Summary
Myosin IIIA (MYO3A) targets actin protrusion tips using a motility mechanism dependent on both motor and tail actin-binding activity [1]. We show that myosin IIIB (MYO3B) lacks tail actin-binding activity and is unable to target COS7 cell filopodia tips, yet is somehow able to target stereocilia tips. Strikingly, when MYO3B is coexpressed with espin-1 (ESPN1), a MYO3A cargo protein endogenously expressed in stereocilia [2], MYO3B targets and carries ESPN1 to COS7 filopodia tips. We show that this tip-localization is lost when we remove the ESPN1 C-terminus actin-binding site. We also demonstrate that, like MYO3A [2], MYO3B can elongate filopodia by transporting ESPN1 to the polymerizing end of actin filaments. The mutual dependence of MYO3B and ESPN1 for tip-localization reveals a novel mechanism for the cell to regulate myosin tip-localization via a reciprocal relationship with cargo that directly participates in actin binding for motility. Our results are consistent with a novel form of motility for class III myosins that requires both motor and tail domain actin-binding activity, and show that the actin-binding tail can be replaced by actin-binding cargo. This study also provides a framework to better understand the late-onset hearing loss phenotype in patients with MYO3A mutations.
Results
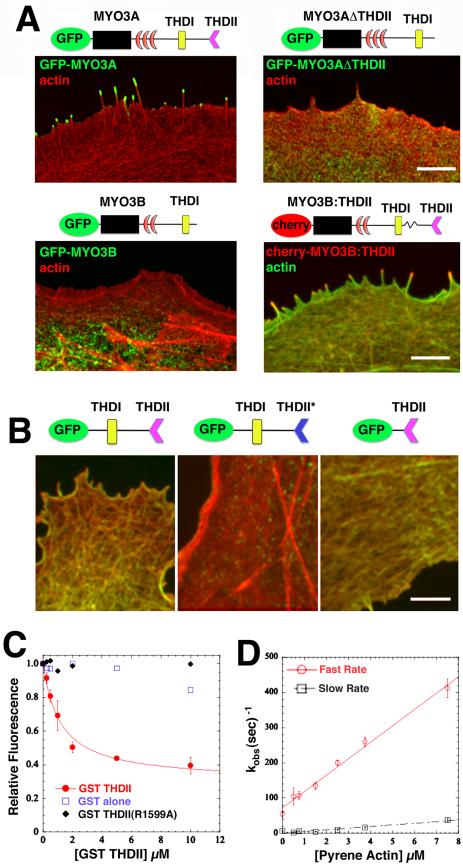
It has been speculated that the motor protein myosin-IIIA (MYO3A) translocates along actin filaments towards actin protrusion tips as a monomer, using an inchworm-like mode of motility that depends on a highly conserved actin-binding site (THDII) in its C-terminus tail [1]. It is unknown whether the myosin-IIIB (MYO3B) tail, which lacks the THDII [3], is capable of binding to actin or whether MYO3B can target the tips of actin protrusions. Consistent with earlier studies [1, 2, 4] we found that GFP-MYO3A localizes to and accumulates at the tips of COS7 cell filopodia (Figure 1A). In contrast, GFP-MYO3B, which lacks the putative second actin-binding domain (THDII, [3]), failed to localize to filopodia tips (Figure 1A). As predicted by the inchworm hypothesis that depends on the actin-binding activity of the THDII [1], MYO3A lacking the THDII (GFP-MYO3AΔTHDII) also failed to localize to filopodia tips (Figure 1A). However, when we fused a THDII from MYO3A onto the C-terminus of MYO3B (cherry-MYO3B:THDII), the hybrid protein localized to the tips of filopodia (Figure 1A).
Figure 1. The MYO3A THDII binds to actin filaments in vivo and in vitro.
