Abstract
Background
Carotid-femoral pulse wave velocity (CFPWV) is a heritable measure of aortic stiffness that is strongly associated with increased risk for major cardiovascular disease events.
Methods and Results
We conducted a meta-analysis of genome-wide association data in 9 community-based European ancestry cohorts consisting of 20,634 participants. Results were replicated in 2 additional European ancestry cohorts involving 5,306 participants. Based on a preliminary analysis of 6 cohorts, we identified a locus on chromosome 14 in the 3′-BCL11B gene desert that is associated with CFPWV (rs7152623, minor allele frequency = 0.42, beta=−0.075±0.012 SD/allele, P = 2.8 x 10−10; replication beta=−0.086±0.020 SD/allele, P = 1.4 x 10−6). Combined results for rs7152623 from 11 cohorts gave beta=−0.076±0.010 SD/allele, P=3.1x10−15. The association persisted when adjusted for mean arterial pressure (beta=−0.060±0.009 SD/allele, P = 1.0 x 10−11). Results were consistent in younger (<55 years, 6 cohorts, N=13,914, beta=−0.081±0.014 SD/allele, P = 2.3 x 10−9) and older (9 cohorts, N=12,026, beta=−0.061±0.014 SD/allele, P=9.4x10−6) participants. In separate meta-analyses, the locus was associated with increased risk for coronary artery disease (hazard ratio [HR]=1.05, confidence interval [CI]=1.02 to 1.08, P=0.0013) and heart failure (HR=1.10, CI=1.03 to 1.16, P=0.004).
Conclusions
Common genetic variation in a locus in the BCL11B gene desert that is thought to harbor one or more gene enhancers is associated with higher CFPWV and increased risk for cardiovascular disease. Elucidation of the role this novel locus plays in aortic stiffness may facilitate development of therapeutic interventions that limit aortic stiffening and related cardiovascular disease events.
Keywords: aorta, arterial stiffness, pulse wave velocity, genetics, cardiovascular disease
Several recent studies have demonstrated that carotid-femoral pulse wave velocity (CFPWV), an important indicator of stiffness of the wall of the thoracic and abdominal aorta, is associated with increased risk for major cardiovascular disease (CVD) events. 1–4 Various risk factors for abnormal CFPWV have been identified, including standard CVD risk factors such as age, glucose intolerance, lipid disorders, and hypertension.5 In addition, CFPWV is a moderately heritable trait with an estimated residual heritability (h2) of approximately 0.40,6 although molecular mechanisms contributing to aortic stiffness remain largely undefined. In order to evaluate associations of common genetic variants with CFPWV, we performed a meta-analysis of genome-wide association study (GWAS) data from 9 community based cohorts, with replication genotyping in 2 additional cohorts. In addition, in light of the association between CFPWV and CVD risk, we interrogated existing clinical endpoint GWAS data to determine whether variants associated with CFPWV are associated with CVD risk.
Methods
Consortium Organization
The AortaGen Consortium includes 9 cohort studies that completed genome-wide genotyping and had measured CFPWV, plus 2 cohort studies that had measured CFPWV and collected DNA for replication genotyping. Each study adopted collaboration guidelines and the consortium established a consensus on phenotype harmonization, covariate selection, and an analytical plan for within-study genome-wide association and prospective meta-analysis of results across studies. Each study received institutional review board approval of its consent procedures, examination and surveillance components, data security measures, and DNA collection and its use for genetic research. All participants in each study gave written informed consent for participation in the study and the conduct of genetic research. Details of study cohort, CFPWV measurement protocols and inclusion and exclusion criteria are provided in the Supplementary Methods and Table 1.
Table 1.
