protein transport in RRCL.
Conclusion
We identified several mechanisms that alter CD20 expression in RRCL and demonstrated that, while CD20 expression is important for rituximab activity, additional factors likely contribute torituximab sensitivityin B-cell lymphoma.
Keywords: CD20 downregulation, B-NHL, rituximab resistance, complement-dependent cytotoxicity, dual resistance to rituximab and chemotherapy
INTRODUCTION
The incorporation of rituximab in the management of non-Hodgkin lymphoma (NHL) has improved the response rate, progression free survival (PFS), and overall survival (OS). In aggressive lymphoma, patientswho do not respond to rituximab-combined chemotherapy in the front-line setting represent a group of patients with a very poor clinical outcome (1, 2).
Rituximab’s primary mechanisms-of-action include: antibody-dependent cellular cytotoxicity (ADCC), complement-dependent cytotoxicity (CMC), induction of apoptosis and anti-proliferation(4). The acquirement of phenotypic changes in cancer cells or host immune cells over time may affect rituximab responsiveness and underscores the complexity of the potential mechanisms-of-resistance to anti-CD20 monoclonal antibodies (mAbs).
Both Martin A et al and Gisselbrecht et al have individually reported that the use of rituximab as frontline therapy in patients with diffuse large B-cell lymphoma results in selection for cells more resistant to subsequent salvage regimens (2, 3), indicating that prior rituximab is changing the biology of relapsed/refractory B-NHL cells. Down-regulation of CD20 has been observed in a number of case reports of patients with relapsed/refractory B-cell lymphoma who became unresponsive to rituximab-based therapies and has been postulated to be one of the most important etiologies contributing to rituximab resistance (5–8). Since this has not been extensively studied in a large number of “paired” patient samples, it is unknown whether CD20 is the most important, if not limiting factor, in the context ofrituximab resistance.
To further define the mechanisms of resistance to rituximab, our group of investigators generated and fully characterized several rituximab-resistant cell lines (RRCL) (9, 10). We observed several changes in RRCL, notably: downregulation of surface CD20 antigen and CD20 mRNA, upregulation of complement inhibitory proteins (CIPs), CD55 and CD59. Concurrent deregulation of pro-apoptotic (Bax/Bak) and anti-apoptotic (Mcl-1, Bcl-XL) proteins were observed and associated withconcomitant chemotherapy resistance(9, 10).
Forced expression of CD20 in a CEM-T cell line model revealed a direct correlation between increasing surface CD20 expression and augmented rituximab-CMC, but not ADCC (11). Similar results were observed when primary chronic lymphocytic leukemia (CLL) and lymphoma samples were studied(11).
Several investigators have studied potential mechanisms that may play a role in CD20 expression changes observed in previously rituximab-treated relapsed/refractory B-cell lymphoma. For instance, Terui et al, found CD20 gene mutations leading to C-terminus truncated forms of CD20 in 22% of patients with rituximab relapsed/refractory lymphomas (12). Beers et al and Beum et al demonstrated CD20 internalization into lysosomes or “shaving” following rituximab exposure, respectively(13, 14).
In this report, we identify two novel mechanisms that regulate CD20 expression in rituximab-refractory lymphoma cells, study the chronological changes ofsurface CD20 in the context of acquired resistance to rituximab, and further evaluate the role of surface CD20 density in the biological activity of rituximab.
Our data suggests that while CD20 levels are important in rituximab sensitivity at an early stage; as lymphoma cells are repeatedly exposed to rituximab, additional gains (i.e. upregulation of Mcl-1, Bcl-XL, CIPs) or losses (down-regulation of Bax and Bak) of other proteins further impair the anti-tumor activity of rituximab alone or in combination with chemotherapeutic agents.
MATERIAL AND METHODS
Cell lines
The generation of RRCLs from RSCLs, and the pre-resistant passages was as previously described (9, 10) and summarized in Supplemental Table S1. Final resistant cell lines (RRCL, 10th passage) and prior passages were utilized for the experiments described below. Single cell clones from RSCL and RRCL were further obtained by limiting dilution.
Primary B-cell isolation from patient specimens
Malignant B cells were isolated by MACS sorting (negative selection) from biopsy tissue obtained from patients with B-NHL under Roswell Park Cancer Institute non-therapeutic protocols I-42804 and I-42904 (Supplemental Table S2). B-cell purity was assessed by flow cytometry using antibodies to CD19 and CD20 (Becton Dickinson). Greater than 95% pure CD19-positive cells were obtained from each biopsy processed. Collected cells were re-suspended in media and used in corresponding experiments.
Chromium51 (51Cr)-release assay
Rituximab-CMC was determined as previously described.(9, 10)Briefly, viable lymphoma cells labeled with 51Cr at 37°C, 5% CO2 for 2-hrs were plated in 96-well plates at a cell concentration of 1×105 cells/well. Cells were then exposed to rituximab or isotype and human serum (1:4 dilution) for 6-hours at 37°C and 5% CO2. 51Cr-release was measured from the supernatant by standard gamma counter. The percentage of lysis was calculated as following: [Test cpm – background cpm]/[Maximum cpm – background cpm]. Pooled human serum was obtained from healthy donors (IRB-approved protocol CIC-016).
ImageStream analysis
Surface CD20 expression density was analyzed by the ImageStream Technology (Amnis Inc.). In brief, 2×106 cells were stained with anti-human CD20-FITC or isotype control and then fixed by 2% paraformaldehyde. Cells were illuminated in the ImageStream system by a bright-field lamp and a 488 nm excitation laser for FITC. Data was analyzed by ImageStream IDEAS image analysis software. Cells were gated for single, focused populations, which were then analyzed for the mean cell size (by mean surface area) and mean surface CD20 expression (by mean CD20-FITC intensity). Mean surface CD20 density was calculated as follows: mean surface CD20-FITC intensity/mean surface area (CD20-FITC per μm2)
CD20 knockdown
48-hours post-transfection using CD20 siRNA duplexes (On-TARGET plus SMARTpool siRNA, Dharmacon/Thremo Scientific), or siRNA control (Qiagen), cells were harvested and subjected to a second transfection using similar experimental conditions. 48-hours after second transfection, cells were harvested for CD20 expression analysis by immunoblotting, ImageStream, andrituximab-CMCbystandard 51Cr-release assay.
CD20 promoter activity analysis by luciferase assay
The PCR fragments of CD20 promoter were amplified by PCR from both RSCL and RRCL genomic DNA by several pairs of specific primers (Supplemental Table S3), ligated individually into the polylinker of pGL3 basic vector (Promega), before being co-transfected with a pEF-RL vector to monitor transfection efficiency (Nucleofector kit V, Lonza). The empty pGL3 basic vector was used to determine the baseline activity. Transfected cells were cultured in complete medium for 48-hoursbefore performing luciferase assay (Promega). In addition, differences in expression of CD20-positive transcription regulatory factors were evaluated by immunoblotting.
Chromatin Immunoprecipitation (ChIP) assay
Cells from RSCL and RRCL were cross-linked, lysed before chromation was sheared to 200–500 bp using Fisher Sonic DIsmembrator (Model 300). Immunoprecipitation (IP) was performed with Magna ChIP G Chromatin Immunoprecipitation kit (Millipore), followed by reverse cross-linking of chromatin, before subjecting to Quantitative-PCR analysis by BioRad iQ5. Antibodies used for immunoprecipitation were listed in the section of Antibodies.