(A) GFP-MYO3A (upper left) targets the tips of filopodia, GFP-MYO3B (bottom left) and GFP-MYO3AΔTHDII (upper right) fail to target filopodia tips, whereas a MYO3B hybrid protein containing the THDII (cherry-MYO3B:THDII, bottom right) successfully targets filopodia tips. Scale bar: 2 μm. Due to the significant attenuation of class III myosin activity by a kinase domain at its N-terminus [1, 2, 4, 9, 10], all of the class III myosin constructs used in our experiments lacked the kinase domain unless otherwise indicated. The cartoon diagrams show the corresponding constructs for each transfection (black box: motor domain, red crescent: IQ domains), emphasizing the lack of an N-terminus kinase domain, and also that MYO3A has three IQ domains while MYO3B only has two. (B) A GFP-tagged construct (green) containing the MYO3A THDI and THDII domains colocalizes with actin (red) in transfected COS7 cells (left). A construct with the R1599A point mutation in the THDII (THDII*) fails to colocalize with actin (middle), while a construct containing only the THDII colocalizes with actin (right). Scale bar: 2 μm. (C) Equilibrium binding experiments monitoring pyrene actin (0.2 μM) quenching were used to examine GST-3THDII (varying concentrations indicated on x-axis) binding to actin (Kd = 1.2 ± 0.2). A GST-3THDII construct with the R1599A point mutation failed to bind to actin, consistent with the colocalization experiments in (B). Error bars represent the standard deviation from three separate experiments. (D) Pyrene quenching (5-10 fold excess over GST-THDII concentrations) was followed in the stopped flow to measure the rate of association (46.6 ± 2.0 μM•sec−1). The rate of dissociation was measured by mixing a GST-THDII:pyrene actin complex with 20-fold excess unlabeled actin (54.8 ± 6.4 sec−1). The association and dissociation rates allowed determination of the overall affinity (Kd = 1.2 ± 0.2 μM) which was similar to that measured by titration in (c). The association transients followed a bi-exponential and the slow rate was likely due to some actin bundling activity (see also Figure S1). Error bars represent the standard error of the fits.
To confirm that the THDII interacts with actin in cells, we transfected COS7 cells with GFP tagged MYO3A tail constructs containing the THDII, and we found that the GFP fluorescence colocalized with actin (Figure 1B). When using a construct containing a point mutation in the THDII (R1599A) known to disrupt its actin binding activity [1], the GFP fluorescence was diffuse with no colocalization with actin (Figure 1B). We used pyrene actin quenching assays to directly examine binding of purified GST-THDII to actin (Figure 1C), thereby allowing us to determine the affinity with which the THDII binds actin. We found in both fluorescence titration and stopped-flow experiments, in which the association and dissociation rate constants were determined (Figure 1D), that the binding affinity was close to one micromolar (Kd = 1.2 ± 0.2 μM). The stopped-flow association transients followed a bi-exponential. The slow rate was likely due to some actin bundling activity, which was also seen in light scatter measurements (Figure S1) using peptides derived from the THDII sequence (MYO3A amino acids 1587-1615). In contrast, a peptide from the C-terminal region of MYO3B (amino acids 1293-1310) did not bind actin (Figure S1).
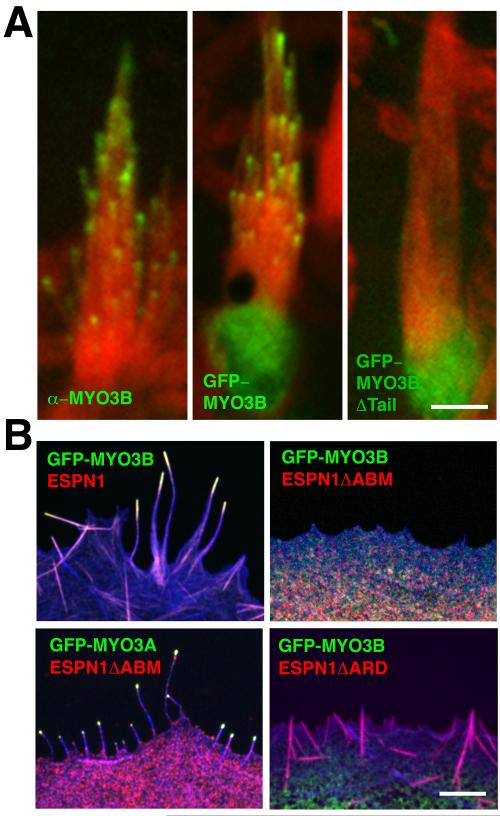
In order to verify MYO3B expression and to compare localizations of MYO3B to ESPN1 and MYO3A in mouse inner ear hair cells, we used affinity purified antibodies specific to these proteins (Figure S2A-B). We found that MYO3B localizes at vestibular (Figure 2A) and cochlear (Figure S2C) hair cell stereocilia tips, similar to the localization of ESPN1 and MYO3A, both of which localize to the tips of stereocilia in a tip-to-base gradient [2, 4]. However, MYO3B labeling in the cochlear hair cells diminished rapidly from postnatal days 0 to 8, while in vestibular hair cells labeling persisted (Figures S2D and 2A, respectively). We also transfected organ of Corti and vestibular hair cells with GFP-tagged MYO3B to corroborate the immunolocalization results, and to test whether, like MYO3A, the MYO3B pattern of distribution at the tips of stereocilia is consistent with a dynamic mode of tip-directed transport [5]. Within 18 hours of transfection GFP-MYO3B localized in a tip-to-base gradient distribution similar to MYO3A (Figure 2A). To test whether the tail domain of MYO3B is required for the stereocilia tip localization we transfected hair cells with a GFP-MYO3B construct lacking the tail domain (GFP-MYO3BΔTail). Hair cells transfected with GFP-MYO3BΔTail showed diffuse fluorescence in the cell body with no apparent enrichment of fluorescence at stereocilia tips (Figure 2A, ncells = 17 hair cells), demonstrating that the MYO3B tail is necessary for tip-localization.
