Abstract
The genetic association of the major histocompatibility complex (MHC) to rheumatoid arthritis risk has commonly been attributed to HLA-DRB1 alleles. Yet controversy persists about the causal variants in HLA-DRB1 and the presence of independent effects elsewhere in the MHC. Using existing genome-wide SNP data in 5,018 seropositive cases and 14,974 controls, we imputed and tested classical alleles and amino acid polymorphisms for HLA-A, B, C, DPA1, DPB1, DQA1, DQB1, and DRB1 along with 3,117 SNPs across the MHC. Conditional and haplotype analyses reveal that three amino acid positions (11, 71 and 74) in HLA-DRβ1, and single amino acid polymorphisms in HLA-B (position 9) and HLA-DPβ1 (position 9), all located in the peptide-binding grooves, almost completely explain the MHC association to disease risk. This study illustrates how imputation of functional variation from large reference panels can help fine-map association signals in the MHC.
Rheumatoid arthritis is a systemic autoimmune disease characterized by intra-articular inflammation1. About 70% of patients have antibodies against cyclic citrullinated peptide (CCP)2. Until now, the strong association of the MHC to anti-CCP disease3,4 has been explained by the presence of consensus amino acid sequences (QRRAA, RRRAA and QKRAA) spanning positions 70 through 74 in the β1 subunit of the HLA-DR molecule. The classical HLA-DRB1 haplotypes carrying these sequences define the “shared epitope” alleles5. The shared epitope association was historically defined by exploring structural differences between HLA-DRB1*04 alleles with allospecific T cell recognition6,7. These reagents focused attention on sequence determinants on the exposed alpha helical rim of the HLA-DR molecule where the shared epitope is located, but left allelic differences at the inaccessible base of the binding groove largely unexplored.
Despite serving as the foundation for rheumatoid arthritis genetic studies, the shared epitope hypothesis does not fully explain the association at DRB1; studies have suggested additional independent associations within the MHC outside DRB13,8–11. However, pinpointing those loci has been challenging, in part due to the complexity and cost of complete HLA genotyping and the broad linkage disequilibrium (LD) characteristic of the MHC12.
To define the association across the region and identify functional and potentially causal variants, we obtained SNP genotype data for 19,992 anti-CCP positive rheumatoid arthritis cases and controls of European descent from six independent genome-wide data sets (Supplementary Table 1)13. We used a large reference panel of 2,767 individuals of European descent14 to impute classical alleles genotypes for HLA-A, HLA-B, HLA-C, DPA1, DPB1, DQA1, DQB1, and DRB1, their corresponding amino acid sequences, and SNPs within the MHC15. In total, we tested 99 classical 2-digit alleles, 164 classical 4-digit alleles, 372 polymorphic amino acid positions, and 3,117 SNPs across the region for association with logistic regression. To control for population stratification, we included as covariates the first five principal components from genome-wide SNP genotypes for each of the six data sets16 (λgc=1.06, see Supplementary Note).
First, to assess imputation accuracy, we compared imputed DRB1 classical alleles to genotyped alleles for a subset of 1,403 individuals from two data sets genotyped to 4-digit resolution (Supplementary Table 2A). Imputations were 95.8% accurate for alleles at 2-digit resolution and 84.0% at 4-digit resolution (see Supplementary Note). We observed high accuracy in frequency estimates and imputation quality for alleles with >2.5% frequency in the reference set (Supplementary Figure 1A). We observed similar accuracy at four other classical loci in a subset of 1958 Birth Cohort samples that were part of the WTCCC controls (Supplementary Table 2B,C). We note that the WTCCC samples have the sparsest SNP coverage across the MHC and that these accuracies probably represent a lower bound (Supplementary Figure 1B).
Next, we compared allelic odds ratios of imputed DRB1 haplotypes in our data with recently reported allelic odds ratios for DRB1 haplotypes in a large study of anti-CCP positive rheumatoid arthritis17. Except the rare *11:02/*11:03 haplotype (<1% frequency), effect sizes from our study were entirely consistent for each of the DRB1 classical haplotypes (Supplementary Figure 2, Supplementary Table 3).