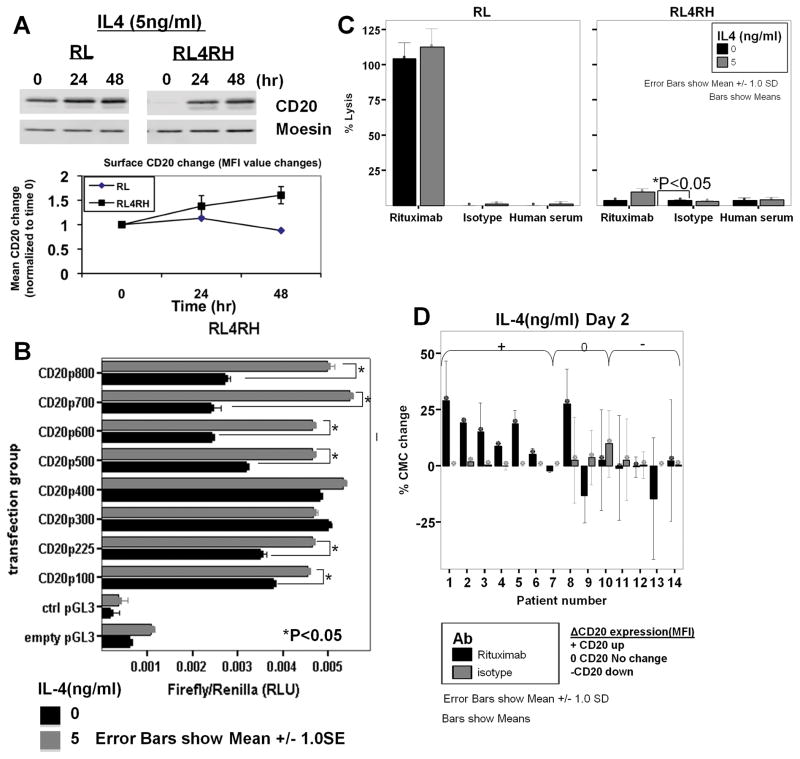
Effect of IL-4 on CD20 promoter activity and CD20 expression
RSCL, RRCL and primary tumor cells isolated from patients with de novo or relapsed/refractory B-cell lymphomas were exposed to IL-4 (5ng/ml) or RPMI-1640 media control, harvested at 24-hour intervals. CD20 expression changes were assessed by immunoblotting and flow cytometry. For CD20 promoter activity analysis, cells were incubated in 5ng/ml IL-4 immediately after co-transfection of the individual CD20 promoter-carrying pGL3 vector and pEF-RL vector. Cells were harvested at 48-hours post-transfection for a Dual-Luciferase Reporter Assay (Promega).
Transient expression of CD20 in RRCL by CD20-BCMGSneo vector
The construct for full-length CD20 on the BCMGSneo backbone was a gift from Julie P. Deans (University of Calgary, Canada)(15, 16). Plasmids were transfected into RRCL using an Amaxa Nucleofector kit V per manufacturer’s protocol. Transfection efficiency was assessed using the pmaxGFP vector (Lonza, Germany). CD20 protein expression was determined by immunoblotting and Amnis ImageStream Analysis.
Immunoblotting
Immunoblotting was performed as previously described (9, 10). Quantification of immunoblots was performed by Image J software as per instructions ((http://rsbweb.nih.gov/ij/download.html)..
Antibodies
GST77, an anti-rabbit antibody which binds to C-terminal region of the intracellular domain of CD20, was gift from Julie P. Deans, University of Calgary, Canada; β-actin (A2066) was from Sigma; Oct-2 (C-20): sc-233, PU.1 (H-135): sc-22805, USF-1 (H-86): sc-8983, USF-2 (C-20): sc-862, and Mcl-1(S-19):sc-819 were from Santa Cruz Biotechnology, Inc. Anti-Bak (B5897) was from Sigma-Aldrich;. Anti-Bax (610982) was from BD Biosciences. IRF4 (F-4) x sc-48338X, normal mouse IgG: sc-2025 were from Santa Cruz Biotechnology. Normal rabbit IgG was from Cell Signaling. CD20-FITC, CD55-FITC, CD59-FITC and FITC mouse IgG1k isotype control were from BD Biosciences. Rituximab (anti-CD20) and trastuzumab (anti-Her-2/neu, as isotype control) were from Genentech Inc., San Francisco.
STATISTICS
All the in vitro experiments were repeated in triplicates, and the results were reported as the mean with standard error determined by SPSS. Significant differences were calculated by Student t-test. P values less than 0.05 were considered statically significant.
RESULTS
Repeated exposure to rituximab led to a gradual reduction of CD20 expression during the development of RRCL
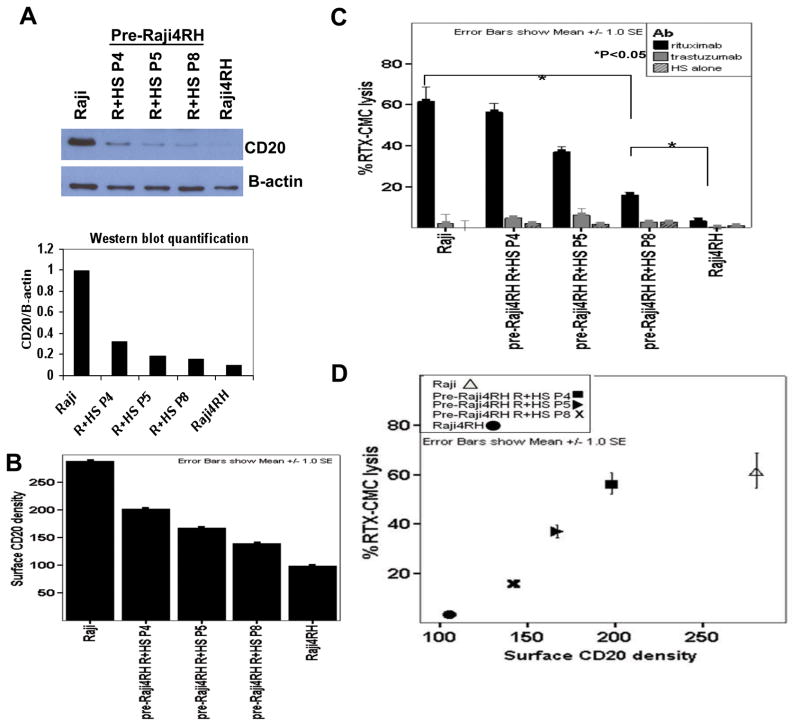
RRCL were generated by exposing sensitive cells to escalating doses of rituximab for 24 hour time periods in the presence or absence of human serum (HS). The process required RSCL to be exposed ten times to rituximab +/− HS, at the end three cell lines isolated: RL4RH, Raji2R and Raji4RH with a rituximab-resistant phenotype and low CD20 surface levels that were further characterized (9, 10). To determine the timing and cumulative-dose of rituximab necessary to negatively affect CD20 expression, changes in CD20 surface expression were studied by Western blotting and ImageStream analysis in Raji, Raji4RH and various pre-Raji4RH passages. At the same time, we correlated rituximab-associated CMC versus surface CD20 expression in RSCL and RRCL.
Overall, there was 70% reduction in total CD20 expression in RRCL even at very early stages in the process of acquiring resistance to rituximab. Significant CD20 downregulation was observed as early as in pre-Raji4RH passage 4 (Figure 1A). Moreover, there was a gradual reduction in surface CD20 density, as determined by the unit of CD20-FITC per μm2 (Figure 1A). Raji had surface CD20 density of approximately 300 CD20-FITC per μm2. As Raji cells were exposed to eight increasing doses of rituximab (pre-Raji4RH passage 8), the surface CD20 density decreased by 50% (150 CD20-FITC per μm2). Raji4RH (10th and final cell passage) was foundto have 67% decrease in surface CD20 density (100 CD20-FITC per μm2); (Figure 1B). Parallel to the changes in CD20 surface expression, we found a gradual decrease in rituximab-CMC. Rituximab activity decreased by approximately 33% after 5 “cycles” of rituximab exposure (pre-Raji4RH passage 5), by 60% in pre-Raji4RH cell passage 8 and by 90% in Raji4RH cells (Figure 1C). There was a direct correlation between the degree of CD20 downregulation and rituximab-CMC. (Figure 1D). Similar results of CD20 reduction and a corresponding loss of rituximab-CMC were seen in Raji cells exposed to rituximab without human serum (Raji 2R); (data not shown).