Figure 2. MYO3B localizes to actin protrusion tips in an ESPN1 dependent fashion.
(A) MYO3B immunolabeling (left) and transfections (middle) shows localization at the tips of vestibular hair cell stereocilia (actin is shown in red) in a tip-to-base gradient (see also Figure S2), with higher amounts in longer stereocilia. In contrast, a construct lacking the C-terminus tail of MYO3B, GFP-MYO3BΔTail, does not target stereocilia tips (right). Scale bar: 5 μm. (B) When co-expressed with ESPN1, GFP-MYO3B targets filopodia (actin is shown in blue) tips (upper left). When GFP-MYO3B is co-expressed with an ESPN1 construct lacking the C-terminus actin-binding site (ESPN1ΔABM), MYO3B fails to target filopodia tips (upper right). GFP-MYO3A targets ESPN1ΔABM to filopodia tips (bottom left). MYO3B fails to target filopodia tips when coexpressed with ESPN1ΔARD (bottom right). Scale bar: 2 μm.
The colocalization of MYO3B and ESPN1, and the fact that the MYO3B tail contains the same THDI that mediates interactions between MYO3A and ESPN1 [2], suggests that a similar interaction could occur between MYO3B and ESPN1. When we coexpressed MYO3B and ESPN1 in COS7 cells, the two proteins colocalized to filopodia and actin bundles (Figure S3A). In order to test whether, like MYO3A, the MYO3B THDI interacts with the ESPN1 ankyrin repeats domain (ARD), we cotransfected COS7 cells with ESPN1 and GFP-tagged constructs encoding either the MYO3B THDI (GFP–MYO3BTHDI) or its immediate flanking regions (GFP-MYO3BpreTHDI or GFP-MYO3BpostTHDI). Colocalization was exclusively observed for GFP-MYO3BTHDI with ESPN1 (Figure S3A). The interaction between the MYO3B THDI and ESPN1 ARD was further confirmed by GST pull-downs in which the GFP-MYO3B fusion protein co-precipitated with a GST-fused ESPN1 ARD (Figure S3B). The MYO3BTHDI:ESPN1 ARD interaction was also verified in which the GST–ESPN1 ARD binds to GFP–MYO3BTHDI, but not to the pre-THDI or post-THDI regions (Figure S3B).
