Abstract
Tumor necrosis factor-α (TNF-α) has important roles in several immunological events by regulating apoptosis and transcriptional activation of cytokine genes. Intracellular signaling mediated by TNF-receptor-type 1 (TNFR1) is constituted by two sequential protein complexes: Complex-I containing the receptor and Complex-II-containing Caspase-8. Protein modifications, particularly ubiquitination, are associated with the regulation of the formation of these complexes. However, the underlying mechanisms remain poorly defined. Here, we identified CLIP-170-related 59 kDa protein (CLIPR-59) as a novel adaptor protein for TNFR1. Experimental reduction of CLIPR-59 levels prevented induction of apoptosis and activation of caspases in the context of TNF-α signaling. CLIPR-59 binds TNFR1 but dissociates in response to TNF-α stimulation. However, CLIPR-59 is also involved in and needed for the formation of Complex-II. Moreover, CLIPR-59 regulates TNF-α-induced ubiquitination of receptor-interacting protein 1 (RIP1) by its association with CYLD, a de-ubiquitinating enzyme. These findings suggest that CLIPR-59 modulates ubiquitination of RIP1, resulting in the formation of Complex-II and thus promoting Caspase-8 activation to induce apoptosis by TNF-α.
Keywords: death receptor, apoptosis, TNF, CLIPR-59
Induction of apoptosis mediated by tumor necrosis factor (TNF) family cytokines has important roles in several immunological processes by killing cells infected with pathogens, eliminating tumor cells and terminating immune responses.1, 2, 3, 4, 5, 6 Members of the death receptor (DR) family, which include TNF-receptor-type 1 (TNFR1), Fas, TRAMP/DR3, TRAILR1/DR4, TRAILR2/DR5 and DR6, are capable of inducing apoptosis in response to stimulation by their specific ligands, which induce higher-order aggregation of DR molecules in the plasma membrane, leading to recruitment of several intracellular proteins to the cytoplasmic domains of these receptors.4, 7
Recently, it was demonstrated that the intracellular signaling mediated by TNFR1 is constituted by formation of two sequential protein complexes.8, 9 Aggregation of TNFR1 mediated by its specific ligand induces the recruitment of adaptor proteins TNF receptor-associated death domain protein (TRADD) and receptor-interacting protein 1 (RIP1) to the cytoplasmic domain of TNFR1 (Complex-I). Subsequently, these proteins dissociate from Complex-I, facilitating the formation of a second protein complex (Complex-II)-containing Fas-associated protein with death domain (FADD) and Caspase-8, which is an initiator of the proteolytic cascade resulting in apoptosis.4, 10, 11 In addition, it is reported that the transition from Complex-I to Complex-II is regulated by protein modifications, particularly ubiquitination of RIP1.12, 13, 14
Early (<30 min) after TNF-α stimulation, RIP1 is conjugated with at least two types ubiquitin chains, K63- and K48-linked.15 Whereas K-48-dependent ubiquitinated RIP1 is degradated by proteasome-dependent mechanisms, K63-dependent ubiquitinated RIP1 functions as a scaffold that assembles signaling complexes that activate kinases responsible for gene expression pathways.16 Later (>60 min), RIP1 is de-ubiquitinated and enters into Complex-II.8, 14, 15 A recent study using a site-specific mutant of RIP1, showed that abolishing ubiquitination of RIP1 facilitates its involvement in Complex-II.12 These results suggest that de-ubiquitination of RIP1 regulates the formation of Complex-II in apoptotic signaling by TNF-α.
Results
CLIP-170-related 59 kDa protein (CLIPR-59) involves in TNFR1-containing Complex-I
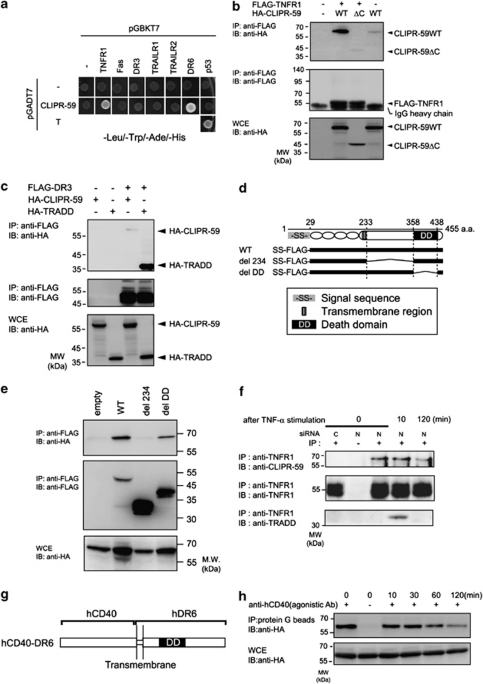
Recently, we identified CLIPR-59 as a novel interacting protein for the cytoplasmic tail of DR6 by yeast two hybrid (Y2H) screening using human fetal brain cDNA library.17 Here, we determined the interaction of CLIPR-59 with other DRs by Y2H assay (Figure 1a). Positive results were detected for DR6 and TNFR1 but not other members of the DR family. Independently, co-immunoprecipitation (co-IP) assays using lysates from cells transfected with plasmids encoding DR family proteins confirmed that CLIPR-59 interacts with TNFR1 (Figure 1b) but not DR3 (Figure 1c). In the case of the interaction of DR6 with CLIPR-59, the membrane proximal region of DR6 was previously found to be essential for this interaction.17 In co-IP assay using various deletion mutants for TNFR1, del 234, lacking the membrane proximal region (234–358 amino acid) or del death domain (DD), lacking the DD (359–438 amino acid; Figure 1d), we observed that the membrane proximal region of TNFR1 is also essential for interaction with CLIPR-59 (Figure 1e).
Figure 1.