Figure 1. Gradual reduction of CD20 expression was observed during the development of rituximab-resistant cell lines (RRCLs).
Raji, rituximab-sensitive cell line, was exposed to repeated, escalating doses of rituximab plus human serum (R+HS) before the rituximab-resistant cell line (RRCL), Raji4RH, was generated at the final 10th passage. Various passages (abbreviated as “P”) were collected during the process of RRCL development and were analyzed. (A) Total CD20 protein expression was analyzed for passages 4, 5, 8 (P4, P5, P8), the final passage (Raji4RH)and the parentalcell line, Raji, by Western blot. (B) Surface CD20 density at different passages were analyzed by Amnis ImageStream technology and calculated according to the following formula: Surface CD20 density=mean surface CD20-FITC intensity/mean surface area (CD20-FITC/μm2). (C) The percentage of rituximab-associated CMC in each passage was determined by 51Cr release assay. (D) Decreasing surface CD20 density was directly correlated with loss of rituximab-associated complement-mediated cytotoxicity (rituximab-CMC).
Additional factors influence rituximab-CMCin RRCL
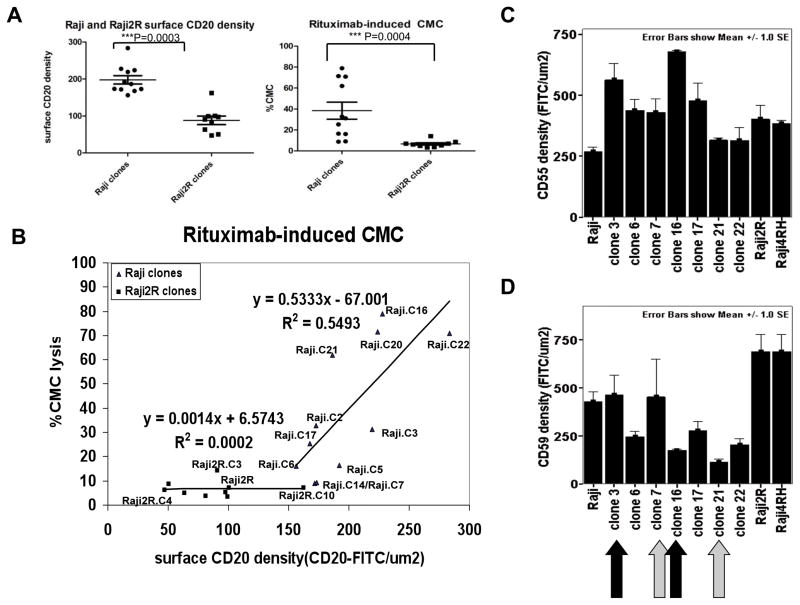
Previously, we demonstrated that RRCL have other phenotypic changes leading to cross-resistance to other mAbs (upregulation of CD55 and CD59) or chemotherapy agents (downregulation of Bax/Bak)(9, 10). While we found a linear correlation between rituximab-CMC and CD20 downregulation as lymphoma cells acquired resistance to rituximab, other factors may have contributed significantly to the acquirement of rituximab resistance. To this end, we isolatedsingle cell clones from the heterogeneous populations of RSCL (Raji and RL) and the RRCL (Raji4RH, Raji2R and RL4RH) by limiting dilution and expanded. Single cell clones were verifiedby ImageStream data(data not shown).
Differences were observed between single cell clones of RSCL and RRCL in terms of surface CD20 expression, cell size, Bcl-2 family member levels and in vitro responsiveness to rituximab. Cell clones isolated from Raji2R cells had a larger size, lower surface CD20 expression (Figure 2A) and lower levels of Bax/Bak when compared to parental Raji cells (Figure S1). Furthermore, taking in consideration both changes in size and CD20 surface expression, the decrease in mean surface CD20 density in Raji2R was magnified when compared to Raji cells (Figure 2B). As previously noted, Raji2R were essentially resistant to rituximab-CMC.
Figure 2. Linear relationship between CD20 expression versus rituximab-associated CMC was observed only in single cell clones from RSCL. A threshold of surface CD20 density appeared to be required for efficient cytotoxicity.
Single cell clones of parentalRajiand the RRCL, Raji2R, were obtained by limiting dilution. ImageStream analysis of approximately 5,000 cells per clone showed: (A) Mean surface CD20 density in each clone was calculated as described earlier. Rituximab-associated CMC in different clones was determined by 51Cr release assay. (B) Surface CD20 expression was plotted against rituximab-associated CMC to determine their correlation. (C, D) Surface CD55 and CD59 expressions of individual clones was determined by ImageStream.
Using ImageStream studies, we defined a threshold of CD20 antigen density expression required for efficient rituximab-CMC at approximately 150 CD20-FITC per μm2. A direct correlation between surface CD20 density and rituximab-CMC was observed in Raji but not in Raji2R cell clones, suggesting the existence of additional factors influencing rituximab activity in RRCL. CD20 antigen density ranged from 150 to 300 CD20-FITC per μm2 in cell clones isolated from Raji cells and rituximab-CMC exhibited a linear correlation with CD20 levels (Figure 2E). In contrast, most of the cells clones isolated from Raji2R cells had a much lower CD20 antigen density (50 to 150 CD20-FITC per μm2) and minimal rituximab-CMC activity (Figure 2B).
We further compared the degree of rituximab-CMC between cell clones isolated from Raji (RSCL) and Raji2R (RRCL) with equivalent levels of CD20 antigen. In some subclones, CD20 antigen levels appeared primarily responsible for rituximab-CMC (e.g. Raji clone 6 and Raji2R). On the other hand, several RSCL and RRCL clones exhibited similar responsiveness to rituximab at different CD20 levels (Raji clone 6 vs. Raji2R clones 3, 4 or 10). Moreover, several pairs of Raji subclones with similar CD20 expression levels demonstrated variable responsiveness to rituximab (Raji clone 3 vs. 20 and 16; 21 vs. 5 and 7, 2 vs. 14). This data suggeststheexistence of other factors affecting rituximab activity (Figure 2B).
To evaluate if the expression of complement inhibitory molecules (CIP), CD55 and CD59, could account for the discrepancies between CD20 expression and rituximab-CMC in Raji clones, we determined their surface expression levels by ImageStream. There were differential surface expression levels of both CIP within Raji single cell clones (Figures 2C, 2D). Raji clone 3 and 7, in particular, expressed higher CD59 than their pair counterparts, clone 16 and 21, respectively. This potentially explained why even though these 2 pairs of clones expressed a similar surface CD20 density, significant differences in rituximab-CMC sensitivity were seen.
While overexpression of CIPs could explain the resistant phenotype observed, we previously demonstrated that RRCL are lysed by mAb targeting CD52 (alemtuzumab) (17). This observation further supports a cause and effect between CD20 expression and a decrease in rituximab activity observed in our in vitro lymphoma model.
Overall, we observed that rituximab-CMC is dependent on CD20 expression in RSCL and may be further affected by the surface expression of CIPs. In RRCL, it appears that CIPs may play a more dominant role. However, CD20 levels observed were below the threshold level for rituximab activity in RRCL clones.