To explain how MYO3B localizes to stereocilia tips we hypothesized that MYO3B compensates for the lack of a THDII by binding to ESPN1, which has a C-terminus actin-binding module (ABM) and is endogenously expressed in hair cells. To test this hypothesis, we cotransfected COS7 cells with MYO3B and ESPN1. In these cotransfected cells, we found that MYO3B colocalized with ESPN1 at filopodia tips (Figure 2B), in a tip-to-base gradient consistent with tip-directed motor-based motility [5]. In order to determine whether the MYO3B:ESPN1 interaction imparts tip-directed motility to MYO3B via the ESPN1 ABM, we generated an ESPN1 construct lacking the ABM (ESPN1ΔABM). When COS7 cells were cotransfected with GFP-MYO3B and ESPN1ΔABM, the two proteins localized diffusely in the cytoplasm and failed to elongate filopodia or target their tips (Figure 2B). In contrast, GFP-MYO3A was still able to transport ESPN1ΔABM to filopodia tips (Figure 2B). In order to determine whether the ESPN1-dependent tip-localization of MYO3B was a secondary effect of ESPN1’s bundling activity, we cotransfected MYO3B and an ESPN1 construct (ESPN1ΔARD) lacking the N-terminus ankyrin repeats domain required for binding the MYO3 THDI, and we found that MYO3B had a diffuse localization in the cell body similar to when MYO3B was expressed alone or with ESPN1ΔABM (Figure 2B). The lack of colocalization also indicates that MYO3B does not interact with the ESPN1 ABM.
MYO3A has been shown to boost the elongation of stereocilia and filopodia via the transport of its cargo protein ESPN1 to the barbed ends of actin filaments, whereupon ESPN1’s WH2 activity enhances actin polymerization in a concentration dependent fashion [2, 5]. To examine whether MYO3B mimics MYO3A activity in stereocilia, we overexpressed GFP-MYO3B (n=13 hair cells) and GFP-MYO3B with the kinase domain (GFP-MYO3BKin, n=6 hair cells) in rat vestibular hair cells. We found that hair cells displayed a 66% increase in stereocilia elongation when transfected with GFP-MYO3B and 51% increase with GFP-MYO3BKin (p<0.005) (Figure S2E), similar to what was found for MYO3A [2].
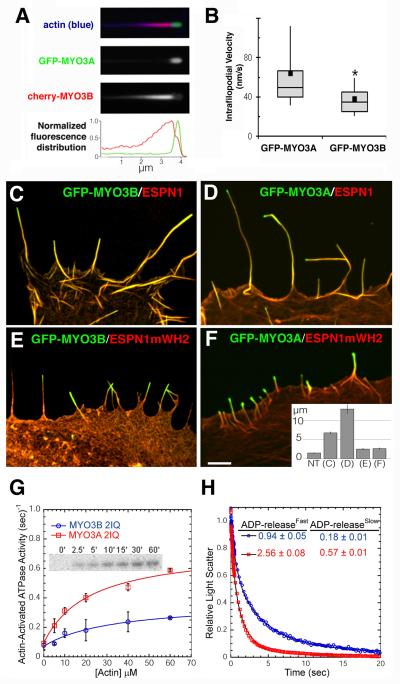
The fact that, like MYO3A, MYO3B can localize to the tips of stereocilia and that MYO3B requires ESPN1 for translocation, led us to further investigate how these proteins behave when all three are coexpressed in a heterologous system. We found that when MYO3A, MYO3B, and ESPN1 are coexpressed in COS7 cells, MYO3A invariably dominated localization at the extreme tips of filopodia, while MYO3B trailed along the shafts of filopodia, and inversely proportional amounts of the two myosins were found in the overlapping regions near the tips of filopodia (Figure 3A). To compare the difference in tip-localization efficiency for each myosin, we measured their tip-to-base gradient distributions and found that MYO3A had an average half-length of Lh = 0.049 ± 0.006 (see Supplementary Methods), while the mean MYO3B half-length was Lh = 0.41 ± 0.04 indicating a nearly ten-fold decrease in tip-localization efficiency for MYO3B compared to MYO3A.
Figure 3. MYO3A outperforms MYO3B in filopodia and in vitro.