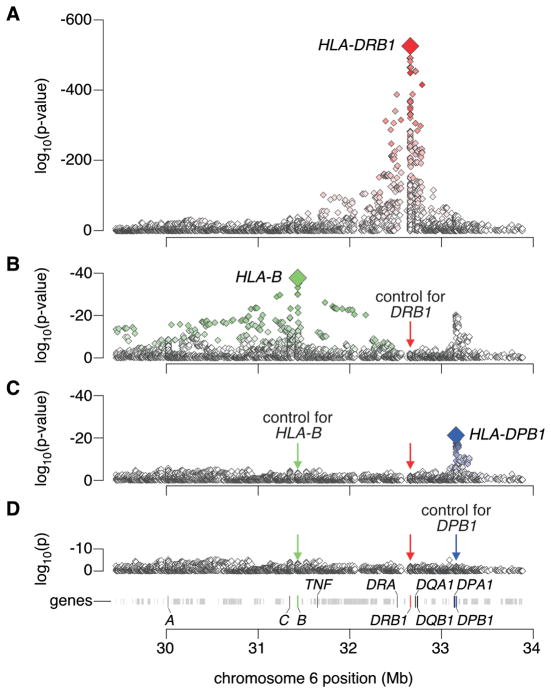
Having demonstrated the validity of our analytic approach, we tested SNPs and HLA alleles across the MHC for association to rheumatoid arthritis. The most significant allele was the A nucleotide at rs17878703, a quadrallelic SNP in the second nucleotide of DRB1 codon 11 (odds ratio (OR) =3.7, p<10−526; Figure 1, Supplementary Table 4). This allele codes for Val-11 or Leu-11 in DRβ1. Thus, the strongest MHC signal mapped to amino acid 11 of DRβ1, and not any of the shared epitope positions (amino acids 70-74).
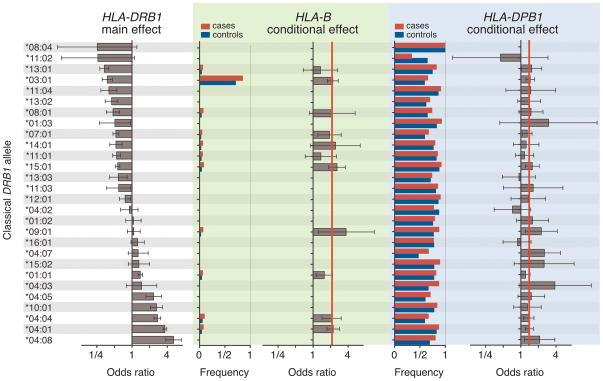
Figure 1. Association tests within the MHC to rheumatoid arthritis.
(A) The major genetic determinants of rheumatoid arthritis risk map to the HLA-DRB1 gene. (B) Subsequent conditional analyses controlling for all classical HLA-DRB1 alleles reveal an independent association at HLA-B corresponding to the B*08 allele or Asp-9 in the protein. (C) Subsequent analyses that condition on HLA-DRB1 alleles and HLA-B*08 reveal an independent association for the HLA-DPβ1 Phe-9 variant. (D) Upon controlling for HLA-DRB1, HLA-B Asp-9 and HLA-DPβ1 Phe-9, no significant association signal is observed.
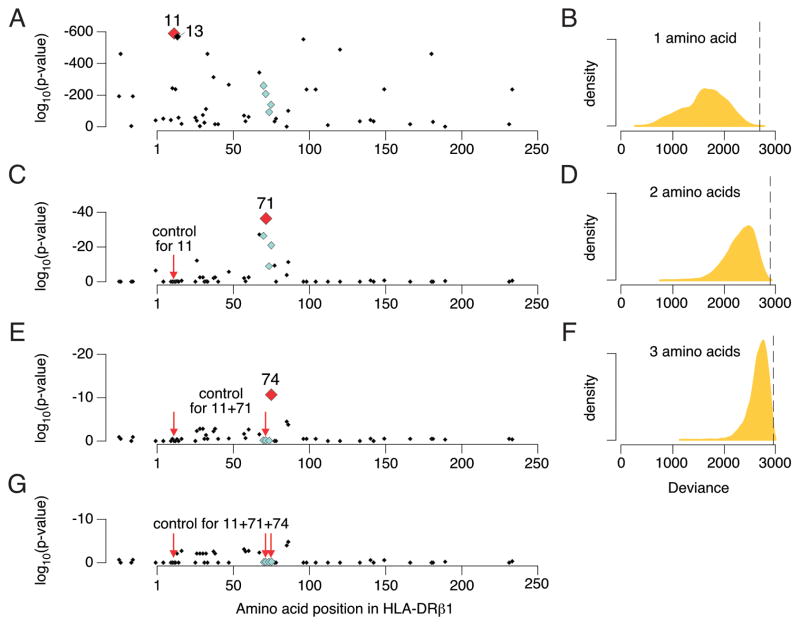
We then tested each of the amino acid positions within DRβ1 for association, by grouping classical DRB1 haplotypes according to the specific amino acid carried at each position (Supplementary Table 5). Amino acid position 11 demonstrated the strongest association (p<10−581; Figure 2). Of the six possible amino acids at this position, the aliphatic residues Val-11 (OR=3.8) and Leu-11 (OR=1.3) confer high risk, whereas other residues confer less risk (Figure 3, Supplementary Table 4). In fact, the polar Ser-11 residue is highly protective against disease (OR=0.38). Amino acid position 13 was similarly statistically significant (p<10−574); its six alleles are in tight LD with those at position 11. Conditioning on position 11 eliminated the effect of position 13 (p=0.57), but conditioning on position 13 did not eliminate the effect of position 11 (p=3.5 × 10−8). While these results favor position 11 over 13, the tight LD between them makes it is difficult to unambiguously assign causality to one position at the exclusion of the other (Table 1). After conditioning on the shared epitope haplotypes amino acid position 11 and 13 remained highly significantly associated (p<10−70 and p<10−63 respectively), and more strongly associated than other amino acid positions.