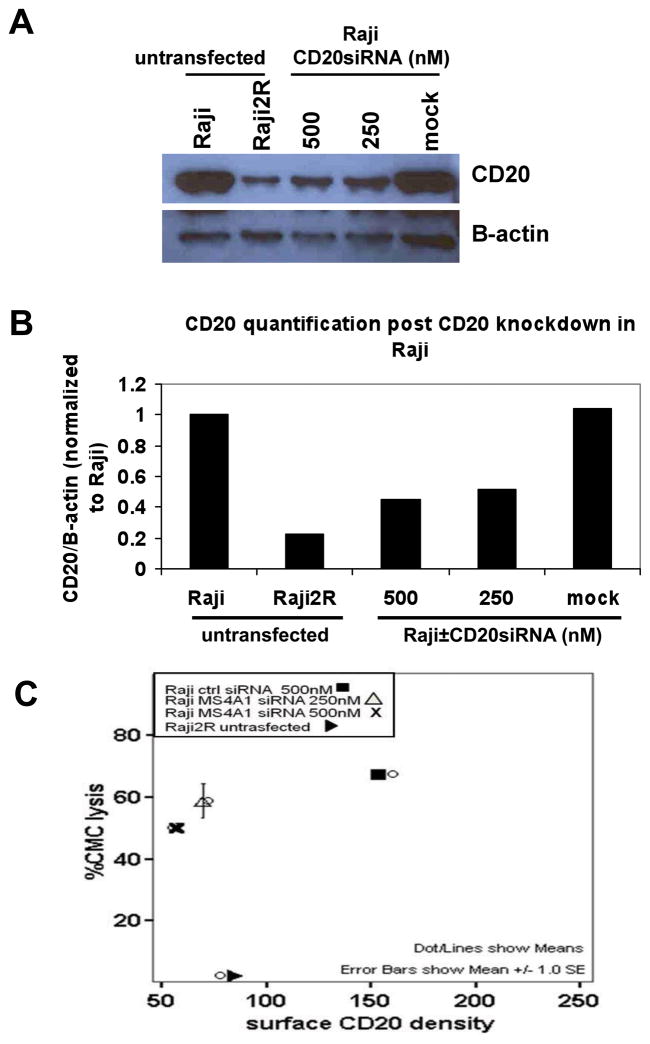
Transient CD20 knockdown in RSCL negatively affected rituximab-CMC
To further investigate the significance of CD20 expression in rituximab activity, and to investigate whether additional mechanisms might be involved in the process of acquired resistance to rituximab, we knocked down endogenous CD20 in RSCL (Raji) using the On-Targetplus SMARTpool siRNA against CD20 (MS4A1), which consisted of 4 siRNA duplexes. Cell lysates from untransfected Raji, Raji2R, as well as mock-and CD20siRNA-transfected Raji were analyzed for endogenous expression of CD20.
CD20siRNA-transfected Raji cells exhibited 60% decrease in endogenous CD20 when compared to untrasfected and mock-transfected Rajicells (Figure 3A–C). Despite a significant reduction of surface CD20 antigen (i.e. comparable to levels observed in Raji2R cells), CD20siRNA-transfected Raji cells had only 15% decrease in rituximab-CMC when compared to rituximab-CMC in Raji control cells (Figure 3C). Together these data suggest that CD20 antigen expression contributes partially to rituximab-CMC and that additional factors such as CIPs, likely affect rituximab biological activity.
Figure 3. CD20 knockdown in Raji only partially decreased rituximab-associated CMC.
Endogenous CD20 in Raji cells were knocked down using On-Targetplus SMARTpool siRNA against CD20 (MS4A1), which consisted of 4 siRNA duplexes. Cell lysates were prepared for (A) Western blot analysis to detect endogenous expression of CD20. (B) Quantification of Western blot analysis by Image J software. (C) Comparison of surface CD20 density and rituximab-associated CMCin Raji and Raji2R.
Mechanisms that regulate CD20 expression in RRCL
Subsequently, we investigated potential mechanisms responsible for the global downregulation of CD20 in RRCL. Previously we demonstrated that the repeated exposure of lymphoma cells to rituximab lead to a decrease in CD20 mRNA (9). In contrast to what has been previously reported (12), we did not find mutations in the CD20 gene to explain changes observed at the mRNA and protein level (data not shown).
Messenger RNA half-life is similar between RSCL and RRCL
CD20 mRNA is a balance between transcription and decay rates. However, using actinomycin-D to stop endogenous gene transcription, we did not observe differences in the mRNA half-life of CD20 between Raji and Raji2R or Raji4RH cells (data not shown).
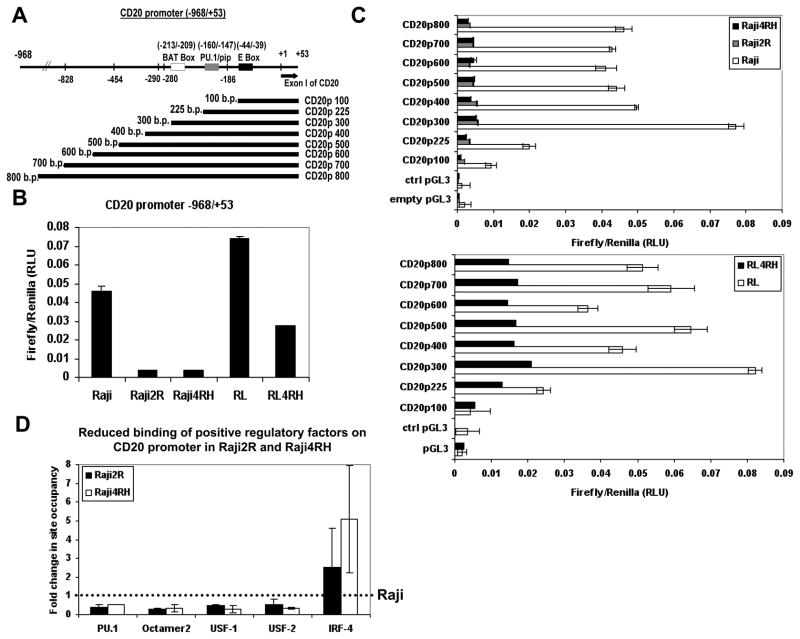
RRCL exhibit a decrease in the CD20 promoteractivity when compared to RSCL
We next tested the possibility that reduced CD20 promoter activity was responsible for lower CD20 mRNA in the RRCL. No mutations were found in the gene sequence of the cloned CD20 promoter (−968/+53) from each RRCL (data not shown), indicating that lower CD20 mRNA levelsin RRCLwas not due to the mutation of the CD20 promoter. On the other hand, a relatively lower CD20 promoter activity in RRCL was observed (Figure 4B), suggesting that a lower CD20 promoter activity in RRCL might be a contributing factor to reduced CD20 expression in rituximab-resistant lymphomas.
Figure 4. Reduced CD20 promoter activity was observed in the rituximab-resistant cell lines.
(A) Schematic diagram of individual CD20 promoter fragments cloned. 5×106 RSCL and RRCL were co-transfected with 2μg of the one individual CD20 promoter-carrying pGL3 plasmid (firefly RLU) and 0.1 μg of pEF-RL control vector (renilla RLU). 48 hours post-transfection, cells were harvested and luciferase activity was measured. Individual promoter activity from each CD20 promoter fragment was calculated by normalizing firefly RLU to renilla RLU. (B) CD20 promoter activity was compared between RSCL (Raji and RL) and RRCL (Raji2R, Raji4RH, and RL4RH) cells. (C) CD20 promoter activity of individual CD20 promoter fragments was compared between RSCL and RRCL. (D) Chromatin Immunoprecipitation study was performed to compare the interaction status of PU.1, IRF-4, Oct-2, USF-1, and USF-2 on CD20 promoter in RSCL (Raji) and RRCL (Raji2R and Raji4RH). Sonicated samples from Raji, Raji2R and Raji4RH cells cross-linked in 1% formaldehyde were immunoprecipitated with the indicated antibodies, as well as isotype IgG antibodies. 10% of lysate was saved as total DNA input. DNA isolated from total DNA input and immunoprecipitated materials were quantified by quantitative-PCR. Fold change in site occupancy was calculated from Ct value generated by quantitative-PCR per instruction from SuperArray ChIP-qPCR Data analysis from SABiosciences.
Three major positive-regulatory binding sites for CD20 promoter have been identified for positive regulatory factors such as USF (which binds to E-box), PU.1/IRF4 and Oct-2(18–21); On the other hand, negative regulators remain unknown. To further dissect the underlying cause for lower CD20 promoter activity in RRCL, we investigated if a particular region of the CD20 promoter was responsible for the overall decrease in CD20 promoter activity observed in RRCL. Different CD20 promoter fragments encompassing various lengths of the CD20 promoter were cloned into pGL3 vector and co-transfected with pEF-RL (Figure 4A). Differences in promoter activity between the constructs transfected into RSCL and RRCL demonstrated that all the 3 positive-regulatory sites on CD20 promoter (E-box, PU.1/pip site, and BAT box) were required for its maximum activity. CD20 promoter activity significantly decreased as the CD20 promoter constructs increased from 300 to 400 pairs, suggesting the existence of a potential negative regulatory site present within this region of the CD20 promoter (Figure 4C).