Interaction of CLIPR-59 with DR. (a) Yeast strain AH109 was transformed with combination of expression plasmids. Cells were selected by prototrophy for leucin, tryptophan, histidine and adenine. The combination of pGBKT7-p53 and pGADT7-large T antigen was used as positive control. (b and c) HEK293T cells were transfected with the indicated combinations of expression plasmids. TRADD interaction with DR3 was included as positive controls. At 17 h after transfection, cells were lysed and subjected to IP with the indicated Ab and analyzed by IB. (e) HEK293T cells were transfected with the expression plasmids for FLAG-tagged WT, deletion mutants, del 234 (Δ234–358 amino acid) or del DD (Δ359–438 amino acid) of TNFR1, schematically shown in (d) and HA-tagged CLIPR-59, and analyzed as described in (b). (f) HeLa cells, transfected with control (N) or CLIPR-59-specific siRNA(C) were stimulated with TNF-α (100 ng/ml) in the presence of CHX (10 μg/ml) during the indicated period. Cells were lysed and immunoprecipitated with anti-TNFR1 antibody(Ab) or control IgG (shown as −). The immunoprecipitated proteins were analyzed as described in (b). (h) NIH3T3 cells (1 × 108 cells) were transfected with expression plasmids encoding hCD40-hDR6 chimeric protein, which schematically shown in (g), and HA-tagged CLIPR-59. At 17 h after transfection, cells were incubated for various times with agonistic anti-human CD40 Ab (clone G28-5). After incubation, cells were lysed, and hCD40-hDR6 complex ligated by clone G28-5 was immunoprecipitated with protein G sepharose. For the sample at 0 time point, clone G28-5(+) or non-specific IgG(−) was added in the cell lysate of non-stimulated cells transfected with hCD40-hDR6 and CLIPR-59, before adding protein G sepharose. Samples were analyzed by IB
The formation of active TNFR1-containing protein complex (Complex-I) is regulated by receptor aggregation.8 Therefore, we examined the effect of the ligand stimulation on the association of TNFR1 with CLIPR-59. In HeLa cells, co-IP assays using anti-TNFR1-specific antibody (Ab) showed that CLIPR-59 was associated with TNFR1 before ligand stimulation and this association was significantly reduced at 120 min after stimulation (Figure 1f). Complex-II formation is reported to be induced >120 min after TNFR1 stimulation.8 Similar results were obtained using a receptor cross-linking assay using a human CD40-specific agonistic Ab with a chimeric CD40/DR6 receptor protein, which consists of the extracellular domain of human CD40 and the transmembrane and cytoplasmic regions of DR6 (Figures 1g and h).
CLIPR-59 is essential for the induction of apoptosis in TNF-α signaling
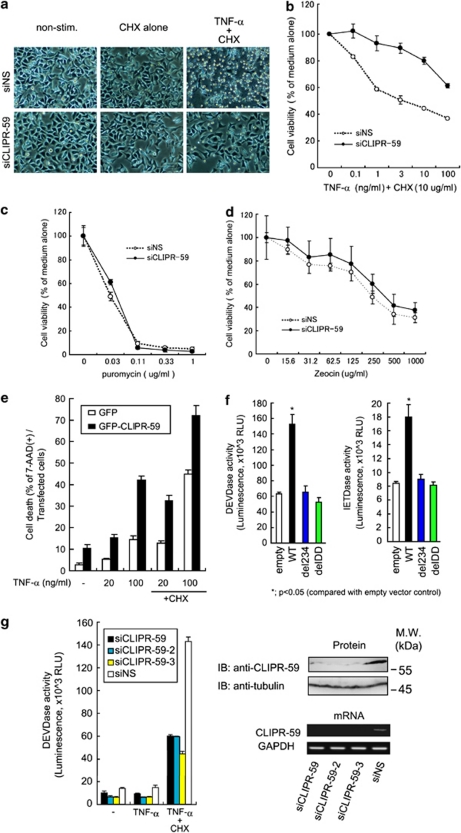
We examined the role of CLIPR-59 in apoptosis induced by TNF-α. Effects of RNAi-mediated knockdown of CLIPR-59 (Supplementary Figure S1) on apoptosis stimulated by TNF-α were assessed by apoptotic morphological changes and by a cell viability assay. HeLa cells transfected with siCLIPR-59 or control siNS were stimulated with TNF-α in the presence of cycloheximide (CHX). Cells transfected with control siNS exhibited typical apoptotic morphological changes (cytoplasmic blebbing and cell shrinking) in response to TNF-α with CHX, but not with CHX alone or with medium alone (Figure 2a). Interestingly, in contrast to these observations, treatment with siCLIPR-59 markedly reduced the number of cells exhibiting these apoptotic morphological changes (Figure 2a). Cell viability assays also showed that treatment with siCLIPR-59 protects cells against cytotoxicity induced by TNF-α (Figure 2b and Supplementary Figure S2), but not by cytotoxic chemical reagents, such as puromycin (Figure 2c), which is a protein synthesis inhibitor, nor zeocin (Figure 2d), which is a member of the bleomycin family of antibiotics and causes cell death by intercalating into and cleaving DNA. In contrast to CLIPR-59 knockdown, overexpression of CLIPR-59 enhanced apoptosis induced by TNF-α alone or TNF-α plus CHX (Figure 2e). In addition, deleting the region of TNFR1 required for CLIPR-59 binding abolished its ability to promote caspase activation, showing that the recruitment of CLIPR-59 into TNFR1 complex is required for TNF-α-mediated apoptosis induction, in addition to TNFR1's DD (Figures 2f and 1d, e). Furthermore, by Caspase-Glo assay using a luminogenic Caspase-3/7 substrate, which contains the tetrapeptide sequence DEVD (Caspase-3/7 Glo reagent, Promega, Madison, WI, USA), it was shown that treatment with siCLIPR-59 markedly reduced the activation of effector caspases induced by TNF-α plus CHX compared with control siNS treatment (Figure 2g). The same data were obtained by the assay using siRNAs (siCLIPR-59-2 and siCLIPR-59-3) targeting different regions within CLIPR-59 mRNA (Figure 2g). Altogether, these findings suggest that CLIPR-59 contributes to the induction of apoptosis by TNF-α.
Figure 2.