(A) Filopodia from COS7 cells co-expressing GFP-MYO3A and cherry-MYO3B consistently display GFP-MYO3A accumulated at their extreme tips, while cherry-MYO3B consistently trails behind MYO3A with a relatively longer tip-to-base decay length (left column, and see also Figure S4). (B) Kymograph analysis of live-cell TIRF time-lapse movies from COS7 cells expressing GFP-MYO3A and ESPN1 (n=15 features) or GFP-MYO3B and ESPN1 (n=13 features) reveal GFP-MYO3A features moving approximately 1.7-fold faster than GFP-MYO3B features. Whiskers represent 90th percentile and 10th percentile. The box represents the 75th and 25th percentile, and the bar across the box indicates the median. The black square indicates the mean. *Mean significantly different from GFP-MYO3A via t-test, p<0.03. (C) GFP-MYO3B targets and elongates filopodia tips (ncells=12, nfilopodia=91) when co-expressed with ESPN1. (D) GFP-MYO3A targets and elongates filopodia tips when coexpressed with ESPN1 (nc = 13, nf = 73). (E) GFP-MYO3B targets, but does not elongate, filopodia tips when co-expressed with ESPN1mWH2 (nc = 15, nf = 61). (F) GFP-MYO3A targets filopodia tips, but fails to elongate them when co-expressed with ESPN1mWH2 (nc = 22, nf =147). Scale bar: 1 μm. (inset) Average filopodia length for non-transfected (NT) cells and for each transfection condition displayed in (C-F). Error bars represent the standard error of the mean. (G) The ATPase activity was measured with the NADH-coupled assay and the maximum ATPase activity (kcat) and actin concentration at which the ATPase activity is one-half maximal (KATPase) was determined by fitting the data to the Michealis-Menton equation. The inset demonstrates the kinase activity of MYO3B 2IQ measured by incorporation of 32P at the indicated time points following the addition of 32P-ATP. (H) The rate ATP-induced dissociation of acto-MYO3B 2IQ and acto-MYO3B 2IQ in the presence of ADP was measured by monitoring light scatter in the stopped-flow. A complex of acto-MYO3B 2IQ (blue) or -MYO3A 2IQ (red) in the presence of ADP was mixed with saturating ATP (final concentrations: 0.6 μM actin, 0.5 μM MYO3A or MYO3B 2IQ, 10 μM ADP, and 2.5 mM ATP) and the decrease in light scatter was monitored at 420 nm. The light scatter transients were best fit to a two exponential function and the fast and slow phases of the fits are shown in the table inset.
To determine if different velocities of intrafilopodial motility (IFM) for MYO3A and MYO3B could be observed, COS7 cells expressing ESPN1 and GFP-MYO3A or MYO3B were imaged using TIRF microscopy, similar to Kerber et al. [6]. The fact that both GFP-MYO3A and GFP-MYO3B label along the length of ESPN1-labelled filopodia resulted in moving features that were less punctate than those observed for MYO10 [6]. GFP-MYO3A tip-directed IFM velocities (64 ± 10 nm/s, n = 15 moving features) were approximately 1.7-fold faster than tip-directed GFP-MYO3B IFM (38 ± 4 nm/s, n = 13 moving features, t-test, p <0.03) (Figure 3B).
Similar to what has been shown for MYO3A [2], we found that COS7 cells cotransfected with GFP-MYO3B and ESPN1 displayed enhanced elongation of filopodia, in which the mean filopodia length was twice as long (6.7 ±0.22 μm; Figure 3C) as those transfected with GFP-ESPN1 alone (2.9 ±0.11 μm; nc = 19, nf = 162, p<0.001). However, MYO3B did not increase filopodia lengths as much as MYO3A (13.0 ± 1.1 μm, p<0.001; Figure 3D).
MYO3A elongation of ESPN1 filopodia depends on the ESPN1 WH2 domain, as shown in a previous study using an ESPN1 construct (ESPN1mWH2) in which the WH2 domain was mutated by substituting two conserved Leu residues (L655A, L656A) essential for WH2 actin-monomer-binding activity [2, 7, 8]. The MYO3A:ESPN1mWH2 co-expression phenotype displays filopodia tip-localization of both MYO3A and ESPN1, but no filopodia elongation. To better understand the similarity between the MYO3A and MYO3B transport function of ESPN1, we tested whether GFP-MYO3B can target filopodia tips with ESPN1mWH2 and mimic the MYO3A:ESPN1mWH2 co-expression phenotype. We found that GFP-MYO3B was able to colocalize with ESPN1mWH2 at filopodia tips, yet it did not enhance filopodia elongation (2.7 ± 0.15 μm; Figure 3E), very similar to the phenotype of MYO3A:ESPN1mWH2 (2.7 ± 0.15 μm; Figure 3F). Since MYO3B tip-localization and synergistic elongation of actin protrusions depends on the ESPN1 ABM, we asked whether it is also required for MYO3A function. Although co-transfection of GFP-MYO3A with ESPN1ΔABM showed filopodia tip targeting and produced filopodia (Figure 2B), these filopodia were significantly shorter (2.5 ± 0.17 μm, nc = 12, nf = 80) than those from cells co-transfected with MYO3A and full-length ESPN1 (13.0 ± 1.1 μm, nc = 13, nf = 73, p<0.001), demonstrating that while ESPN1 WH2 activity is necessary for filopodia elongation [2], it is not sufficient.