When CD20 promoter activity of each construct was compared between RSCL and RRCL, we observed decreased CD20 promoter activity in RRCL regardless of the CD20 promoter fragment analyzed (Figure 4C). However, the analysis of changes in expression of USF, PU.1, IRF-4 and Oct-2 between RSCL and RRCL revealed no significant changes as determined by Western blotting (data not shown). As we further compared the association status of these transcription factors with CD20 promoter in RSCL and RRCL by ChIP assay, we observed an approximately 50% reduction in binding of PU.1 (but not IRF-4), Oct-2, USF-1 and USF-2 to CD20 promoter in both Raji2R and Raji4RH, as compared to Raji (Figure 4D). Our findings suggest alternative mechanisms, such as the methylation status of the promoter affecting the binding of positive transcription factors, and acetylation of known transcription factors that regulated CD20 promoter activity.
CD20 promoter activity and surface CD20 expression can be enhanced by IL-4 in RRCL and in primary tumor cells derived from patients with NHL-B lymphoma
The use of IL-4 has been shown to modulate the level of CD20 expression in malignant cells isolated from patients with CLL (22). Subsequently, we investigated whether CD20 expression level in RRCL could be modulated by IL-4. Of interest, total and surface CD20 expression increased in RRCL following IL-4 exposure in vitro (Figure 5A).
Figure 5. IL-4-associated increase in intracellular and surface CD20 is associated with enhanced CD20 promoter activity and an increase in rituximab-associated CMC.
(A) Cells were incubated with IL-4 (5ng/ml) and harvested at indicated time points for total CD20 expression by Western blot analysis and surface CD20 expression by flow cytometry. Cells were incubated with RPMI 1640-10 media alone, or with 5ng/ml IL-4 for 48 hours before cells were: (B) lysed for CD20 promoter activity analysis in RRCL by luciferase assay; or (C) used to analyze rituximab-associated CMC by 51Cr release assay. (D) B-NHL cells were isolated by negative selection and treated with 5ng/ml of IL-4 for 48 hours before subjected to flow cytometric analysis for CD20 expression and 51Cr release assay for rituximab-associated CMC.
We further investigated whether IL-4-induced higher CD20 expression in RRCL was associated with effects on CD20 promoter activity. RRCL co-transfected CD20 promoter-fragments carrying vector and pEF-RL were incubated with IL-4 or control for 48-hours. In vitro exposure of RRCL to IL-4 increased the CD20 promoter activity in RRCL (Figure 5B). Moreover, 51Cr release assay demonstrated a 1.5 to 2-fold increase in rituximab- CMC in RRCL exposed to IL-4(Figure 5C). While this result suggests that IL-4is an attractive strategy to improve CD20 antigen expression and rituximab activity in RRCL, the absolute percentage of rituximab-CMC following IL-4 exposure was only 1/10th of rituximab anti-tumor activity observed in RSCL. As noted before, it is possible that a specific CD20 surface expression threshold is required to fully restore rituximab sensitivity (Figure 2B).
We further investigated if IL-4 could modulate CD20 expression and rituximab activity in vitro in primary tumor cells isolated from patients with B-cell lymphoma (Figure 5D). Ex vivo IL-4 exposure of primary tumor cells isolated from six lymphoma patients induced CD20 expression that correlated with an improvement in rituximab-CMC. Alternatively, IL-4 did not have an effect on CD20 expression and rituximab-CMC in seven other NHL patient specimens. As patients represented different tumor histologies and disease status (e.g. de novo vs. relapsed/refractory), it is difficult to conclude any strong correlation between the observed effects of IL-4and our preliminary results(Supplemental Table S2).
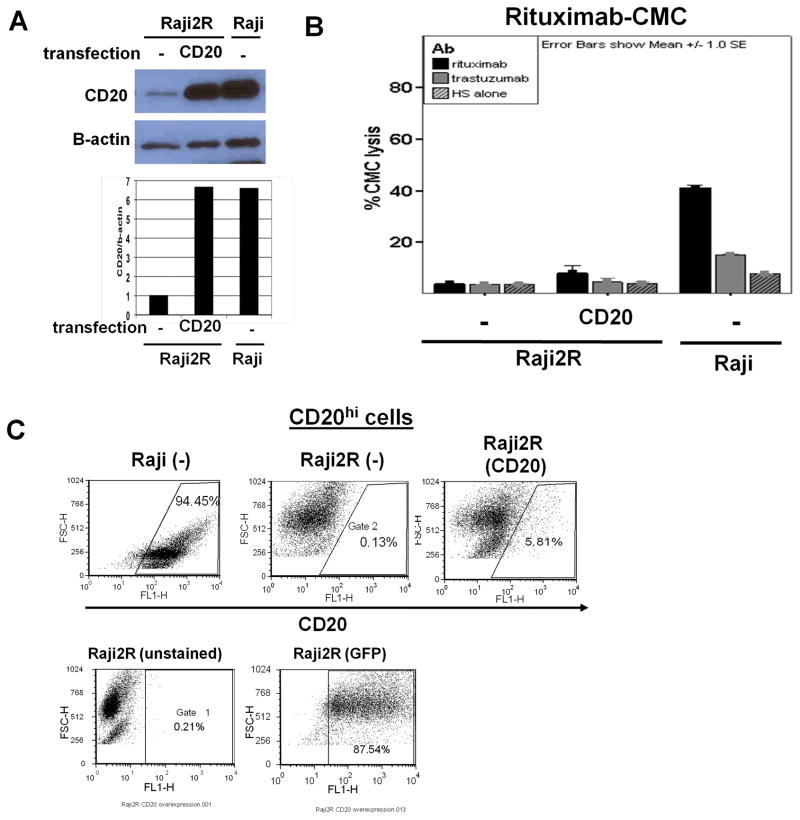
Re-expression of CD20 expression level in RRCLrevealed a potential defect in CD20 protein transport to the cell surface
Finally, we force-expressed CD20 in RRCL in an attempt to restore rituximab activity. CD20-carrying BCMGSneo vector was transfected into RRCL. Subsequently, CD20 expression was determined by Western blotting, quantitative flow cytometry and correlated to rituximab activity. Western blot analysis showed that CD20-BCMGSneo-transfected Raji2R cells over-expressed cytoplasmic CD20 (6-fold increase) to similar levels as seen in Raji cells (Figure 6A). However, 51Cr-release assay showed no significant increase in rituximab-CMC (<5% increase in CMC) (Figure 6B). Of interest, while CD20 forced expression was “successful” in Raji2R as determined by Western blotting, no significant changes in surface CD20 antigen were noted in CD20BCMGSneo-transfected Raji2R cells. Detailed flow cytometric analysis showed that the majority of the CD20-transfected Raji2R failed to express CD20 on the cell surface despite adequate transfection and CD20 synthesis in the cytoplasmic compartment (Figure 6C). Hela and Jurkat T cells, which do not express endogenous CD20, and Raji were used as controls to verify whether this transport defect was RRCL-specific (Figure S2). Unlike Raji2R, which expressed maximum total CD20 by Western blotting 4 days post-transfection, Hela cells had maximum CD20 expression on day 2, while Jurkat T cells failed to express CD20 up to 2days post-transfection. Detailed analysis of the percentage of cells expressing surface CD20 by Flow Cytometry showed that Hela cells had higher percentages of cells expressing surface CD20 than Raji2R (38% vs. 2%). We were unable to further increase CD20 expression in Raji, likely due to an inability to further upregulate CD20 expression regulatory factors, which were operating at “maximum” levels (Figure S2). This finding suggests the existence of a defect in the transport of CD20 onto the cell surface in RRCL.