CLIPR-59 contributes to TNF-α-mediated pro-apoptotic signaling. (a) HeLa cells transfected with siCLIPR-59 or siNS were stimulated with medium alone, (CHX, 10 μg/ml) or TNF-α (10 ng/ml) with CHX (10 μg/ml). At 6 h after stimulation, morphological changes were observed by light microscopy. HeLa cells transfected with siNS or siCLIPR-59 were stimulated with various amounts of TNF-α in the presence of CHX (10 μg/ml; b), or chemical cytotoxic drug, puromycin (c) or zeocin (d). At 6 h (in the case for TNF-α), 24 h (for puromycin) or 48 h (for zeocin) after stimulation, cell viability was assessed by WST assay (error bar means±S.D.; n=3). (e) HeLa cells, transfected with the plasmid for expressing GFP-tagged CLIPR-59 or GFP alone, were stimulated with TNF-α (the indicated concentration, ng/ml) with or without CHX (10 μg/ml). At 24 h after stimulation, the percentage of cell death of the transfected cells was assessed as described in Materials and Methods (error bar means±S.D.; n=3). (f) HeLa cells were transfected with the indicated expression vector plasmids. At 24 h after transfection, cell-viability, IETDase or DEVDase activity of these cells were assessed by WST assay, Caspase-8 or Caspase-3/7 Glo reagents (Promega) measuring relative fluorescent units (RFU), respectively, (error bar means±S.D.; n=3). (g) HeLa cells transfected with the indicated siRNAs directed to three different target sites on CLIPR-59 mRNA were stimulated with TNF-α (10 ng/ml) with or without CHX (10 μg/ml). At 4.5 h after stimulation, DEVDase activity was assessed as described in (f) (error bar means±S.D.; n=3). To determine the effects of siRNA treatment on the protein and mRNA expression level of CLIPR-59 were assessed by western blot using the indicated Abs and RT-PCR using primers specific for CLIPR-59 or GAPDH (lower panel), respectively
CLIPR-59 regulates the activation of Caspase-8 in TNF-α signaling
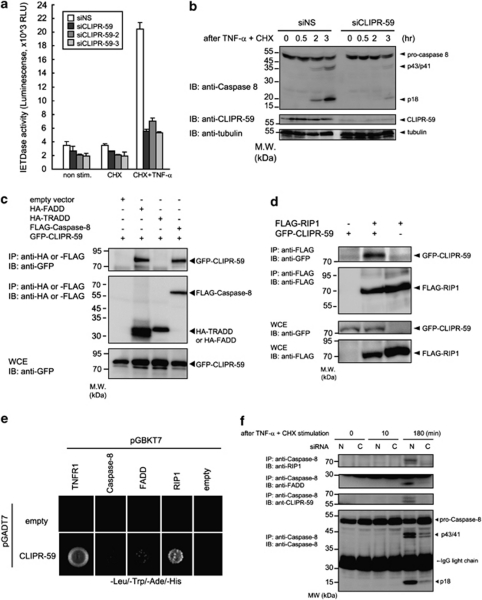
We also evaluated the role of CLIPR-59 in TNF-α-induced activation of Caspase-8, an upstream initiator caspase that is recruited to Complex II. HeLa cells treated with siNS or siCLIPR-59 were stimulated with TNF-α plus CHX, then Caspase-8 activity was measured 3 h later by luminogenic substrate, Z-IETD-aminoluciferin (Caspase-8 Glo reagent, Promega). Treatment with siCLIPR-59 markedly reduced Caspase-8 activity induced by TNF-α plus CHX, compared with siNS treatment (Figure 3a). By immunoblot assay using anti-Caspase-8-specific Ab, we also observed that CLIPR-59 is required for TNF-α-induced Caspase-8 processing (Figure 3b). These findings suggested that CLIPR-59 participates in the activation of Caspase-8 in TNF-α signaling.
Figure 3.
CLIPR-59 promotes the activation of Caspase-8 by regulating the formation of Complex-II in TNF-α signaling. (a) HeLa cells treated with the indicated siRNAs were stimulated by TNF-α (10 ng/ml) plus CHX(10 μg/ml). At 3 h after stimulation, Caspase-8 activity was measured as described in Figure 2f. Data are mean±S.D.; n=3. (b) HeLa cells transfected with siNS or siCLIPR-59 were stimulated with TNF-α (10 ng/ml) plus CHX (10 μg/ml) for the indicated periods. Cells were lysed and analyzed by IB using the indicated Abs. (c and d) HEK293T cells were transfected with the indicated combination of expression plasmids. At 24 h after transfection, cells were analyzed as described in Figure 1b. (e) Yeast two-hybrid assays using the combination of plasmids for the indicated genes were performed as described in Figure 1a. (f) HeLa cells transfected with siNS (N) or siCLIPR-59 (C) were stimulated with TNF-α (100 ng/ml) in the presence of CHX (10 μg/ml) for the indicated periods. After stimulation, cells were lysed and immunoprecpitated using anti-Caspase-8-specific Ab. Precipitated materials were analyzed by IB using the indicated Abs
In previous report, CLIPR-59 was associated with Akt activation in the context of adipocyte differentiation.18 In addition, some reports have suggested that Akt has anti-apoptotic role by directly phosphorylating the pro-apoptotic protein BAD at position Ser 136. Thus, we tested the effect of CLIPR-59 knockdown on Akt-mediated phosphorylation of BAD in the setting of TNF-α signaling (Supplementary Figure S3A). In this assay, we did not observe a role for CLIPR-59 in regulating phosphorylation of this Akt substrate during TNF-α signaling. In addition, siCLIPR-59 did not affect apoptosis induced by overexpression of either Caspase-8 or BAD (Supplementary Figure S3B). Taken together, our findings suggest that CLIPR-59 operates upstream of Caspase-8 in TNF-α signaling.
CLIPR-59 regulates the formation of Complex-II-containing Caspase-8
Several adaptor proteins, such as FADD, TRADD and RIP1, regulate the activation of Caspase-8 during TNFR1 signaling,4, 7, 11, 14 prompting us to hypothesize that CLIPR-59 might exert its influences by these adaptor proteins. We first examined the association of CLIPR-59 with adaptor proteins by co-IP assay using plasmid-transfected cells. These experiments provided evidence that CLIPR-59 associates with complexes containing FADD and Caspase-8, but not TRADD (Figure 3c). Another adaptor protein RIP1 was also immunoprecipitated with CLIPR-59 (Figure 3d). Moreover, based on Y2H assays, the interaction of CLIPR-59 with RIP1 but not Caspase-8 or FADD, appears to be direct (Figure 3e). These findings thus suggested that CLIPR-59 could be involved in the protein complex containing Caspase-8, FADD and RIP1, which is known as Complex II. To evaluate these protein interactions in the context of TNF-α signaling, co-IP assays using anti-Caspase-8-specific Ab were performed, showing that CLIPR-59 was co-precipitated with Caspase-8 and revealing that this interaction was enhanced by TNF-α stimulation (Figure 3f). Moreover, CLIPR-59 knockdown disrupted the interaction of Caspase-8 with both FADD and RIP1 (Figure 3f). These findings suggest that CLIPR-59 is involved in and regulates the formation of Complex-II, containing Caspase-8, FADD and RIP1 in the TNF-α signal transduction pathway.