Clinical characteristics of study participants.
| Cohort | N | Percent women | Age (yrs) | Percent <55 yrs of age | Height (cm) | Weight (kg) | CFPWV (m/s) | Inverse CFPWV (ms/m) | Years of CFPWV assessment | Years of DNA collection |
|---|---|---|---|---|---|---|---|---|---|---|
| AGES | 967 | 58 | 75±5 | 0 | 168±9 | 75±14 | 13.1±4.3 | 83±24 | 2005 | 2002–2006 |
| BLSA | 618 | 49 | 62±18 | 34 | 170±10 | 75±16 | 7.2±2.5 | 154±49 | 1989–2008 | 1995–2006 |
| ERF | 1970 | 57 | 48±14 | 55 | 167±9 | 75±15 | 9.5±2.1 | 110±21 | 2002–2005 | 2002–2005 |
| FHS | 6033 | 54 | 49±15 | 68 | 169±10 | 77±17 | 8.5±3.5 | 131±35 | 1999–2001* | 1996–1999* |
| 1998–2001† | 1996–1999† | |||||||||
| 2002–2005‡ | 2002–2005‡ | |||||||||
| HABC | 1354 | 49 | 74±3 | 0 | 166±9 | 74±14 | 8.8±3.7 | 131±47 | 1997–1998 | 1997–1998 |
| HAPI | 808 | 46 | 46±15 | 67 | 167±9 | 74±13 | 5.5±1.4 | 193±42 | 2003–2008 | 2000–2008 |
| RS-I | 3011 | 57 | 72±7 | 0 | 167±9 | 74±12 | 13.6±3.0 | 77±17 | 1997–1999 | 1990–1993 |
| RS-II | 1657 | 54 | 64±8 | 0 | 169±9 | 77±13 | 12.6±3.2 | 84±18 | 2000–2001 | 2000–2001 |
| SARDINIA | 4216 | 56 | 43±17 | 78 | 160±9 | 65±13 | 6.7±2.1 | 163±44 | 2001–2004 | 2001–2004 |
| Replication Cohorts | ||||||||||
| ACCT | 2932 | 52 | 34±19 | 77 | 171±10 | 72±14 | 6.7±2.2 | 161±40 | 2001–2009 | 2001–2009 |
| Asklepios | 2374 | 52 | 46±6 | 93 | 169±9 | 74±14 | 6.6±1.5 | 157±29 | 2002–2004 | 2002–2004 |
Mean values ± SD except as noted;
Original cohort;
Offspring cohort;
Third Generation cohort;
AGES, Age, Gene/Environment Susceptibility-Reykjavik Study; BLSA, Baltimore Longitudinal Study of Aging; ERF, Erasmus Rucphen Family Study; FHS, Framingham Heart Study; HABC, Health, Aging and Body Composition; HAPI, Heredity and Phenotype Intervention; RS, Rotterdam Study; ACCT, Anglo Cardiff Collaborative Trial.
Phenotyping
Only cohorts that measured CFPWV based on the carotid-to-femoral transit time and distance were included in the meta-analysis. CFPWV increases nonlinearly and exhibits marked variance inflation with advancing age, resulting in a strongly right skewed distribution. In addition, differences in the method used to ascertain transit distance can alter values by up to 30% and the amount of error may be influenced by sex and other anthropomorphic factors such as height and weight. Thus, genetic association analyses were performed using a sex-specific standardized residual that was based on the inverse of CFPWV, which normalizes the distribution, and that was further adjusted for age, age2, height and weight. As a result of these transformations, the cohorts had a highly comparable distribution of the phenotype (mean of 0 and standard deviation of 1 with a normal distribution).
Genotyping and Imputation
Genotyping and imputation methods have been described previously and are summarized in Supplementary Table S1. For genome-wide SNP sets, genotyping was carried out using commercially available arrays. Prior to imputation, quality control measures were applied as outlined in Supplementary Table S1. MACH was used by all cohorts for imputation of genotypes to the HapMap set of approximately 2.5 million SNPs. For replication cohorts, genotyping was carried out on the platforms noted in Supplementary Table S1. All genetic coordinates in tables and figures refer to HapMap release 22 build 36.
Expression Methods
Details of expression studies, including RNA extraction, cDNA preparation, PCR amplification and sequencing are provided in the Supplementary Methods.