Figure 6. CD20 overexpression in RRCL indicated a potential defect in CD20 transport to cell surface.
20×106 Raji2R, Hela or Jurkat (data not shown) Tcells were transfected with 4μg of CD20-BCMGSneo vector by Amaxa Nucleofection. On day 2 or 4, cells were harvested for CD20 expression analysis by (A) Western blot. CD20 and β-actin control expression on Western blot were quantified by Image Jsoftware. (B) 51Crrelease assay was performed for rituximab-associated CMC. (C) Flow cytometry for percentage of cells with CD20 overexpression in mock-transfected and CD20-BCMGSneo vector-transfected Raji2R cells. Raji cells were used as positive control. Transfection efficiency was demonstrated post pmaxGFP transfection by GFP-expressing population.
In summary, our data demonstrates that surface CD20 antigen expression plays a role in rituximab-CMC. We provide data from several experiments studying the possible mechanisms leading to CD20 downregulation in RRCL and the impact that CD20 expression has on rituximab activity. While reduced CD20 promoter activity appeared to account forthe relatively lower CD20 mRNA expression, an altered CD20 protein transport may also contribute to the global downregulation of CD20 in RRCL. Our data stresses the complexity of the mechanisms affecting rituximab activity in B-cell lymphoma.
DISCUSSION
The development of rituximab-resistance is an emerging clinical problem and its frequency may increase as a consequence of repeated exposure to rituximab, especially over prolonged periods of time (e.g. maintenance programs) in B-cell lymphoma. A formal study of the frequency of phenotypic changes occurring in rituximab resistant lymphoma is needed to better understand this clinical concern. The Rituximab Extended Schedule or Retreatment (RESORT) trial is an ongoing ECOG clinical study aiming to define the optimal duration of treatment with rituximab single agent in patients with indolent CD20+ B-cell lymphoma and low tumor burden (23). This study will potentially provide important information on the incidence of rituximab resistance and the phenotypic changes occurring in rituximab-resistant low-grade lymphomas.
Downregulation of CD20 has been observed in patients with relapsed/refractory B-cell lymphoma following prior rituximab therapy (5–7). Based on initial studies, it appears that CD20 surface expression is a limiting step for rituximab-CMC, but not ADCC (11). In our model, we observed that the repeated exposure of RSCL to rituximab lead to CD20 downregulation and a decrease in rituximab biological activity as determined by standard apoptosis, CMC and ADCC assays (9). From the laboratory standpoint, CMC assays are easily reproducible with less confounding variables than ADCC or apoptosis assays and thus are a valuable tool with which to study the activity of rituximab in vitro. Detailed examination of cell passages demonstrated an incremental loss in CD20 surface expression and corresponding rituximab-CMC activity. Furthermore, we defined a CD20 surface expression “threshold” level necessary for effective rituximab-CMC.
Given the multiple mechanisms of action of rituximab (i.e. CMC, ADCC and apoptosis), the overall clinical relevance of variable CD20 surface antigen expression on lymphoma cells in vivo is difficult to ascertain. While CD20 levels appear to play an important role in rituximab-CMC, it appears to be less relevant for rituximab-ADCC. Our group and other investigators have demonstrated that immune effector cells and ADCC play a pivotal role in the anti-tumor activity of mAbs, including rituximab (25, 26). RRCL exhibited not only resistance to rituximab-CMC, but also rituximab-mediated calcium influx, apoptosis and ADCC (9, 10). It is possible that a gradual decrease in CD20 antigen expression may not affect rituximab-ADCC at early stages, but once CD20 levels drop below a certain “threshold” (i.e. 150 CD20-FITC per μm2), the capacity of the innate immune cells to kill rituximab-coated lymphoma cells may also be significantly impaired. Our findings are clinically relevant since novelmAbs capable of effectively inducing anti-CD20 CMC or ADCC at even low CD20 levels may be an attractive therapeutic strategy by which to kill rituximab-resistant lymphoma.
Cancer cells develop multiple phenotypic changes in response to selective pressure; therefore, additional alterations, besides CD20 downregulation likely contribute to rituximab resistance. Our data suggests that CD20 expression plays an important role, but also suggests the existence of additional factors affecting rituximab-associated biological activity (e.g. CIPs). Pre-resistant cell passages demonstrated a gradual loss of CD20 expression that directly correlated with a gradual loss of rituximab-CMC and single cell clones obtained from RSCL (but not RRCL) exhibited a linear correlation between CD20 expression and rituximab-CMC.
We previously identified additional phenotypic changes in RRCL that can impair rituximab activity. Similar to the findings from Bonavida’s group (27), we demonstrated that RRCL had a deregulation of the ubiquitin-proteasome system and in Bcl-2 family members leading to cross-resistance to chemotherapy agents (9, 10). In addition, we demonstrated differential expressions of CIPs (CD55 and CD59) within the RSCL cell populations, which together with surface CD20 modulated rituximab-associated activity. This observation could potentially extrapolate to a “selective elimination” of rituximab-sensitive clones (CD20hi, CD55low, CD59low), finally leading to RRCL with altered phenotypes (CD20low, CD55hi, CD59hi). Additional studies are necessary to define what the relative contribution of each of these factors is to rituximab resistance in B-cell lymphoma.
The regulation of CD20 expression in normal and malignant B-cells is poorly-defined. A limiting factor in understanding CD20 regulation is the absence of immunological defects in CD20-knockout mouse models(28). Recently, a group of investigators reported the case of a pediatric patient lacking CD20 expression in B-cells showing signs/symptoms compatible with immunodeficiency which has alerted the scientific community to renew studies regarding the function and regulation of CD20 in normal B-cell differentiation(29).
In contrast to the report from Terui et al., which demonstrated a point mutation in CD20 gene in some rituximab-resistant lymphoma patients (12), we found no CD20 gene mutations affecting the primary structure of CD20. Moreover, no changes in the CD20 mRNA stability were found in RRCL. On the other hand, we identified two additional mechanisms that negatively regulate CD20 in RRCL and lead to a decreased rituximab activity: a lower CD20 promoter activity and/or defect impairing the transport of CD20 to the cell surface.
ChIP analysis showed 50% reduction in association of PU.1, USF, and Oct-2 with CD20 promoter in RRCL, potentially explained reduced CD20 promoter activity in RRCL. “Pharmacologic strategies” to increase CD20 promoter activity can potentially lead to clinically applicable strategies to restore rituximab-sensitivity in rituximab-resistant lymphoma. The use of IL-4 has been shown to modulate CD20 expression in various B-cell lymphoma cell lines(22). In our RRCL, in vitro exposure to IL-4 increased CD20 promoter activity, CD20 antigen expression and enhanced rituximab activity. Similar findings were found in a subset of primary tumor cells isolated from patients with B-cell lymphoma. IL-4 mediated increase in CD20 promoter activity was observed in CD20 promoter fragments containing positive or negative regulatory binding sites, suggesting that IL-4 effects may be mediated by altering the binding status of these regulatory transcription factors. Ongoing studies are seeking to study differences in the methylation status of the CD20 promoter between RSCL and RRCL that could affect the binding of transcription factors and thereby CD20 expression.
Surface protein transport can be regulated by general or specific factors. These include: chaperone proteins that ensure properprotein folding (30–34); the presence of endoplasmic reticulum (ER) retention signals which need to be cleaved before protein can be transported out of ER (35, 36); post-translational modification such as glycosylation and phosphorylation which ensure proper folding and structure stability (37); and/or protein-specific accessory proteins in the ER before surface transport(36, 38, 39). In B-cells, the requirement for immunoglobulin M (IgM) surface protein expression has been well-defined (36, 40, 41). Specifically, the presence of the heterodimers of the co-receptors, Igα and Igβ, has been found to berequired for IgM to be transported from ER to the surface transport of normal B-cells(36). Similarly, the proper assembly of MHC Class I and II molecules requires calnexin and calreticulinaccessory protein(39, 42, 43). It is unknown whether any accessory proteins are required for CD20 surface expression. Thus, future research is focused in delineating factors responsible for CD20 transport defects.