CLIPR-59 negatively regulates ubiquitination on RIP1 in TNF-α signaling
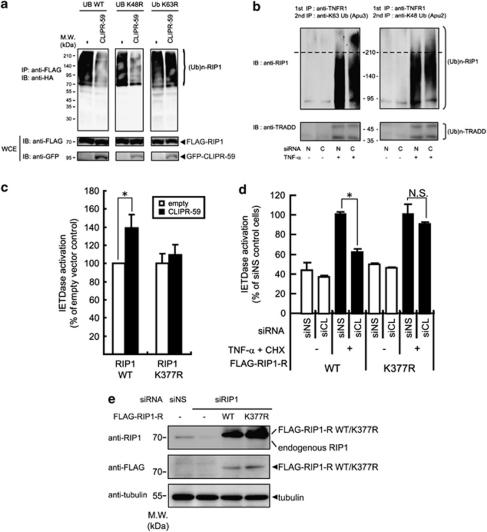
An early event of TNF-α signaling (<30 min after stimulation) entails the formation of Complex I containing the ligand, its receptor TNFR1, and associating proteins, including RIP1 and TRADD, resulting in the ubiquitination of RIP1. Progression to Complex II requires de-ubiquitination of RIP1.8, 12, 13, 14, 16 Ectopic expression of RIP1 induces auto-ubiquitination in mammalian cells. Therefore, we explored the impact of CLIPR-59 in this system by using ubiquitin mutant, K63R or K48R, lysine at the position 63 or 48 is mutated to arginine, respectively. In the cells expressing wild-type (WT) or K48R ubiquitin, expression of CLIPR-59 remarkably decreased ubiquitination on RIP1 compared with control cells (Figure 4a). On the other hands, in the cells expressing K63R ubiquitin, this effect by the expressed CLIPR-59 was not observed. These findings suggested that CLIPR-59 should specifically regulate lysine-63-dependent ubiquitination on RIP1. Regarding to the ubiquitination on RIP1 mediated by TNF-α stimulation, it is known that in the cells, only a small fraction of RIP1 recruited into TNFR1 complex, is ubiquitinated.15 To clarify the role of CLIPR-59 for the ubiquitination on the receptor associated RIP1, K63- or K48-dependent ubiquitination states of receptor associated RIP1 by CLIPR-59 knockdown were compared with control siRNA treatment (Figure 4b). In the control cells (N), K63-dependent ubiquitinated RIP1 was observed as smear bands with 95–210 kDa molecular weight after TNF-α stimulation. Contrary to this observation, an additional high-molecular weight smear bands (>210 kDa) were observed in CLIPR-59 knock-down cells (C; Figure 4b, upper left panel). In the case of K48-dependent ubiquitination, the ubiquitinated RIP1 was observed as smear bands of the same molecular weight (95–210kDa) in both control and CLIPR-59 knock-down cells (Figure 4b, upper right panel). These effects by the reduction of CLIPR-59 expression could not affect on both K63- and K48-dependent ubiquitination of TRADD involved in the same receptor complex (Figure 4b, lower left and right panels). Same effects on K63-linked ubiquitination of RIP1 were observed in the experiments using the other siRNAs against CLIPR-59 mRNA (siCLIPR-59 -2 or -3; Supplementary Figure S4). These findings suggest that CLIPR-59 regulates K63-linked ubiquitination of RIP1 in TNF-α signaling.
Figure 4.
CLIPR-59 regulates the ubiquitination on RIP1 in TNF-α signaling. (a) HEK293T cells were transfected with the expression plasmids for GFP alone (−) or GFP-CLIPR-59 (CLIPR-59)(2 μg/well) with the combination of the plasmids for FLAG-RIP1 (2 μg/well) and HA-ubiquitin WT, K63R or K48R mutant (UB WT, K48R or K63R, respectively; 0.1 μg/well). At 24 h after transfection, cells were analyzed as described in Figure 1b. (b) HeLa cells transfected with siCLIPR-59(C) or control siNS(N) were pretreated with proteasome inhibitor MG132 (21 μM) for 10 min before stimulation with TNF-α (100 ng/ml) or medium for additional 10 min. After cell lysing, the cell lysates were subjected to sequential IP methods for detecting ubiquitination of RIP1 associated with TNR1 as described in Materials and Methods. (c) HeLa cells were transfected with the combination of expression plasmids for the indicated genes. At 24 h after the transfection, Caspase-8 activity was assessed as described in Figure 3a. Data are mean±S.D.; n=3. (d) HeLa cells were transfected with the expression plasmid for siRNA-resistant RIP1 (RIP1-R) WT or mutated form (K377R), by electropolation, and the cells were selected by G418 (600 μg/ml). These cells were transfected with the indicated combination of siRNAs plus human RIP1-specific siRNA. At 72 h after transfection, the cells were stimulated with TNF-α (10 ng/ml) plus CHX (10 μg/ml). At 3 h after the stimulation, Caspase-8 activity was assessed as described in Figure 3a. Error bar means±S.D.; n=3. *P<0.05 by unpaired t-test. NS, not significant. (e) The protein expression level of endogenous RIP1 and siRNA-resistant FLAG-tagged RIP1 WT or K377R mutant (FLAG-RIP1-R WT or FLAG-RIP1-R K377R, respectively) was assessed by IB using the indicated Ab
Recently, it is reported that site-specific mutation of Lysine at position 377 on RIP1 prevents its K63-linked ubiquitination and enhances the formation of RIP1 with Caspase-8 in TNF-α signaling.12 Consistent with a role for CLIPR-59 in modulating ubiquitin-dependent RIP1 signaling, CLIPR-59 expression enhanced Caspase-8 activation induced by WT RIP1 but not RIP1 K377R (Figure 4c). More importantly, it is observed that in HeLa cells expressing mutated RIP1 (RIP1-R K377R), CLIPR-59 knockdown did not affect the Caspase-8 activation by TNF-α, unlike in the cells expressing WT (RIP1-R WT) RIP1 (Figures 4d and e). These findings suggest that CLIPR-59 negatively regulates RIP1 ubiquitination to promote Caspase-8 activation in the context of TNF-α signaling.