Figure 2. Association results for amino acids in HLA-DRβ1.
(A) Amino acid position 11 represents the strongest association with rheumatoid arthritis (p < 10−581), followed by position 13 (p < 10−574). Shared epitope positions (70 through 74) are indicated by light-blue diamonds. (B) Distribution of deviance in 10,000 permutations of amino acid sequences across classical HLA-DRB1 alleles, where deviance is calculated as −2 times the log-likelihood for the best amino acid position. The vertical dashed line indicates the deviance for position 11 in the actual data (p = 0.0002). (C) Controlling for position 11, position 71 is significantly associated with rheumatoid arthritis (p = 5.6 × 10−38). (D) Deviance of the best two amino acid positions in 10,000 permutations. The vertical dashed line indicates the deviance for positions 11 and 71 in the actual data (p = 0.0002). (E) Controlling for positions 11 and 71, position 74 is significantly associated with rheumatoid arthritis (p = 1.5 × 10−11). (F) Deviance of the best three amino acid positions in 10,000 permutations. The vertical dashed line indicates the deviance for positions 11, 71 and 74 in the actual data (p = 0.004). (G) After controlling for positions 11, 71 and 74, no amino acid position is significant (p > 8 × 10−4).
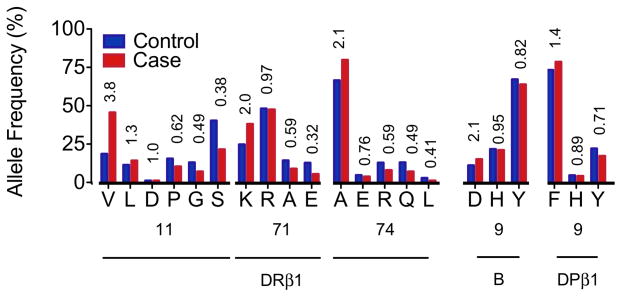
Figure 3. Effect of individual amino acids within HLA proteins.
For amino acid positions 11, 71 and 74 in HLA-DRβ1, 9 in HLA-B, and 9 in HLA-DPβ1, the allele frequencies in cases (red) and controls (blue) are plotted and univariate odds ratios listed. The HLA-B and DPβ1 effects are adjusted for HLA-DRB1 alleles.
Table 1.
Effect Estimates for the Five Amino Acids Associated with RA Risk.
Estimated effects for haplotypes of HLA-DRB1, HLA-B and HLA-DPB1.
Classical alleles of HLA-DRB1 are grouped based on amino acid residues at positions 11 (or 13), 71, and 74 within DRβ1. We have bolded the classical shared epitope alleles. For each haplotype, the multivariate effect is given as an odds ratio, taking the most frequent haplotype (PRAA) in the control samples as the reference (that is, odds ratio = 1). All effects are conditional on Asp-9 in HLA-B and Phe-9 in HLA-DPβ1. Unadjusted haplotype frequencies are given for cases and controls. HLA-DRB1 haplotypes in aggregate explain 9.7% of the phenotypic variance of rheumatoid arthritis. The multivariate effect sizes, allele frequencies, and classical alleles corresponding to Asp-9 in HLA-B and Phe-9 in HLA-DPβ1 are also listed.