We hypothesized that this difference in tip-localization efficiency and filopodia elongation activity might correlate with the motor activity of each myosin. Using an NADH-coupled ATPase assay [9-11] with an expressed MYO3B construct containing the kinase, motor, and two IQ domains (MYO3B 2IQ), we found that the maximum actin-activated ATPase rate (kcat) was 0.30 ± 0.07s−1 (Figure 3G), which corresponded to about half that of MYO3A 2IQ [9-11]. In addition, we observed two phases in our ATP-induced dissociation experiments in the presence of ADP - the slow phase with acto-MYO3B 2IQ was similar to kcat (Figure 3H) indicating that the ADP release or detachment is rate-limiting, as was found with MYO3A 2IQ [3]. In myosins that are rate limited by ADP release or detachment, the in vitro motility rates have been found to correlate well with maximal ATPase activity [12-14]. The actin concentration at which the ATPase rate is one-half maximal (KATPase = 34 ± 20μM), which is a relative measure of actin affinity, was found to be slightly larger in MYO3B 2IQ than MYO3A 2IQ. Radiolabeled (32-P) ATP assays indicated that MYO3B 2IQ had kinase activity (Figure 3G - inset), as was found in earlier MYO3A 2IQ studies [9, 11]. We have previously demonstrated that fully phosphorylated MYO3A 2IQ has reduced motor ATPase activity [9]. However, our MYO3B 2IQ kinase activity measurements suggest that the relatively slower motor ATPase activity for MYO3B compared to MYO3A is not a result of phosphorylation-mediated downregulation of MYO3B 2IQ.
Discussion
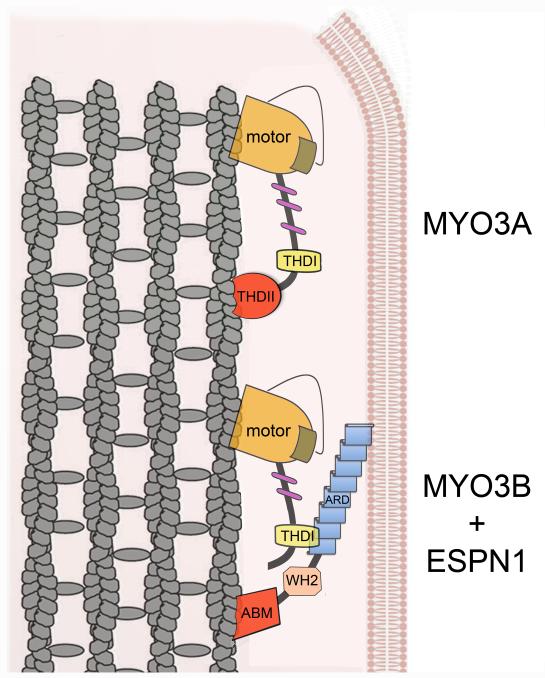
MYO3A has been presumed to be a single-headed monomeric motor protein with barbed-end directed motility that relies on its THDII-dependent actin-binding activity [1]. Although direct measurements of single molecule MYO3A motility have yet to be performed, our data are consistent with a model of “inchworm” based movement along actin bundles. The relatively high affinity (KActin = 7μM) of the MYO3A motor for actin in the presence of ATP [11] combined with THDII actin binding may prevent dissociation of MYO3A even in the weak actin binding states. In addition, the MYO3A motor rapidly transitions into the actomyosin-ADP state [11], which may prevent dissociation even though the THDII rapidly cycles on and off (koff ~ 50 sec−1) actin. Therefore, the rapid on-off rates of THDII may facilitate movement along actin without interfering with motor stepping. In contrast, MYO3B does not contain the THDII. However, we show that the absence of an actin-binding domain in the C-terminus of MYO3B can be compensated for by the ESPN1 cargo’s actin-binding activity, or by adding the actin-binding THDII to its tail. Thus, ESPN1 cargo could serve as an actin-binding replacement or “crutch” (Figure 4). It will be interesting to compare the actin-binding properties of the THDII and the ESPN1 ABM, as well as to determine whether ESPN1 enhances MYO3A motility due to the additional actin-binding activity of its ABM, perhaps analogously to what has been shown for the microtubule-binding dynein:dynactin complex [15].