Functional association of CLIPR-59 with CYLD in TNF-α signaling
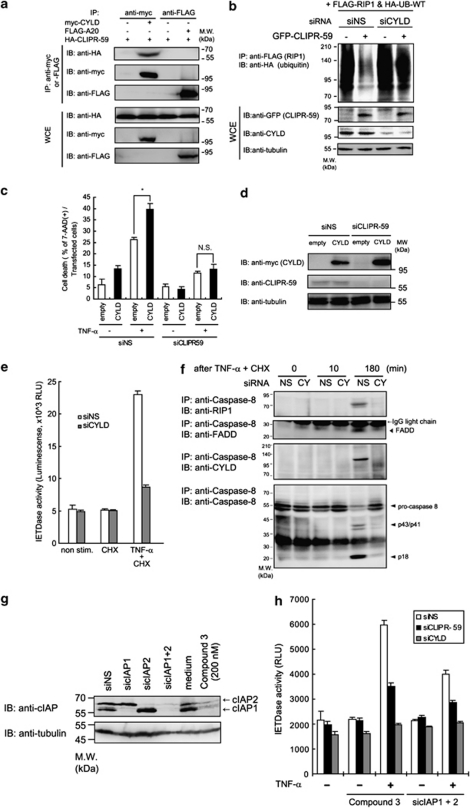
Ubiquitination of RIP1 is regulated by de-ubiquitinating enzymes (DUBs), such as CYLD, which is a K63-specific DUB, and A20 protein, which is a K63- to K48-ubiquitination-converting enzyme involved in TNF-α signaling.16, 19 Because CLIPR-59 has no predicted enzyme domains,20 we hypothesized that CLIPR-59 might exert its function by association with such DUBs. To test this hypothesis, we first determined whether CLIPR-59 could form protein complexes with CYLD or A20. In co-IP assays using transfected cells, CLIPR-59 co-IP with CYLD but not A20 protein (Figure 5a). The carboxy-terminal region of CLIPR-59, was determined to be essential for its interaction with CYLD (Supplementary Figures S5A and B) and this interactive domain is needed for de-ubiquitinating function of CLIPR-59 (Supplementary Figure S5C, upper panel). In contrast, C-terminal deletion of CLIPR-59 did not impair its binding to RIP1 (Supplementary Figure S5C, lower panel). In addition, CYLD mRNA-specific siRNA (siCYLD) treatment suppressed de-ubiquitinating activity of CLIPR-59 (Figure 5b). These findings suggested that CYLD associates with the de-ubiquitinating function of CLIPR-59.
Figure 5.
Functional association of CLIPR-59 with CYLD in TNF-α-mediated pro-apoptotic signaling. (a) HEK293T cells were transfected with the indicated combinations of expression plasmids. At 24 h after transfection, cells were lysed and analyzed as described in Figure 1b. (b) HEK293T cells were transfected with the indicated siRNA. At 48 h after first transfection, cells were reseeded into 6-well culture plate. At 24 h after incubation, cells were re-transfected with the expression plasmids for GFP alone (−) or GFP-CLIPR-59 (2 μg/well) with the combination of the plasmids for FLAG-RIP1 (2 μg/well) and HA-ubiquitin (UB-WT; 0.1 μg/well). At 24 h after second transfection, cells were analyzed as described in Figure 1b. (c) HeLa cells were transfected with the indicated siRNA. At 24 h after transfection, the cells were reseeded into 12-well culture plate. At 20 h after incubation, the cells were transfected with empty or CYLD-expressing plasmids plus GFP-expressing plasmid. At 20 h after second transfection, the cells were stimulated with TNF-α (20 ng/ml) in the presence of CHX (10 μg/ml). The percentage of the dead cells of transfected GFP-positive cells was assessed as described in Materials and Methods. Data are mean±S.D.; n=3. *P<0.05 by unpaired t-test. NS, not significant. (d) Expression of CYLD or CLIPR-59 was assessed by IB. (e) HeLa cells were transfected with the indicated siRNA. At 72 h after transfection, the cells were stimulated by the indicated reagents. At 3 h after stimulation, the activity of Caspase-8 was measured as described in Figure 3a (error bar means±S.D.; n=3). (f) HeLa cells transfected with control siRNA (NS) or CYLD gene-specific (CY) siRNA were stimulated by TNF-α in the presence of CHX. At various times thereafter, the cells were lysed and analyzed as described in Figure 3f. (h) HeLa cells were transfected with control siNS or siRNAs against the indicated genes (72 h) or treated with medium alone or smac mimetic, Compound 3 (200 nM, 4 h). After treatments, cells were stimulated by TNF-α (10 ng/ml, 3 h) and Caspase-8 activity was assessed as described in Figure 3a. Effect of these treatments on the protein expression level of cIAP1 or cIAP2 was assessed as described in Figure 1b (g)
Regarding the effect of TNF-α stimulation on the formation of CLIPR-59/CYLD/RIP1 protein complex, it is shown in co-IP assay that co-precipitated CYLD with CLIPR-59 was observed more abundantly at late phase (180 min) after TNF-α stimulation than at 0 time or early phase (10 min; Supplementary Figure S5D, upper panel). As it was also shown in the same condition that TNF-α stimulation could not affect on the interaction of RIP1 with CLIPR-59 (Supplementary Figure S5D, middle panel), it is suggested that the formation of CLIPR-59/RIP1/CYLD should be regulated at the point on the interaction of CLIPR-59 with CYLD during TNF-α stimulation. To clarify the role of CLIPR-59 in the formation of RIP1 and CYLD-containing protein complex, we assessed the effect of siCLIPR-59 on the association of CYLD with RIP1 in TNF-α-stimulated cells by co-IP assays using anti-RIP1-specific Ab. In control siNS-treated HeLa cells, co-IP of CYLD with RIP1 was detected after TNF-α plus CHX stimulation. In contrast, in siCLIPR-59-treated cells, the association of CYLD with RIP1 was reduced (Supplementary Figure S5E). These co-IP assays provided evidence that CLIPR-59 is needed for the formation of protein complex containing CYLD and RIP1. Altogether, these findings suggest that CLIPR-59 scaffolds CYLD into the complex containing RIP1 and thus facilitates de-ubiquitination on RIP1 in TNF-α signaling.