| HLA-DRβ1 Amino Acid Position | Multivariate Odds Ratio (95% Confidence Interval) | Unadjusted Allele Frequency | Classical HLA-DRB1 Alleles | |||||
|---|---|---|---|---|---|---|---|---|
| 11 | 13 | 71 | 74 | Controls | Cases | |||
| V | H | K | A | 4.44 | (4.02 – 4.91) | 0.106 | 0.316 | *04:01 |
| V | H | R | A | 4.22 | (3.75 – 4.75) | 0.056 | 0.141 | *04:08, *04:05, *04:04 |
| F | *10:01 | |||||||
| L | F | R | A | 2.17 | (1.94 – 2.42) | 0.109 | 0.143 | *01:02, *01:01 |
| P | R | R | A | 2.04 | (1.59 – 2.62) | 0.013 | 0.012 | *16:01 |
| V | H | R | E | 1.65 | (1.24 – 2.19) | 0.010 | 0.009 | *04:03, *04:07 |
| D | F | R | E | 1.65 | (1.29 – 2.10) | 0.011 | 0.013 | *09:01 |
| V | H | E | A | 1.43 | (1.04 – 1.96) | 0.011 | 0.006 | *04:02 |
| S | S | K | A | 1.04 | (0.76 – 1.41) | 0.012 | 0.006 | *13:03 |
| P | R | A | A | 1 | REF | 0.142 | 0.092 | *15:01, *15:02 |
| G | Y | R | Q | 0.91 | (0.80 – 1.03) | 0.133 | 0.064 | *07:01 |
| S | S | R | A | 0.88 | (0.77 – 1.00) | 0.103 | 0.049 | *11:01, *11:04 |
| G | *12:01 | |||||||
| S | S | R | E | 0.84 | (0.67 – 1.05) | 0.025 | 0.012 | *14:01 |
| L | F | E | A | 0.73 | (0.42 – 1.27) | 0.004 | 0.002 | *01:03 |
| S | G | R | L | 0.71 | (0.57 – 0.89) | 0.028 | 0.013 | *08:01, *08:04 |
| S | S | K | R | 0.63 | (0.54 – 0.73) | 0.128 | 0.083 | *03:01 |
| S | S | E | A | 0.59 | (0.51 – 0.68) | 0.112 | 0.041 | *11:02, *11:03, *13:01, *13:02 |
| HLA-B Amino Acid Position 9 | Classical HLA-B Allele | |||||||
|
| ||||||||
| D | 2.12 | (1.89 – 2.38) | 0.118 | 0.130 | *08 | |||
| H,Y | 1 | REF | 0.882 | 0.870 | *07, *13, *14, *15, *18, *27, *35, *37, *38, *39, *40, *41, *44, *45, *47, *49, *50, *51, *52, *53, *55, *56, *57, *58, *73 | |||
| HLA-DPβ1 Amino Acid Position 9 | Classical HLA-DPB1 Alleles | |||||||
|
| ||||||||
| F | 1.40 | (1.31 – 1.50) | 0.728 | 0.799 | *02:01, *02:02, *04:01, *04:02, *05:01, *16:01, *19:01, *23:01 | |||
| H,Y | 1 | REF | 0.272 | 0.201 | *01:01, *03:01, *06:01, *09:01, *10:01, *11:01, *13:01, *14:01, *15:01, *17:01, *20:01 | |||
To replicate these DRβ1 effects without imputed genotypes, we analyzed an independent South Korean data set of 616 anti-CCP positive cases and 675 controls with genome-wide SNP data18 and sequencing-based classical HLA-DRB1 genotypes at 4-digit resolution19. We used the first five principal components as covariates to correct for population stratification (λgc = 1.01). Of all amino acids tested in HLA-DRβ1, the strongest associations mapped to amino acid positions 11 (p=6.1×10−36) and 13 (p=3.1×10−36), with statistically indistinguishable effects (p>0.08; Supplementary Table 3, Supplementary Table 6). Thus, amino acids 11 and 13 in DRβ1 are the strongest associations in two different continental populations.
Given the polymorphic nature of HLA-DRB1, we evaluated whether a similarly significant result could emerge by chance, by “tagging” classical alleles of differential risk. To test this possibility, we preserved classical HLA genotypes and case-control status in all samples, and permuted the amino acid sequence defined by each classical HLA-DRB1 allele 10,000 times. We found that a single amino acid position only rarely resulted in a better model goodness-of-fit (measured by the deviance) as compared to amino acid position 11 in the actual data (p=0.0002; Figure 2B). Therefore, the degree to which the six alleles at amino acid position 11 divide the classical alleles of HLA-DRB1 into differential risk groups is extremely unlikely to occur by chance.
After accounting for the amino acid 11 effects in DRβ1 with conditional haplotype analysis, we observed an independent association at position 71 (p<10−37; Figure 2C, Supplementary Table 5A). We tested all possible pairs of polymorphic amino acid positions in DRβ1; of the 1,275 pairs of amino acids tested, none achieved a better goodness-of-fit than positions 11 and 71 (p~4×10−615). Using the same permutation strategy described above, we found that the degree to which amino acid positions 11 and 71 divide the classical alleles of HLA-DRB1 into differential risk groups is unlikely to occur by chance (p=0.0002) (Figure 2D). At HLA-DRβ1 position 71, the positively charged Lys-71 and Arg-71 residues confer greater odds of disease (OR=2.0 and 0.97, respectively) than the small aliphatic Ala-71 (OR=0.59); the negatively charged Glu-71 confers the least odds of disease (OR=0.32, Figure 3).