In summary, our data suggests that several factors contribute to acquired resistance to rituximab. For the first time we link an abnormal CD20 promoter activity and/or a defect in the Golgi-to-surface protein transport with a decrease in CD20 expression and resistance to rituximab in B-cell lymphoma. While CD20 antigen expression plays a role in rituximab-resistance in B-cell lymphoma, additional factors likely affect anti-CD20 activity (e.g. differences in Bcl-2 family proteins, complement inhibitory proteins, etc.). Based on our pre-clinical studies, it appears that surface CD20 expression may play a role in rituximab sensitivity at early stages of resistance. As lymphoma cells gain additional genetic/phenotypic changes, the role of CD20 expression in rituximab biological activity may be less relevant at later stages of rituximab resistance. A better understanding in the mechanisms that regulate CD20 antigen expression have the potential to someday contribute to the development of novel therapeutic strategies to overcome rituximab-resistance in patients with B-cell lymphoma.
Supplementary Material
Statement of Translational Relevance.
Rituximab resistance is a clinical problem that has arose as a result of the incorporation of rituximab in the management of B-cell lymphoma. The incidence of rituximab resistance may significantly increase in the near future following the adoption of prolonged rituximab maintenance schedules by a significant percentage of practicing oncologists. CD20 down-regulation has been reported to be associated with a decrease in rituximab sensitivity in B-NHL. However, many other alterations, such as altered pro-versus anti-apoptotic molecules, have been reported to contribute to rituximab resistance. It is currently unknown whether CD20 expression is the “major” factor responsible for the decrease in rituximab’s biological activity. Our current work investigated, by several approaches, the contribution of CD20 expression to rituximab sensitivity in a well-defined rituximab resistant cell line model. We have identified reduced CD20 promoter activity and a defect in CD20 transport as two novel mechanisms responsible for CD20 down-regulation in RRCL.
Acknowledgments
Grant support: This work was supported, in part, by grants from the National Cancer Institute (Targeting the proteasome to overcome therapy resistance; Sponsor Award number 5RO1CA136907-02); The Eugene and Connie Corasanti Lymphoma Research Fund; and the New York State Empire Clinical Research Investigator Program (ECRIP)
We thank Dr. Julie P. Deans for sharing GST77 antibody and CD20-BCMGSneo vector; PhD committee members: Drs. Kelvin Lee, Sandra Gollnick, and Lee Ann Garrett-Sinha for their input; and additional members of the Department of Immunology at Roswell Park Cancer Institute for their suggestions.
Footnotes
Conflict-of-interest disclosure: M.S.C. has served on advisory boards and has received clinical research support from Genentech, BiogenIdec, and GSK Pharmaceuticals. The remaining authors declare no competing financial interest.
References
- 1.Calvo-Villas JM, Martin A, Conde E, Pascual A, Heras I, Varela R, et al. Effect of addition of rituximab to salvage chemotherapy on outcome of patients with diffuse large B-cell lymphoma relapsing after an autologous stem-cell transplantation. Ann Oncol. 2010;21:1891–7. doi: 10.1093/annonc/mdq035. [DOI] [PubMed] [Google Scholar]
- 2.Gisselbrecht C, Glass B, Mounier N, Singh Gill D, Linch DC, Trneny M, et al. Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28:4184–90. doi: 10.1200/JCO.2010.28.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin A, Conde E, Arnan M, Canales MA, Deben G, Sancho JM, et al. R-ESHAP as salvage therapy for patients with relapsed or refractory diffuse large B-cell lymphoma: the influence of prior exposure to rituximab on outcome. A GEL/TAMO study. Haematologica. 2008;93:1829–36. doi: 10.3324/haematol.13440. [DOI] [PubMed] [Google Scholar]
- 4.Dalle S, Dumontet C. Rituximab: mechanism of action and resistance. Bulletin du cancer. 2007;94:198–202. [PubMed] [Google Scholar]
- 5.Davis TA, Czerwinski DK, Levy R. Therapy of B-cell lymphoma with anti-CD20 antibodies can result in the loss of CD20 antigen expression. Clin Cancer Res. 1999;5:611–5. [PubMed] [Google Scholar]
- 6.Jilani I, O’Brien S, Manshuri T, Thomas DA, Thomazy VA, Imam M, et al. Transient down-modulation of CD20 by rituximab in patients with chronic lymphocytic leukemia. Blood. 2003;102:3514–20. doi: 10.1182/blood-2003-01-0055. [DOI] [PubMed] [Google Scholar]
- 7.Rawal YB, Nuovo GJ, Frambach GE, Porcu P, Baiocchi RA, Magro CM. The absence of CD20 messenger RNA in recurrent cutaneous B-cell lymphoma following rituximab therapy. Journal of cutaneous pathology. 2005;32:616–21. doi: 10.1111/j.0303-6987.2005.00305.x. [DOI] [PubMed] [Google Scholar]
- 8.Haidar JH, Shamseddine A, Salem Z, Mrad YA, Nasr MR, Zaatari G, et al. Loss of CD20 expression in relapsed lymphomas after rituximab therapy. European journal of haematology. 2003;70:330–2. doi: 10.1034/j.1600-0609.2003.00007.x. [DOI] [PubMed] [Google Scholar]
- 9.Czuczman MS, Olejniczak S, Gowda A, Kotowski A, Binder A, Kaur H, et al. Acquirement of rituximab resistance in lymphoma cell lines is associated with both global CD20 gene and protein down-regulation regulated at the pretranscriptional and posttranscriptional levels. Clin Cancer Res. 2008;14:1561–70. doi: 10.1158/1078-0432.CCR-07-1254. [DOI] [PubMed] [Google Scholar]
- 10.Olejniczak SH, Hernandez-Ilizaliturri FJ, Clements JL, Czuczman MS. Acquired resistance to rituximab is associated with chemotherapy resistance resulting from decreased Bax and Bak expression. Clin Cancer Res. 2008;14:1550–60. doi: 10.1158/1078-0432.CCR-07-1255. [DOI] [PubMed] [Google Scholar]
- 11.van Meerten T, van Rijn RS, Hol S, Hagenbeek A, Ebeling SB. Complement-induced cell death by rituximab depends on CD20 expression level and acts complementary to antibody-dependent cellular cytotoxicity. Clin Cancer Res. 2006;12:4027–35. doi: 10.1158/1078-0432.CCR-06-0066. [DOI] [PubMed] [Google Scholar]
- 12.Terui Y, Mishima Y, Sugimura N, Kojima K, Sakurai T, Kuniyoshi R, et al. Identification of CD20 C-terminal deletion mutations associated with loss of CD20 expression in non-Hodgkin’s lymphoma. Clin Cancer Res. 2009;15:2523–30. doi: 10.1158/1078-0432.CCR-08-1403. [DOI] [PubMed] [Google Scholar]
- 13.Beers SA, French RR, Chan HT, Lim SH, Jarrett TC, Vidal RM, et al. Antigenic modulation limits the efficacy of anti-CD20 antibodies: implications for antibody selection. Blood. 2010;115:5191–201. doi: 10.1182/blood-2010-01-263533. [DOI] [PubMed] [Google Scholar]
- 14.Beum PV, Kennedy AD, Williams ME, Lindorfer MA, Taylor RP. The shaving reaction: rituximab/CD20 complexes are removed from mantle cell lymphoma and chronic lymphocytic leukemia cells by THP-1 monocytes. J Immunol. 2006;176:2600–9. doi: 10.4049/jimmunol.176.4.2600. [DOI] [PubMed] [Google Scholar]
- 15.Polyak MJ, Tailor SH, Deans JP. Identification of a cytoplasmic region of CD20 required for its redistribution to a detergent-insoluble membrane compartment. J Immunol. 1998;161:3242–8. [PubMed] [Google Scholar]