CYLD has been identified as the responsible gene for familial cylindromatosis.21 Previously, it was reported that CYLD promotes induction of cell death by TNF-α.22, 23, 24, 25 In cell death assays, overexpression of CYLD significantly enhanced induction of cell death stimulated by TNF-α, but this effect was abolished in CLIPR-59 knock-down cells (Figure 5c). Immunoblotting (IB) showed that CLIPR-59 knockdown did not affect expression of CYLD (Figure 5d). Similar with these observations, we found that CYLD is also essential for the enhancement of TNF-α-induced apoptosis caused by CLIPR-59 overexpression (Supplementary Figure S6A) and that CLIPR-59 is also needed for the recruitment of CYLD into Caspase-8 complex (Supplementary Figure S6B). In addition, siCYLD-treatment reduced the activation of Caspase-8 (Figure 5e) and the formation of Complex-II (Figure 5f) stimulated by TNF-α, further suggesting that CLIPR-59 modulates CYLD-dependent events of TNFR1 signaling.
In a recent study, it was demonstrated that CYLD is also essential for the induction of apoptosis and Complex-II formation by TNF-α in the presence of a Smac mimetic compounds that induces auto-degradation of endogenous anti-apoptotic proteins, c-inhibitor of apoptosis (IAP)1/2.26 We examined the effect of CLIPR-59 and CYLD knockdown on the induction of apoptosis by treatment of tumor cell lines with TNF-α plus Smac mimetics or plus c-IAPs knockdown. In control siNS-treated HeLa cells, stimulation with TNF-α robustly activated Caspase-8 in the presence of Smac mimetic (Compound-3)27 and also in c-IAP1/2 double knock-down cells (Supplementary Figure S7 and Figures 5g and h). As expected from prior reports,28 the TNF-α-induced activation of Caspase-8 was reduced by CYLD knockdown (Figure 5h). Importantly, CLIPR-59 knockdown also prevented the activation of Caspase-8 by TNF-α in the presence of Compound-3 or cIAPs knock-down treatment (Figure 5h). These changes in sensitivity to TNF-α cytoxicity, mediated by the treatment of siCLIPR-59 or siCYLD, correlated with differences in RIP1 ubiquitination (Supplementary Figure S8) These findings thus suggest that both CLIPR-59 and CYLD are essential for activation of Caspase-8 by TNF-α in the presence of Compound-3.
A cellular model for CLIPR-59-mediated regulation of sensitivity to TNF-α
Loss of sensitivity to apoptosis mediated by DRs has a role in the escape of tumors from host immune defenses. To evaluate the association of CLIPR-59 with this phenomenon, we first examined CLIPR-59 gene expression in experimental immune-resistant tumor cells (TC-1 P3 cells) compared with its parental immune-susceptible cells (TC-1 P0 cells).29 Quantitative RT-PCR and IB assays indicated that expression of CLIPR-59 was markedly reduced in TC-1 P3 cells compared with TC-1 P0 cells (Supplementary Figures S9A and B). TNF-α stimulated less cell death and less caspase activation in TC-1 P3 cells compared with TC-1 P0 cells (Supplementary Figures S9C and D). Transient overexpression of CLIPR-59 in TC-1 P3 cells restored sensitivity to TNF-α (Supplementary Figure S9E) whereas reduction of CLIPR-59 expression by shRNA prevented the activation of Caspas-8 by TNF-α stimulation in TC-1 P0 cells (Supplementary Figure S9F).
Discussion
We have identified CLIPR-59 as a novel modulator of TNF-α signaling. CLIPR-59 interacts with the membrane proximal region of TNFR1 in resting cells. Stimulation of ligand induces the recruitment of several interacting proteins to TNFR1, including RIP1 and TRADD to form Complex-I, facilitating the ubiquitination of RIP1. CLIPR-59 subsequently associates with CYLD and mediates de-ubiquitination of RIP1, thus promoting the recruitment of Caspase-8 and FADD to create Complex-II. In Complex-II, Caspase-8 is activated and promotes apoptosis (Supplementary Figure S10). In parallel, we observed that both CLIPR-59 and CYLD were involved in Complex-I (Supplementary Figure S11) and Complex-II (Figures 3f and 5f), and K63-linked ubiquitination of RIP1 induced by TNF-α stimulation was significantly increased by CLIPR-59 knock-down treatment (Figure 4b). Thus, we propose that CLIPR-59 collaborates with CYLD to achieve de-ubiquitination of RIP1 that thus promote formation of Complex-II and apoptosis.
We observed that CLIPR-59 dissociates from TNFR1 in response to receptor aggregation mediated by ligand or agonistic Ab stimulation, like as other receptor associating proteins, TRADD and RIP1.8 In this regard, it has been suggested that ligand stimulation promotes TNFR1 cleavage (receptor shedding).30 Consistent with destruction or shedding of TNFR1 accounting for release of associated cytosolic proteins, we found that TNFR1 present in cytoplasmic vesicles at 30 min after stimulation (Supplementary Figures S12 E–G, L, M, R, S, X and Y). As CLIPR-59 did not modulate TNFR1 expression on the cell surface or ligand-dependent internalization of TNFR1 (Supplementary Figure S12), CLIPR-59 probably does not modulate this step.
CLIPR-59 appears to interact directly with RIP1 based on the results from Y2H assay (Figure 3e). The domains within CLIPR-59 required for binding RIP1 versus CYLD are different, as a C-terminal mutant of CLIPR-59 (ΔC60) still interacted with RIP1 but not CYLD (Supplementary Figures S5A–C). It is suggested that interaction of CLIPR-59 with CYLD but not RIP1 is regulated during TNF-α stimulation (Supplementary Figure S5D). Moreover, siCLIPR-59 treatment disrupted the formation of protein complex of CYLD with RIP1 (Supplementary Figure S5E). We propose therefore that CLIPR-59 functions as an adapter that bridges CYLD to RIP1.
Several proteins involved in DR signaling have conserved domains, such as the DD, which is present in TNFR1, FADD, TRADD and RIP1, or the death effector domain found in Caspase-8 and FADD.7 Both CLIPR-59 and CYLD also have a conserved domain, the CAP-Gly domain, containing two or three such domains, respectively.19, 20, 23 Here, we show that the second CAP-Gly of CLIPR-59 is essential for its interactions with TNFR1 (Figure 1b). Previously, it was revealed that the first and third CAP-Gly domains of CYLD are responsible for interaction with microtubules and NEMO/IKKγ, respectively.19, 23, 31 Although CLIPR-59 binds DR617 and TNFR1, CYLD has been demonstrated to associate with several intracellular signaling molecules, including TNFR1, EDAR, CD40, TLRs, IL1R, TCR, LMP1 and BCL3.19, 23 Thus, CYLD may have a broader role in receptor-mediated signaling than CLIPR-59.