Conditioning on positions 11 and 71 revealed an additional association at position 74 (p=1.5×10−11; Figure 2E, Supplementary Table 5A). When we tested all possible combinations of three amino acid positions in DRβ1, we found that only one combination of amino acids sites (37, 67 and 74, p=2×10−624) out of 20,825 tested outperformed the combination of amino acid sites 11, 71 and 74 (p=1.6×10−622). However, even that combination did not outperform the 11, 71 and 74 combination by a statistically superior margin (p>0.01). As before, we permuted amino acid sequences, and only rarely were we able to pick three amino acid positions that obtained a better goodness-of-fit in the permuted data than positions 11, 71 and 74 in the actual data (p=0.004; Figure 2F). Addition of each of these three amino acid positions yielded improved model fit, even after accounting for the increased number of parameters (Supplementary Table 5B). No residual association was observed at other DRβ1 amino acids after conditioning on positions 11, 71 and 74 (p>8×10−4; Figure 2G, Supplementary Table 5A).
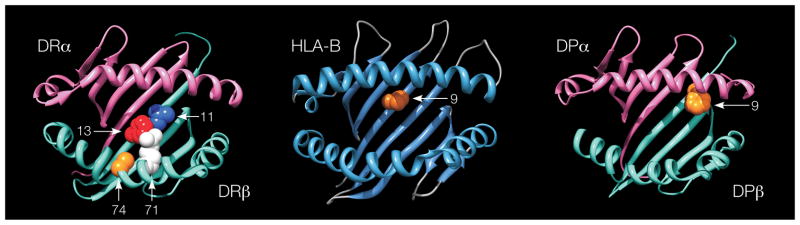
The amino acids at positions 11, 71 and 74 in DRβ1 define 16 haplotypes (Table 1). In fact, individual disease risk predicted by a full model where each classical DRB1 allele confers its own unique risk, and a simpler model where risk is defined by amino acid positions 11, 71 and 74, are nearly perfectly correlated (r=0.994). Hence, the model based on the amino acid residues at positions 11, 71 and 74 provides a parsimonious explanation for the effects of the classical DRB1 haplotypes, and suggests an important role for these amino acids in DRβ1 function in rheumatoid arthritis etiology. This is underscored by their central location in the peptide-binding groove of the HLA-DR structure (Figure 4). Positions 11 and 13 are located on the beta-sheet floor with their side chains oriented into the peptide-binding groove. Positions 71 and 74 are separated by a single turn along the α-helix, and their side chains are spatially close to those of positions 11 and 13.
Figure 4. Three-dimensional ribbon models for the HLA-DR, HLA-B and HLA-DP proteins.
These structures are based on Protein Data Bank entries 3pdo, 2bvp and 3lqz, respectively, with a direct view of the peptide-binding groove. Key amino acid positions identified by the association analysis are highlighted. This figure was prepared with UCSF Chimera25.
In order to assess if there were other independent MHC associations outside of HLA-DRB1, we conditioned on DRβ1 amino acids 11, 71 and 74 and tested all MHC SNPs and HLA alleles. We observed the most significant association at HLA-B in the class I region (p<2×10−37; Figure 1B). This association maps to Asp-9 in HLA-B (OR=2.12 relative to His-9 or Tyr-9; Table 1, Figure 3, Supplementary Table 4), although we could not statistically distinguish this effect from the classical B*08 allele (p>0.68). Like positions 11, 71 and 74 in DRβ1, position 9 in HLA-B is also located within the binding groove (Figure 4). Many of the previously described associations across the MHC, including markers in the TNF region, are in LD with Asp-910.
Since previously observed B*08 associations to autoimmune diseases, including rheumatoid arthritis, have been attributed specifically to the long ancestral 8.1 haplotype, containing B*08 on the DRB1*03 background9,11, we tested whether the B*08/Asp-9 effect is general to all DRB1 backgrounds. Since B*08 and DRB1*03 are not in perfect LD and both are seen independent of the 8.1 haplotype, we were able to apply conditional haplotype analysis to demonstrate that B*08/Asp-9 increases risk roughly two-fold regardless of DRB1 background (Figure 5). Therefore, this risk effect is not restricted to the 8.1 haplotype. Risk alleles for HLA-B and DRB1 contribute risk additively (on a log-odds scale) even though they are in strong (but incomplete) LD.
Figure 5. Conditional haplotype analysis.
Each row refers to a single classical HLA-DRB1 allele. In the left box, the main (univariate) effect is plotted as an odds ratio (with 95% confidence intervals) for each DRB1 allele (versus not having that allele), sorted in order of rheumatoid arthritis risk. In the middle box (in green), case and control allele frequencies and odds ratios are plotted for the HLA-B Asp-9 allele. In the right box (in blue), case and control allele frequencies and odds ratios (with 95% confidence internvals) are plotted for the HLA-DPβ1 Phe-9 allele. The red vertical lines indicate the aggregate effects for HLA-B and HLA-DPβ1 across all DRB1 haplotypes. The Asp-9 allele in HLA-B and the Phe-9allele in HLA-DPβ1 both have consistent effects across all HLA-DRB1 haplotype backgrounds. This suggests that these three effects are additive and independent, and not the consequence of any individual extended haplotype.