Figure 4. Cartoon of the cargo-assisted motility model of MYO3B -ESPN1 complexes.
MYO3A (top) uses coordination between the motor head activity (regulated by the kinase domain, brown) and the tail THDII actin-binding site in its C-terminus (red) to translocate along actin filaments (gray). In contrast, MYO3B (bottom), which lacks the THDII but is otherwise very similar to MYO3A, binds to the ESPN1 ARD via its THDI, allowing it to use the C-terminus ABM of ESPN1 as a substitute for its missing THDII to translocate along actin filaments towards the protrusion tip.
One mechanism by which myosins are activated by their cargo is the cargo-binding induced dimerization effect, as has been reported for myosin VI [8, 16]. Another mechanism is the cargo-binding dependent activation of an otherwise auto-inhibited folded state of the motor, as has been shown to be the case for other molecular motors [17-20]. The fact that we did not observe MYO3B localization to filopodia tips when we removed the C-terminus actin-binding domain of ESPN1 (ESPN1ΔABM) is consistent the model in which the ESPN1 actin-binding activity via its C-terminus ABM provides a second actin-binding site that cooperates with the MYO3B actin-binding motor domain for motility, similarly to the way the MYO3A THDII coordinates with the MYO3A motor domain. However, we cannot rule out the possibility that the full-length ESPN1 protein somehow activates MYO3B motility via some other mechanism, e.g. cargo-mediated dimerization [8, 16] or relief of auto-inhibition [17-20]. In any case, the dependence of MYO3B motility on the presence of ESPN1 demonstrates that the cell can regulate the efficiency and selectivity of MYO3B targeting to stereocilia via regulation of its cargo.
The ability of MYO3B to enhance elongation of ESPN1-induced filopodia in COS7 cells mimics MYO3A:ESPN1 activity only partially, since MYO3BΔK:ESPN1 produced filopodia only half the length of MYO3AΔK:ESPN1. Since MYO3A elongation has been directly correlated with MYO3A ATPase rates [2], which should correlate well with motor velocity in class III myosins, we propose that the less enhanced elongation activity of MYO3B is a direct reflection of the ~50% slower ATPase rates and tip-directed motility for MYO3B versus MYO3A.
MYO3A was first implicated in hearing loss via its association with late-onset non-syndromic hearing loss (DFNB30) [21]. The persistence of proper vestibular function along with the late-onset hearing loss phenotype in DFNB30 patients [21] as well as a mouse model for DFNB30 [22] suggests that another protein (i.e. MYO3B) may be partially compensating for MYO3A. Here we show that MYO3B is expressed in hair cell stereocilia in the same spatial pattern of localization as MYO3A and its cargo ESPN1. The similar localization of MYO3B, MYO3A and ESPN1 in stereocilia suggests that MYO3B is similar in function and activity to MYO3A. The fact that the THDI domains of both MYO3B and MYO3A interact with ESPN1, and that both proteins exhibit ESPN1-dependent elongation activity in filopodia strongly indicate some level of redundancy in the roles of these proteins in hair cell stereocilia length regulation [23, 24] and function [25, 26].
Supplementary Material
Acknowledgments
We thank Drs. Aurea de Sousa and Mark Schneider for help with preliminary experiments and Dr. Ronald Petralia for helpful suggestions with the manuscript. We thank Michael Rose for assistance with the equilibrium binding experiments with GST-THDII. This work was supported by National Institutes of Health Intramural Research Fund Z01-DC000002-22 (B.K.) and NIH awards EY018141 and HL093531 (C.M.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information Supplemental Information includes 3 figures, one table, and Supplemental Experimental Procedures.