RIP1 has multiple roles in regulating cell death signaling.14, 16 Gene disruption of RIP1 prevents the activation of NF-κB but not the induction of cell death in TNFα-stimulated murine embryonic fibroblast (MEF) cells.32 In contrast, RIP1 appears to be essential for induction of apoptosis mediated by TNF-α in several tumor cell lines, including HeLa and Jurkat cells.33, 34 Recent reports using RIP1 gene knock-out mice, also suggest that RIP1 is dispensable for activating NF-κB but required for the induction of apoptosis in response to chemical antagonists of IAPs, where it has been suggested that the abolishment of cIAP1 or cIAP2 is needed to unmask RIP1-induced apoptosis.35 We assessed the effects of CLIPR-59 knockdown on apoptosis induction by TNF-α in several tumor cell lines (Supplementary Figure S13), finding that CLIPR-59 knockdown was more effective in HeLa cells (human cervical cancer cells) than A549 (human alveolar basal epithelial cells) or HT1080 (human fibrosarcoma cells). These findings suggest that the role of CLIPR-59 and RIP1 on TNF-α-mediated pro-apoptotic signaling may be dependent on cell type and context.
In previous reports,12, 36 it was suggested that post translational modification, such as ubiquitination, of RIP1 switches its function from pro-apoptotic to anti-apoptotic. Interestingly, constitutive RIP1 ubiquitination is detected in several tumor cell lines, but was not observed in primary cells such as MEFs.13 Interestingly, CLIPR-59 gene expression is reportedly epigenetically silenced in tumor-conditioned human umbilical vein endothelial cells.37 Also, surveying the Oncomine data set suggests that CLIPR-59 gene expression is decreased in some tumors compared with normal tissues. Taken together, these findings suggest that CLIPR-59 may have a tumor suppressor role, which merits further investigation.
Materials and Methods
Yeast two-hybrid assay
Yeast strain AH109 was transformed with combination of expression plasmids for yeast, pGBKT7- or pGADT7-containing cDNA encoding the indicated gene. Positive yeast clones were selected by prototrophy for leucine, tryptophan, histidine and adenine.
Immunoprecipitation and IB
Cells were lysed in RIPA buffer (150 mM NaCl, 10 mM Tris (pH 7.2), 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate and 5 mM EDTA) with serine and threonine phosphatase inhibitor cocktail (Sigma, St. Louis, MO, USA) and protease inhibitor cocktail (Roche, Heidelberg, Germany). In some experiments, the lysate was directly analyzed by SDS-PAGE (WCE), while in others IP with various Abs, was performed. After washing four times with lysis buffer, the immunoprecipitated beads were suspended in SDS-sample buffer, boiled and the eluted material was analyzed by SDS-PAGE. Proteins were transferred to nitrocellulose membrane (Millipore, Bradford, MA, USA). The membranes were incubated with the indicated Abs (IB). Visualization of immunoblots was performed by enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ, USA).
siRNA preparation and transfection
All siRNAs used in this study were obtained from Ambion Inc. (Austin, TX, USA). The target sequences of siRNAs for the human CLIPR-59 were CLIPR-59; 5′-GGACTACGCTTTCACCTTC-3′, CLIPR-59-2; 5′-GGTCACGCTACCCAACTAT-3′ and CLIPR-59-3; 5′-CCAACATGAACGCGCTTCA-3′, for human CYLD was 5′-GATTGTTACTTCTATCAAA-3′. For human cIAP1 and human cIAP2, the sequences were described in previously.38 For human RIP1 the sequences were described previously.39 Non-specific siRNA (Silencer Negative Control #1 siRNA) was used as control (Ambion). For gene-specific knockdown, HeLa cells (1 × 106 cells in 10 cm tissue culture dish) were transfected with various siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufactures. At 24 h after transfection, cells were trypsinized and re-seeded into 6-, 24- or 96-well culture plates. After 24 h of culture, cells were analyzed.
Measurement for caspase activity
Cells (1 × 104 cells) were seeded in 96-well white wall culture plates. At 24 h after incubation, cells were stimulated and at 3 h (for Caspase-8) or 4.5 h (for Caspase-3/7) after stimulation, Caspase-3/7 or -8 activity was measured by using Caspase Glo-3/7 or -8 reagents (Promega).
Cell viability assay
siRNA-transfected HeLa cells were seeded into 96-well culture plates. The following day, cells were stimulated with the indicated amount of TNF-α with CHX (10 μg/ml), puromycin (Cayla, Toulouse, France), or zeocin (Invitrogen). At 6 h (for TNF-α with CHX), 24 h (for puromycin) or 48 h (for zeocin) after stimulation, cell viability was assessed using the cell counting kit-8 (Dojindo, Tokyo, Japan).
Immunoprecipitation assay for detecting ubiquitination on TNFR1-associated RIP1
Detail of IP assay for detecting ubiquitination on TNFR1-associated RIP1 has been described previously.15 Briefly, cells were pretreated with 21 μM MG132 proteasome inhibitor for 10 min before stimulation with TNF-α (100 ng/ml). These cells were lysed with TNFR1 immunoprcipitation buffer (20 mM Tris-HCl (pH7.5), 150 mM NaCl, 1% Triton X-100 and 1 mM EDTA) containing protease inhibitor cocktail and 25 μM MG132 at 4°C. After centrifugation for removing debris at 15 000 r.p.m. at 4°C 30 min, cell lysate was precleared with protein G sepharose and then incubated with anti-TNFR1 Ab with protein G sepharose at 4°C overnight. After four times washes with the lysis buffer, precipitated proteins were eluted in lysis buffer containing 6 M urea. Each eluted sample was diluted 25-fold with lysis buffer for reducing the concentration of urea and divided two samples. One aliquot was subjected to second IP using anti-K48-linked ubiquitin chain-specific Ab (clone Apu2) and the other was subjected to anti-K63-linked ubiquitin chain-specific Ab (clone Apu3) with protein G sepharose for 2 h at 4°C. After four times washes with lysis buffer, the precipitated materials were analyzed by western blot method using the indicated Abs.