Conditioning on the HLA-DRB1 and HLA-B effects, we observed the most significant association at HLA-DPB1 in the class II region (p<10−20; Figure 1C), which corresponds to Phe-9 in DPβ1 (OR=1.40 relative to His-9 and Tyr-9; Table 1, Figure 3, Supplementary Table 4). This effect is significantly stronger than any 2- or 4-digit HLA-DPB1 classical allele, but in LD with and indistinguishable from the Val-8 allele. Amino acid position 9 is within the binding groove of HLA-DP (Figure 4).
We observed no residual signals across the MHC after conditioning on DRB1, B*08/Asp-9 in B, and Phe-9 in DPβ1 effects (p>3×10−6; Figure 1D). Nor did we observe any evidence of epistatic interactions between known risk loci13,20,21 and any of the HLA alleles described here (p>0.0003, see Supplementary Note). These results are consistent with a disease model where classical HLA genes/proteins are the dominant factors in rheumatoid arthritis pathogenesis with only a minor contribution from non-HLA loci in the MHC.
A key finding of this study is the major influence of amino acids 11 and 13 within DRβ1, but outside of the well-described shared epitope region. It is possible that one position is driving the effect and the other is in tight LD. Alternatively, there may be a joint effect involving both amino acids, driven by combined selection. This is plausible given the important role of natural selection22 in the MHC and the physical proximity of these two positions. To disentangle these effects, larger studies including multiple ethnicities, and many more examples of alleles where the LD between 11 and 13 is discordant will be necessary. Alternatively, if candidate rheumatoid arthritis auto-antigens can be determined, then these effects might be disentangled by comparing T-cell responses to these antigens presented in the context of DRB1 molecules engineered to contain distinct combinations of amino acids at positions 11 and 13.
This study implicates three amino acid positions in the HLA-DRβ1, and two additional amino acid positions in HLA-B and HLA-DP in conferring rheumatoid arthritis risk. These variants account for 12.7% of the phenotypic variance, whereas common validated alleles outside the MHC explain ~4%13 (see Supplementary Note). The location of these positions within the peptide-binding grooves implies a functional impact on antigenic peptide presentation to T-cells, either during early thymic development or peripheral immune responses. The presence of class I and II alleles implicate both CD8+ cytotoxic and CD4+ helper T-cells in pathogenesis. Besides rheumatoid arthritis, type 1 diabetes has also been shown to have strong HLA class I and II associations23. We also note that the HLA-B*08 allele, carrying Asp-9, has been documented in many autoimmune diseases, including myasthenia gravis, immunoglobulin-A deficiency, and systemic lupus erythematosus24.
The pathogenic auto-antigens in most autoimmune disorders remain controversial. For rheumatoid arthritis, these results could facilitate evaluation of specific citrullinated polypeptides with molecular modeling and binding assays, and in doing so will guide our understanding of how HLA risk alleles influence the immune repertoire and disease susceptibility.
METHODS
Sample Collections
All cases met 1987 American College of Rheumatology diagnostic criteria26, were diagnosed by a board-certified rheumatologist, and were confirmed as anti-CCP positive. Samples came from multiple studies, each receiving approval from the appropriate institutional review boards; all participants signed informed consent.
For primary analysis, we used six sample collections (Supplementary Table 1) from the United Kingdom (WTCCC), Sweden (EIRA), Canada (CANADA), United States (NARAC-I and NARAC-III), and Boston (BRASS), from a recent rheumatoid arthritis GWAS meta-analysis13. We followed the quality control steps outlined in the original publication. Additionally, we excluded WTCCC cases that were not confirmed as anti-CCP positive, WTCCC shared controls used to study other phenotypes, and individuals that failed HLA-DRB1 phasing (n=57 individuals). All individuals were self-described white and of European descent. In total, there were 5,018 cases and 14,974 controls.
For secondary analysis, we used a South Korean collection of 616 cases and 675 controls recruited at the Hanyang University Hospital for Rheumatic Diseases in Seoul, described in detail elsewhere18. Our study followed quality control steps outlined in the original publication. We excluded cases not confirmed as anti-CCP positive, and individuals not successfully genotyped for HLA-DRB1 classical alleles.
For all samples, we had access to genome-wide SNP data. The European samples were genotyped on different platforms (Supplementary Table 1). South Korean samples were genotyped with Illumina HumanHap-550v3 or 660W platforms. All South Korean samples, a subset of WTCCC samples (n = 700, all controls), and a subset of NARAC-I (n = 450) samples, had full genotype data to 4-digit resolution at the HLA-DRB1 locus. Korean samples were genotyped with polymerase chain reaction sequence-based typing (PCR-SBT); NARAC samples were genotyped with sequence specific oligonucleotide (SSO) genotyping9,19. Some WTCCC controls were part of the 1958 British Birth Cohort27, and were HLA typed at the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory; that data was made available through the European Genome-phenome Archive (EGA).