Statistical Analyses
The phenotype for meta-analysis was a sex-specific (in Framingham, cohort- and sex-specific) standardized regression residual for 1000/CFPWV, adjusted for age, age2, height and weight. Genome-wide association analyses were conducted within each cohort using an additive gene-dose model. Linear mixed effects models were fitted to account for relatedness in pedigrees. Within-study associations were combined by prospective meta-analysis using inverse-variance weighting. Meta-analyses were performed using the software program MetABEL (http://www.genabel.org/packages/MetABEL). During meta-analysis, SNPs were excluded if weighted mean minor allele frequency was <1%, resulting in 2.41 million SNPs for analysis. The genomic control parameter was calculated to adjust each study and after meta-analysis, was recalculated to adjust for among-study heterogeneity. For the initial meta-analysis, a predetermined threshold of 4.0 x 10−7 (stage 1) was used to select SNPs for attempted replication.7 Based on a preliminary analysis of 6 cohorts, we selected SNPs from 2 loci (the SNP with the lowest P and 1 or 2 proxy SNPs to accommodate differing genotyping platforms) for attempted replication. SNPs were genotyped in 2 additional cohorts and analyzed within cohort using a similar analysis plan except that observed rather than imputed genotypes were used in the analyses. Results from the 2 replication cohorts were then combined by meta-analysis. We considered a P<0.025 (0.05/2) and same direction of effect for the replication meta-analysis as indicative of successful replication.
To assess possible effect modification by age, we performed an age-stratified analysis based on the approximate overall median age of 55 years. For cohorts that spanned this age cutoff (FHS, ERF, Sardinia, ACCT), analyses were repeated in subgroups <55 and ≥55 years of age. Cohorts with predominantly older (AGES, BLSA, HABC, RS-I, RS-II) or younger (HAPI, Asklepios) participants were included in the older or younger group in their entirety to preserve adequate sample size. These groupings resulted in 9 sets of data consisting of predominantly older participants and 6 sets of data consisting of predominantly younger participants. In addition, because of known associations between CFPWV and clinical events,1–4;8 we performed lookups of the top result from our CFPWV GWAS in separate GWAS meta-analyses for clinical endpoints thought to be related to arterial stiffness, including coronary artery disease, heart failure, stroke and kidney disease. We also performed a lookup of our top result in an ongoing pulse pressure GWAS meta-analysis in order to determine whether genetic effects on aortic stiffness were detectable as an increase in blood pressure pulsatility in additional cohorts (see Supplement for details of clinical GWAS meta-analyses).
Results
Characteristics of participants at the time of CFPWV measurement in the 11 (9 discovery, 2 replication) AortaGen Consortium cohorts are presented in Table 1. Cohort mean age varied from 34 to 75 years whereas cohort mean CFPWV varied from 5.5 to 13.6 m/s, corresponding to inverse CFPWV of 193 to 77 ms/m, respectively. Sample sizes varied from 618 to 6,033 participants, with an aggregate of 20,634 and 5,306 participants in the discovery and replication phases, respectively.