- 16.Deans JP, Kalt L, Ledbetter JA, Schieven GL, Bolen JB, Johnson P. Association of 75/80-kDa phosphoproteins and the tyrosine kinases Lyn, Fyn, and Lck with the B cell molecule CD20. Evidence against involvement of the cytoplasmic regions of CD20. The Journal of biological chemistry. 1995;270:22632–8. doi: 10.1074/jbc.270.38.22632. [DOI] [PubMed] [Google Scholar]
- 17.Cruz RI, Hernandez-Ilizaliturri FJ, Olejniczak S, Deeb G, Knight J, Wallace P, et al. CD52 over-expression affects rituximab-associated complement-mediated cytotoxicity but not antibody-dependent cellular cytotoxicity: preclinical evidence that targeting CD52 with alemtuzumab may reverse acquired resistance to rituximab in non-Hodgkin lymphoma. Leukemia & lymphoma. 2007;48:2424–36. doi: 10.1080/10428190701647879. [DOI] [PubMed] [Google Scholar]
- 18.Mankai A, Bordron A, Renaudineau Y, Martins-Carvalho C, Takahashi S, Ghedira I, et al. Purine-rich box-1-mediated reduced expression of CD20 alters rituximab-induced lysis of chronic lymphocytic leukemia B cells. Cancer research. 2008;68:7512–9. doi: 10.1158/0008-5472.CAN-07-6446. [DOI] [PubMed] [Google Scholar]
- 19.Himmelmann A, Riva A, Wilson GL, Lucas BP, Thevenin C, Kehrl JH. PU. 1/Pip and basic helix loop helix zipper transcription factors interact with binding sites in the CD20 promoter to help confer lineage- and stage-specific expression of CD20 in B lymphocytes. Blood. 1997;90:3984–95. [PubMed] [Google Scholar]
- 20.Thevenin C, Lucas BP, Kozlow EJ, Kehrl JH. Cell type- and stage-specific expression of the CD20/B1 antigen correlates with the activity of a diverged octamer DNA motif present in its promoter. The Journal of biological chemistry. 1993;268:5949–56. [PubMed] [Google Scholar]
- 21.Thevenin C, Rieckmann P, Kozlow EJ, Kehrl JH. Identification of a diverged octamer binding site important in the B cell-specific expression of the CD20 gene. Transactions of the Association of American Physicians. 1992;105:15–24. [PubMed] [Google Scholar]
- 22.Venugopal P, Sivaraman S, Huang XK, Nayini J, Gregory SA, Preisler HD. Effects of cytokines on CD20 antigen expression on tumor cells from patients with chronic lymphocytic leukemia. Leukemia research. 2000;24:411–5. doi: 10.1016/s0145-2126(99)00206-4. [DOI] [PubMed] [Google Scholar]
- 23.Kahl BS. Eastern Cooperative Oncology Group 4402: Rituximab Extended Schedule or Retreatment Trial (RESORT) Clin Lymphoma Myeloma. 2006;6:423–6. doi: 10.3816/CLM.2006.n.024. [DOI] [PubMed] [Google Scholar]
- 24.Foran JM, Norton AJ, Micallef IN, Taussig DC, Amess JA, Rohatiner AZ, et al. Loss of CD20 expression following treatment with rituximab (chimaeric monoclonal anti-CD20): a retrospective cohort analysis. British journal of haematology. 2001;114:881–3. doi: 10.1046/j.1365-2141.2001.03019.x. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Ilizaliturri FJ, Jupudy V, Ostberg J, Oflazoglu E, Huberman A, Repasky E, et al. Neutrophils contribute to the biological antitumor activity of rituximab in a non-Hodgkin’s lymphoma severe combined immunodeficiency mouse model. Clin Cancer Res. 2003;9:5866–73. [PubMed] [Google Scholar]
- 26.Lefebvre ML, Krause SW, Salcedo M, Nardin A. Ex vivo-activated human macrophages kill chronic lymphocytic leukemia cells in the presence of rituximab: mechanism of antibody-dependent cellular cytotoxicity and impact of human serum. J Immunother. 2006;29:388–97. doi: 10.1097/01.cji.0000203081.43235.d7. [DOI] [PubMed] [Google Scholar]
- 27.Jazirehi AR, Vega MI, Bonavida B. Development of rituximab-resistant lymphoma clones with altered cell signaling and cross-resistance to chemotherapy. Cancer research. 2007;67:1270–81. doi: 10.1158/0008-5472.CAN-06-2184. [DOI] [PubMed] [Google Scholar]
- 28.Uchida J, Lee Y, Hasegawa M, Liang Y, Bradney A, Oliver JA, et al. Mouse CD20 expression and function. Int Immunol. 2004;16:119–29. doi: 10.1093/intimm/dxh009. [DOI] [PubMed] [Google Scholar]
- 29.Kuijpers TW, Bende RJ, Baars PA, Grummels A, Derks IA, Dolman KM, et al. CD20 deficiency in humans results in impaired T cell-independent antibody responses. The Journal of clinical investigation. 2010;120:214–22. doi: 10.1172/JCI40231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YJ, Inouye M. The intramolecular chaperone-mediated protein folding. Curr Opin Struct Biol. 2008;18:765–70. doi: 10.1016/j.sbi.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–49. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 32.Hartl FU. Principles of chaperone-mediated protein folding. Philos Trans R Soc Lond B Biol Sci. 1995;348:107–12. doi: 10.1098/rstb.1995.0051. [DOI] [PubMed] [Google Scholar]
- 33.Hubbard TJ, Sander C. The role of heat-shock and chaperone proteins in protein folding: possible molecular mechanisms. Protein Eng. 1991;4:711–7. doi: 10.1093/protein/4.7.711. [DOI] [PubMed] [Google Scholar]
- 34.Willardson BM, Howlett AC. Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell Signal. 2007;19:2417–27. doi: 10.1016/j.cellsig.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagny S, Cabanes-Macheteau M, Gillikin JW, Leborgne-Castel N, Lerouge P, Boston RS, et al. Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell. 2000;12:739–56. doi: 10.1105/tpc.12.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens TL, Blum JH, Foy SP, Matsuuchi L, DeFranco AL. A mutation of the mu transmembrane that disrupts endoplasmic reticulum retention. Effects on association with accessory proteins and signal transduction. J Immunol. 1994;152:4397–406. [PubMed] [Google Scholar]
- 37.Guan JL, Machamer CE, Rose JK. Glycosylation allows cell-surface transport of an anchored secretory protein. Cell. 1985;42:489–96. doi: 10.1016/0092-8674(85)90106-0. [DOI] [PubMed] [Google Scholar]
- 38.Spiliotis ET, Pentcheva T, Edidin M. Probing for membrane domains in the endoplasmic reticulum: retention and degradation of unassembled MHC class I molecules. Mol Biol Cell. 2002;13:1566–81. doi: 10.1091/mbc.01-07-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiliotis ET, Manley H, Osorio M, Zuniga C, Edidin M. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity. 2001;14:205. doi: 10.1016/s1074-7613(01)00102-9. [DOI] [PubMed] [Google Scholar]
- 40.Venkitaraman AR, Williams GT, Dariavach P, Neuberger MS. The B-cell antigen receptor of the five immunoglobulin classes. Nature. 1991;352:777–81. doi: 10.1038/352777a0. [DOI] [PubMed] [Google Scholar]
- 41.Polyak MJ, Li H, Shariat N, Deans JP. CD20 homo-oligomers physically associate with the B cell antigen receptor. Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. The Journal of biological chemistry. 2008;283:18545–52. doi: 10.1074/jbc.M800784200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Salter RD. Distinct patterns of folding and interactions with calnexin and calreticulin in human class I MHC proteins with altered N-glycosylation. J Immunol. 1998;160:831–7. [PubMed] [Google Scholar]
- 43.Anderson KS, Cresswell P. A role for calnexin (IP90) in the assembly of class II MHC molecules. EMBO J. 1994;13:675–82. doi: 10.1002/j.1460-2075.1994.tb06306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.