Imputing HLA Genotypes
As previously published15, we imputed classical HLA alleles and corresponding amino acid sequences using reference data collected by the Type 1 Diabetes Genetics Consortium (T1DGC). This reference data contains genotype data for 2,537 SNPs, selected to tag the entire MHC, and classical types for HLA-A, B, C, DRB1, DQA1, DQB1, DPA1 and DPB1 at 4-digit resolution in 2767 unrelated individuals of European descent14. Overlapping SNPs between the GWAS and the T1DGC samples ranged from 219 to 674 (Supplementary Table 1). We encoded all variants in the reference panel as biallelic markers, which facilitated application of BEAGLE for imputation (using default parameters)28. For each data set, we imputed cases and controls together.
Statistical framework for association testing
To test markers for an effect on risk that was fixed (consistent across data sets) and additive on the log odds scale we used logistic regression. To account for population stratification, we included as covariates five principal components for each individual data set (see Supplementary Methods). We also included five indicator variables to account for cohort-specific effects or differences in the proportion of cases and controls between GWAS data sets. This resulted in the following logistic regression model:
where a indicates the specific allele being tested, ga,i is the dosage (imputed or genotyped) of allele a in individual i. The βα parameter represents the additive effect per allele. For testing a multi-allelic locus with m possible alleles (for example amino acid residues at a specific position), we include m-1β parameters, one for each allele, where one allele is arbitrarily selected as a reference. We use the most frequent allele in controls as the reference allele. Here δi,j is an indicator variable that is 1 only if individual i is in patient collection j. The γj parameter is the effect for the jth patient collection, and for one arbitrarily selected reference cohort is set to 0. The πj,k parameter is the effect for each of the principal components and pi,k is the value for individual i for the kth principal component.
Testing across the MHC locus
We defined a series of binary markers across the region using SNPs, classical HLA alleles, and amino acid residues15, listed in Supplementary Table 4. For biallelic SNPs, the binary marker was simply the alternate (minor) allele. For classical HLA alleles, the binary marker was simply the presence of the allele versus the absence of the allele. For binary amino acid residues, the binary marker was simply the presence of the less frequent amino acid in lieu of the more frequent one. For multi-allelic amino acid positions and SNP residues, we defined composite markers for testing where each possible individual allele and combination of alleles was tested for association. For example, a biallelic SNP allele induces a single variable, a triallelic SNP induces three variables, a quadrallelic SNP induces six variables. Across the MHC, we applied the logistic regression framework above to test each of these binary markers for association, controlling for collection effects and population stratification. For each marker we used probabilistic genotypes that take uncertainty in imputation into account.
Conditional analysis outside of the DRB1 locus
To assess whether there were independent effects outside of the DRB1 locus we used the same additive logistic regression approach defined above to test all markers across the MHC. We included DRB1 alleles as covariates, taking either all 4-digit classical DRB1 alleles (which is more conservative) or the DRB1 haplotypes defined by amino acid positions 11, 71 and 74 (listed in Table 1). Both approaches yielded very similar results. If we identified other independently associated markers, we included them as covariates in subsequent conditional analyses to identify additional independent effects.
Analysis of DRB1 amino acid sites
To test amino acid effects within DRB1, we applied conditional haplotypic analysis. We tested each single amino acid position by first identifying the m amino acid residues occurring at that position, and then partitioning the classical alleles into m groups of alleles with identical residues at that position. We estimated the effect of each of the m groups using logistic regression model (including covariates as above), and calculated the log-likelihood (LL) improvement in model fit over a null model. We assessed the significance of the improvement in fit by calculating the deviance (defined as −2×LL), which is distributed as a chi-squared distribution with m−1 degrees of freedom. This is equivalent to testing a single multi-allelic locus for association with m alleles.
For conditional analyses, we assumed the null model consists of haplotypes as defined by residues at previously defined amino acid positions. Addition of another position with m residues, if the amino acid is independent, may result in k additional unique haplotypes. We tested if the addition of those amino acid positions, and the creation of k additional haplotype groups, improve upon the previous set. We assessed the significance in improvement in log-likelihood over the previous model (with fewer haplotype groupings) by calculating the deviance (which is chi-squared distributed with k degrees of freedom).
We also used logistic regression with probabilistic dosages of amino acids, taking into account imputation uncertainty, and confirmed that the same amino acids emerged in the exact same order.