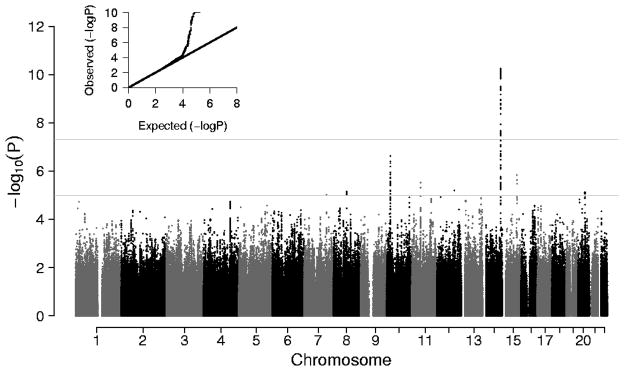
GWAS meta-analysis results from 9 cohorts are summarized in Figure 1. The quantile-quantile (Q-Q) plot shows minimal evidence of test statistic inflation (λgc = 1.03) and a sharp divergence from a slope near unity at a P-value of approximately 1 x 10−4. The negative log P (Manhattan) plot reveals a region of genome-wide significant association on the distal long arm of chromosome 14 (14q32.2, rs1381289, beta=−0.073±0.011 SD/allele, P = 5.6 x 10−11). Imputation quality for the genome-wide significant SNP’s in this region was high with median expected/observed variance ratio of 0.99–1.00. In addition, there is a suggestive region of association on the short arm of chromosome 10 (10p12.32, rs10764094, beta=−0.057±0.011 SD/allele, P=2.4x10−7). A listing of top SNPs from the 9-cohort meta-analysis with a P < 1 x 10−5 is presented in Table 2. The table provides results for the top SNP from separate loci defined by LD structure (r2<0.80). Results of analyses that further adjusted for mean arterial pressure at the time of CFPWV measurement are presented in Supplementary Table S3. Comparison of Tables 2 and S3 reveals that several associations were relatively stronger following adjustment for mean arterial pressure (CFDP1, FGFR2, NMUR2, ADAMTS9, OCA2, VPS54), others were unaffected (C10orf112, EFTUD1, CKAP5) and a few were weakened (ELK3, SLCO5A1, MAFB, CADPS2). For the locus on chromosome 14 (C14orf64), associations for some SNPs were weaker (rs1381289) while others were stronger (rs9323989) after adjustment for mean arterial pressure.
Figure 1.
Q-Q and signal intensity (Manhattan) plots of genome-wide association data for CFPWV. The upper horizontal line corresponds to P = 5.0 x 10−8, which was the threshold for genome-wide significance, whereas the lower line corresponds to P = 1.0 x 10−5, which was the threshold used to prepare Table 2.
Table 2.
Genome wide association results for CFPWV in 9 cohorts.
| SNP | Chromosome
|
Allele
|
Meta-analysis* |
Closest Gene | ||||
|---|---|---|---|---|---|---|---|---|
| Number | Position | Coded | Freq | Beta | SE | P | ||
| rs1381289† | 14 | 97,662,117 | T | 0.436 | −0.073 | 0.011 | 5.6 x 10−11 | C14orf64 |
| rs987514 | 14 | 97,698,696 | T | 0.436 | −0.069 | 0.011 | 4.5 x 10−10 | C14orf64 |
| rs10782490 | 14 | 97,619,136 | C | 0.471 | −0.066 | 0.011 | 2.7 x 10−9 | C14orf64 |
| rs22225442 | 14 | 97,692,347 | C | 0.323 | −0.071 | 0.012 | 1.2 x 10−8 | C14orf64 |
| rs17773233 | 14 | 97,652,412 | T | 0.225 | −0.074 | 0.013 | 2.1 x 10−8 | C14orf64 |
| rs1461587 | 14 | 97,673,604 | G | 0.256 | −0.070 | 0.013 | 1.5 x 10−7 | C14orf64 |
| rs1381273 | 14 | 97,718,813 | T | 0.469 | −0.059 | 0.011 | 1.9 x 10−7 | C14orf64 |
| rs10764094 | 10 | 19,950,544 | C | 0.473 | 0.057 | 0.011 | 2.4 x 10−7 | C10orf112 |
| rs8015529 | 14 | 97,571,972 | G | 0.359 | −0.066 | 0.013 | 2.5 x 10−7 | C14orf64 |
| rs4778983 | 15 | 80,290,133 | C | 0.301 | 0.057 | 0.012 | 1.5 x 10−6 | EFTUD1 |
| rs7161307 | 14 | 97,677,436 | T | 0.215 | −0.065 | 0.013 | 1.7 x 10−6 | C14orf64 |
| rs6485690 | 11 | 46,755,207 | A | 0.308 | −0.056 | 0.012 | 3.0 x 10−6 | CKAP5‡ |
| rs10740923 | 10 | 19,907,637 | G | 0.464 | −0.052 | 0.011 | 3.9 x 10−6 | C10orf112 |
| rs7959220 | 12 | 95,117,079 | G | 0.027 | 0.266 | 0.059 | 6.3 x 10−6 | ELK3 |
| rs6472483 | 8 | 70,791,920 | T | 0.452 | −0.050 | 0.011 | 7.1 x 10−6 | SLCO5A1 |
| rs6101837 | 20 | 38,155,981 | C | 0.416 | −0.050 | 0.011 | 7.5 x 10−6 | MAFB |
| rs10827649 | 10 | 19,949,776 | G | 0.436 | −0.049 | 0.011 | 8.6 x 10−6 | C10orf112 |
| rs6947805 | 7 | 121,844,471 | T | 0.050 | 0.117 | 0.026 | 9.5 x 10−6 | CADPS2 |
Individual analyses were adjusted for age, age2, sex, height and weight.