Cell death assay using flow cytometry
The cells were transfected with the indicated plasmids. At 24 h after transfection, cells were washed two times by PBS for removing the stimulation-independent dead cells, and then stimulated by the reagents in culture medium. At 6 h after treatment, the cells were trypsinized and resuspended into PBS. Dead cells in each sample were stained with 7-AAD according to the manufactures.
Fluorescence activities of the cells were detected by Flow cytometer (BD Biosciences, Heidelberg, Germany). The data were shown as the percentage of 7-AAD(+) cells in GFP(+)-gated cells (at least 104 cell counts).
Acknowledgments
We thank Dr. TC Wu for kindly providing cell line TC-1 P0 and P3 cells, Drs. N Inohara, JI Inoue, S Hatakeyama and KI Nakayama for the gift of plasmids, Dr. X Wang for kindly providing smac mimetic compound ‘Compound 3′ and Dr. M Maruyama for comments and discussion, HM Li for technical assistance. This work was supported by Grants-in-Aid for Scientific Research and for Young Scientists, Grant of the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from MEXT, Grants from Japan Health Sciences Foundation and a grant from the National Institution of Health (CA69381).
Glossary
- TNF
tumor necrosis factor
- DR
death receptor
- TRADD
TNF receptor-associated death domain protein
- RIP1
receptor-interacting protein 1
- FADD
Fas-associated protein with death domain
- CLIPR-59
CLIP-170-related 59 kDa protein
- DD
death domain
- CHX
cycloheximide
- IAP
inhibitor of apoptosis
- MEF
murine embryonic fibroblast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by P Salomoni
Supplementary Material
References
- Benedict CA, Banks TA, Ware CF. Death and survival: viral regulation of TNF signaling pathways. Curr Opin Immunol. 2003;15:59–65. doi: 10.1016/s0952-7915(02)00018-3. [DOI] [PubMed] [Google Scholar]
- Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CF. Network communications: Lymphotoxins, LIGHT, and TNF. Annu Rev Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Aggarwal BB. Receptor-mediated choreography of life and death. J Clin Immunol. 2003;23:317–332. doi: 10.1023/a:1025319031417. [DOI] [PubMed] [Google Scholar]
- Vassalli P. The pathophysiology of tumor necrosis factors. Annu Rev Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat Immunol. 2002;3:1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- Reed JC. Mechanisms of apoptosis. Am J Pathol. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Muppidi JR, Tschopp J, Siegel RM. Life and death decisions: secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity. 2004;21:461–465. doi: 10.1016/j.immuni.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Reed JC. Apoptosis-regulating proteins as targets for drug discovery. Trends Mol Med. 2001;7:314–319. doi: 10.1016/s1471-4914(01)02026-3. [DOI] [PubMed] [Google Scholar]
- Green DR. Overview: apoptotic signaling pathways in the immune system. Immunol Rev. 2003;193:5–9. doi: 10.1034/j.1600-065x.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappa B independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJM, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Fujikura D. [Analysis of the mechanism involved in intra-cellular information transmission through death receptor 6 (DR6), a new apoptosis inducing receptor] Hokkaido Igaku Zasshi. 2006;81:399–407. [PubMed] [Google Scholar]
- Ding JX, Du KY. ClipR-59 interacts with Akt and regulates Akt cellular compartmentalization. Mol Cell Biol. 2009;29:1459–1471. doi: 10.1128/MCB.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F, Pernet-Gallay K, Nizak C, Goodson HV, Kreis TE, Goud B. CLIPR-59, a new trans-Golgi/TGN cytoplasmic linker protein belonging to the CLIP-170 family. J Cell Biol. 2002;156:631–642. doi: 10.1083/jcb.200111003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell G, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SMB, Dirac AMG, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappa B. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappa B signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappa B activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Hovelmeyer N, Wunderlich FT, Massoumi R, Jakobsen CG, Song J, Worns MA, et al. Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J Exp Med. 2007;204:2615–2627. doi: 10.1084/jem.20070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- Li L, Thomas RM, Suzuki H, De Brabander JK, Wang XD, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNF alpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- Wang L, Du FH, Wang XD. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Lin KY, Lu D, Hung CF, Peng SW, Huang LQ, Jie CF, et al. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer Res. 2007;67:1832–1841. doi: 10.1158/0008-5472.CAN-06-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dri P, Gasparini C, Menegazzi R, Cramer R, Alberi L, Presani G, et al. TNF-induced shedding of TNF receptors in human polymorphonuclear leukocytes: role of the 55-kDa TNF receptor and involvement of a membrane-bound and non-matrix metalloproteinase. J Immunol. 2000;165:2165–2172. doi: 10.4049/jimmunol.165.4.2165. [DOI] [PubMed] [Google Scholar]
- Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, Sakamoto A, et al. The CAP-Gly domain with the proline-rich of CYLD associates sequence in NEMO/IKK gamma. Structure. 2004;12:1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappa B signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Jin ZY, El-Deiry WS. Distinct signaling pathways in TRAIL- versus tumor necrosis factor-induced apoptosis. Mol Cell Biol. 2006;26:8136–8148. doi: 10.1128/MCB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel-Muinos FX, Seed B. Regulated commitment of TNF receptor signaling: a molecular switch for death or activation. Immunity. 1999;11:783–793. doi: 10.1016/s1074-7613(00)80152-1. [DOI] [PubMed] [Google Scholar]
- Wong WWL, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappa B. Cell Death Differ. 2010;17:482–487. doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJJ. Activation of IKK by TNF alpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Hellebrekers D, Melotte V, Viré E, Langenkamp E, Molema G, Fuks F, et al. Identification of epigenetically silenced genes in tumor endothelial cells. Cancer Res. 2007;67:4138–4148. doi: 10.1158/0008-5472.CAN-06-3032. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Peyton M, Minna JD, Wang XD. Overcoming cancer cell resistance to Smac mimetic induced apoptosis by modulating cIAP-2 expression. Proc Natl Acad Sci USA. 2010;107:11936–11941. doi: 10.1073/pnas.1005667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Ohoka N, Okuhira K, Sai K, Nishimaki-Mogami T, Naito M. Modulation of RIP1 ubiquitylation and distribution by MeBS to sensitize cancer cells to tumor necrosis factor alpha-induced apoptosis. Cancer Sci. 2010;101:2425–2429. doi: 10.1111/j.1349-7006.2010.01697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.