HLA allele permutations to determine significance
Given the polymorphic nature of HLA genes and the strong DRB1effect sizes, we wanted to assess whether the observed associations at positions 11, 71 and 74 could emerge by chance, just by “tagging” classical alleles of differential risk. To test this, we repeatedly reassigned amino acid sequences to each of the classical HLA-DRB1 alleles (as defined by the standard HLA dictionary29). In each permutation, we selected amino acids sequentially and assessed the improvement in deviance. We conducted 10,000 such permutations, in each case selecting three polymorphic amino acids sequentially that most improves the model deviance. We compared the improvement achieved by fitting randomized amino acid sequences to the observed improvement by fitting the actual data.
Exhaustively testing combinations of amino acids
We tested all possible amino acid pairs and triplets for association to disease risk. For each set of amino acid positions, we defined groups of classical DRB1 alleles with consistent residues at those positions. We used those groups to predict rheumatoid arthritis risk, and calculated for each of these models the log-likelihood improvement in risk prediction (and its significance) over the null model.
Conditional haplotype analysis
With multiple effects observed across the MHC region, we were concerned that they might be driven by LD to other classical DRB1 alleles. We obtained fully phased haplotypes across the MHC (from the imputed data). Using the statistical framework and covariates as defined above, we individually tested each of the classical DRB1 alleles. For each DRB1 allele we included a variable that represented its dosage (0, 1, or 2). We also included a variable that indicated the dosage of Asp-9 (or *08) alleles of HLA-B in phase with the DRB1 classical allele being tested, and similarly included a variable that indicated the dosage of the Phe-9 allele of HLA-DPβ1 in phase with the DRB1 classical allele being tested.
Availability of Software
Available from authors upon request.
Supplementary Material
Acknowledgments
We thank the Type 1 Diabetes Genetics Consortium and Wellcome Trust Case Control Consortium for data access. We acknowledge use of the HLA genotyping data in the British 1958 Birth Cohort DNA collection, performed by the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory. This project is supported by grants from the National Institutes of Health (K08AR055688, S.R.; R01-AR44422, P.K.G.; R01-AR057108, R.M.P.; and U01-GM092691, R.M.P.), the Korea Healthcare Technology R&D Project (A102065, A111218-11-GM01, H.-S.L. and S.-C.B.), a Career Award for Medical Scientists from the Burroughs Wellcome Fund (R.M.P.), and by the Eileen Ludwig Greenland Center for Rheumatoid Arthritis (P.K.G.).
Footnotes
CONTRIBUTIONS
SR and PIWdB conceptualized and coordinated this study, oversaw the statistical analyses, and wrote the initial manuscript. SR, PIWdB, CS, EAS, JF, and XJ conducted all of the statistical analyses. H-SL, S-CB, LA, LP, LK, JW, KAS, RMP, and PKG organized and contributed patient samples, and collected genome-wide SNP data. S-CB, H-SL, and PKG provided classical HLA genotype data. All authors contributed to writing the final manuscript.
References
- 1.Isenberg D. Oxford textbook of rheumatology. Oxford University Press; Oxford; New York: 2004. p. 1278. [Google Scholar]
- 2.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–72. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 3.Ding B, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–8. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Woude D, et al. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60:916–23. doi: 10.1002/art.24385. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 6.Stastny P. HLA-D and Ia antigens in rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 1978;21:S139–43. doi: 10.1002/art.1780210921. [DOI] [PubMed] [Google Scholar]
- 7.Reinsmoen NL, Bach FH. Five HLA-D clusters associated with HLA-DR4. Hum Immunol. 1982;4:249–58. doi: 10.1016/0198-8859(82)90040-4. [DOI] [PubMed] [Google Scholar]
- 8.Vignal C, et al. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2009;60:53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 9.Lee HS, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14:293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton JL, Harney SM, Wordsworth BP, Brown MA. A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 2004;5:151–7. doi: 10.1038/sj.gene.6364045. [DOI] [PubMed] [Google Scholar]
- 11.Jawaheer D, et al. Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet. 2002;71:585–94. doi: 10.1086/342407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bakker PI, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown WM, et al. Overview of the MHC fine mapping data. Diabetes Obes Metab. 2009;11 (Suppl 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.van der Woude D, et al. Protection against anti-citrullinated protein antibody-positive rheumatoid arthritis is predominantly associated with HLA-DRB1*1301: a meta-analysis of HLA-DRB1 associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in four European populations. Arthritis Rheum. 2010;62:1236–45. doi: 10.1002/art.27366. [DOI] [PubMed] [Google Scholar]
- 18.Freudenberg J, et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63:884–93. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, et al. Microsatellite typing for DRB1 alleles: application to the analysis of HLA associations with rheumatoid arthritis. Genes Immun. 2006;7:533–43. doi: 10.1038/sj.gene.6364325. [DOI] [PubMed] [Google Scholar]
- 20.Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22:109–18. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhernakova A, et al. Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137:1–8. doi: 10.1016/j.imlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Nejentsev S, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price P, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–74. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Arnett FC, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 28.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.