References
- 1.Les Erickson F, Corsa AC, Dose AC, Burnside B. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Molecular biology of the cell. 2003;14:4173–4180. doi: 10.1091/mbc.E02-10-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salles FT, Merritt RC, Jr., Manor U, Dougherty GW, Sousa AD, Moore JE, Yengo CM, Dose AC, Kachar B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nature cell biology. 2009;11:443–450. doi: 10.1038/ncb1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dose AC, Hillman DW, Wong C, Sohlberg L, Lin-Jones J, Burnside B. Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Molecular biology of the cell. 2003;14:1058–1073. doi: 10.1091/mbc.E02-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider ME, Dose AC, Salles FT, Chang W, Erickson FL, Burnside B, Kachar B. A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J Neurosci. 2006;26:10243–10252. doi: 10.1523/JNEUROSCI.2812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naoz M, Manor U, Sakaguchi H, Kachar B, Gov NS. Protein localization by actin treadmilling and molecular motors regulates stereocilia shape and treadmilling rate. Biophysical journal. 2008;95:5706–5718. doi: 10.1529/biophysj.108.143453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, Quintero OA, Cheney RE. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomis PA, Kelly AE, Zheng L, Changyaleket B, Sekerkova G, Mugnaini E, Ferreira A, Mullins RD, Bartles JR. Targeted wild-type and jerker espins reveal a novel, WH2-domain-dependent way to make actin bundles in cells. Journal of cell science. 2006;119:1655–1665. doi: 10.1242/jcs.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 9.Quintero OA, Moore JE, Unrath WC, Manor U, Salles FT, Grati M, Kachar B, Yengo CM. Intermolecular autophosphorylation regulates myosin IIIa activity and localization in parallel actin bundles. The Journal of biological chemistry. 2010;285:35770–35782. doi: 10.1074/jbc.M110.144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dose AC, Ananthanarayanan S, Moore JE, Corsa AC, Burnside B, Yengo CM. The kinase domain alters the kinetic properties of the myosin IIIA motor. Biochemistry. 2008;47:2485–2496. doi: 10.1021/bi7021574. [DOI] [PubMed] [Google Scholar]
- 11.Dose AC, Ananthanarayanan S, Moore JE, Burnside B, Yengo CM. Kinetic mechanism of human myosin IIIA. The Journal of biological chemistry. 2007;282:216–231. doi: 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris CA, Wells AL, Yang Z, Chen LQ, Baldacchino CV, Sweeney HL. Calcium functionally uncouples the heads of myosin VI. The Journal of biological chemistry. 2003;278:23324–23330. doi: 10.1074/jbc.M208957200. [DOI] [PubMed] [Google Scholar]
- 14.Nagy NT, Sakamoto T, Takacs B, Gyimesi M, Hazai E, Bikadi Z, Sellers JR, Kovacs M. Functional adaptation of the switch-2 nucleotide sensor enables rapid processive translocation by myosin-5. Faseb J. 2010;24:4480–4490. doi: 10.1096/fj.10-163998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nature cell biology. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 16.Phichith D, Travaglia M, Yang Z, Liu X, Zong AB, Safer D, Sweeney HL. Cargo binding induces dimerization of myosin VI. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai T, Umeki N, Ikebe R, Ikebe M. Cargo binding activates myosin VIIA motor function in cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7028–7033. doi: 10.1073/pnas.1009188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umeki N, Jung HS, Sakai T, Sato O, Ikebe R, Ikebe M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 20.Thirumurugan K, Sakamoto T, Hammer JA, 3rd, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh VL, Raviv D, Dror AA, Shahin H, Walsh T, Kanaan MN, Avraham KB, King MC. A mouse model for human hearing loss DFNB30 due to loss of function of myosin IIIA. Mamm Genome. 2011;22:170–177. doi: 10.1007/s00335-010-9310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manor U, Kachar B. Dynamic length regulation of sensory stereocilia. Semin Cell Dev Biol. 2008;19:502–510. doi: 10.1016/j.semcdb.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin HW, Schneider ME, Kachar B. When size matters: the dynamic regulation of stereocilia lengths. Current opinion in cell biology. 2005;17:55–61. doi: 10.1016/j.ceb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Schwander M, Kachar B, Muller U. Review series: The cell biology of hearing. The Journal of cell biology. 2010;190:9–20. doi: 10.1083/jcb.201001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waguespack J, Salles FT, Kachar B, Ricci AJ. Stepwise morphological and functional maturation of mechanotransduction in rat outer hair cells. J Neurosci. 2007;27:13890–13902. doi: 10.1523/JNEUROSCI.2159-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.