R2 = 0.93 for rs1381289 and rs7152623.
LD block includes ARHGAP1, ZNF408, F2, CKAP5 and LRP4.
Based on a preliminary meta-analysis of early GWAS results from 6 cohorts (Table 1; AGES, FHS, ERF, RS-I, RS-II, and Sardinia; 17,854 participants), we selected rs7152623 on chromosome 14 (results from 6 cohorts: beta=−0.075±0.012 SD/allele, P = 2.8 x 10−10) and rs17729837 on chromosome 10 (results from 6 cohorts: beta=0.062±0.012 SD/allele, P=3.6x10−7) for attempted replication. Note that rs7152623, the SNP on chromosome 14 selected for replication, falls within the LD block that includes rs1381289 as the SNP with the lowest P-value in the block (Table 2). The two SNPs are closely linked (R2 = 0.93 for rs1381289 and rs7152623). We successfully replicated the association with rs7152623 on chromosome 14 (replication beta=−0.086±0.020 SD/allele, P=1.4x10−6). Results for rs7152623 from the full set of 11 cohorts gave a combined beta=−0.076±0.010 SD/allele, P=3.1x10−15 (Figure 2). The effect was attenuated modestly and remained significant when we further adjusted for mean arterial pressure at the time of measurement of CFPWV (beta=−0.060±0.009 SD/allele, P=1.0 x 10−11). In addition, results were consistent when evaluated separately in subgroups defined by median age, remaining associated in both younger (<55 years of age, 6 cohorts, N=13,914, beta=−0.081±0.014 SD/allele, P=2.3 x 10−9) and older (≥55 years of age, 9 cohorts, N=12,026, beta=−0.061±0.014 SD/allele, P=9.4x10−6) participants. The association with rs17729837 on chromosome 10 did not replicate (P = 0.97).
Figure 2.
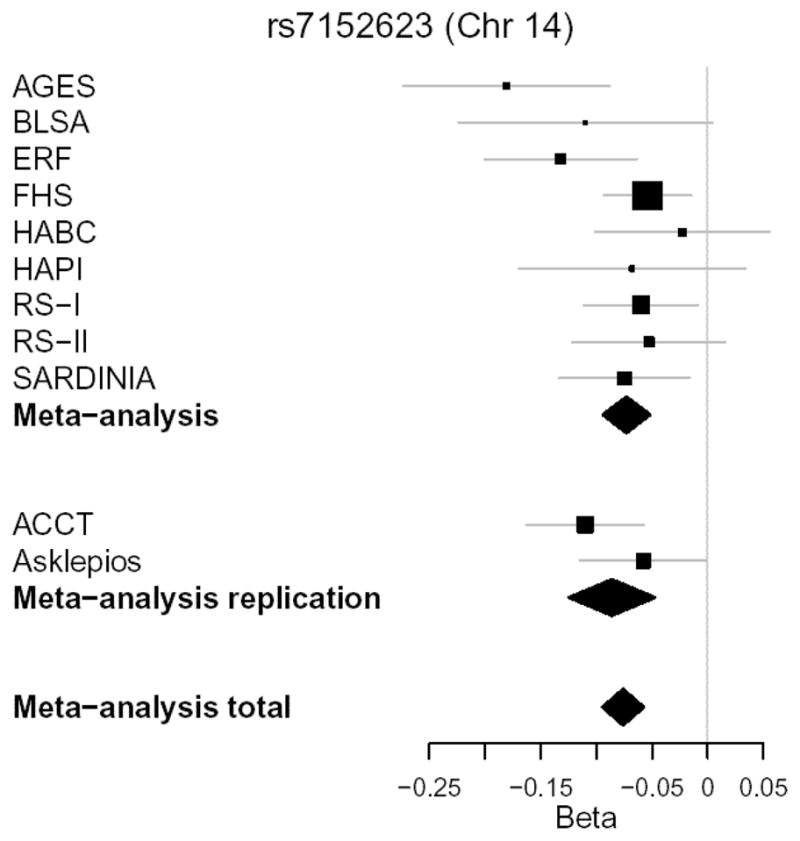
Forest plot of association results for rs7152623 on chromosome 14. Results for individual cohorts are plotted against the cohort effect size (beta coefficient). The size of the box is proportional to the study’s weight in the meta-analysis (inversely proportional to estimated variance of the effect-size estimator). Horizontal lines are the 95% confidence intervals. Diamonds represent the results of meta-analyses; the center denotes overall estimate and the width denotes 95% confidence interval.
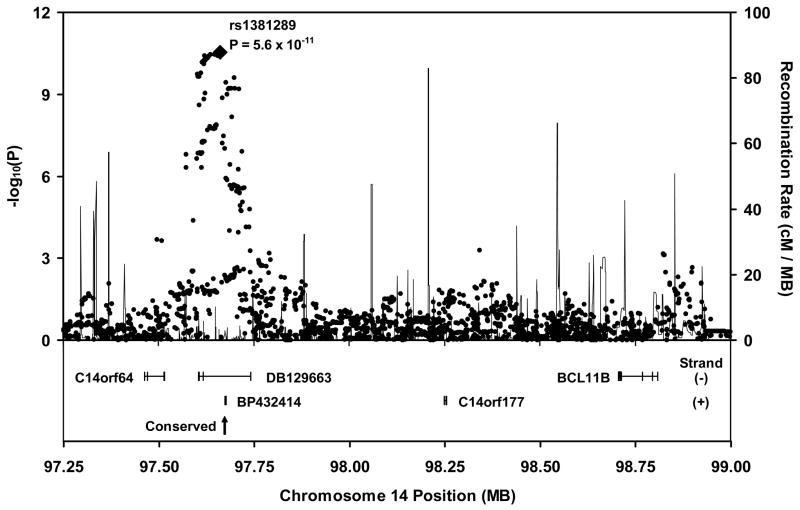
Details of the region of significant association on chromosome 14 are presented in Figure 3. The closest known gene (3′ BCL11B) is nearly 1 MB telomeric to this locus. The associated SNPs are located in a block of linkage disequilibrium (LD) spanning from approximately 97.60 to 97.74 MB, which corresponds closely with the location of a cluster of previously documented, overlapping, spliced expressed sequence tags (ESTs), including DB129663 and BP432414, which are on the minus and plus strand, respectively. One of our highly associated SNPs (rs710285, P=5.1 x 10−11) is located in exon 3 of DB129663. In addition, there is a conserved sequence near the center of the chromosome 14 locus (97.67 MB, Figure 3), in overlapping intronic regions of DB129663 and BP432414.
Figure 3.
Regional association plot and surrounding genomic neighborhood for the locus on chromosome 14. P-values are based on the meta-analysis of 9 cohorts. The SNP with the lowest overall P-value in the meta-analysis (rs1381289) is highlighted. The LD block of highest associations spans from approximately 97.6 to 97.7 MB, corresponding to the length of DB129663. A conserved sequence (arrow) spanning approximately 550 bases lies in an intron common to DB129663 and BP432414.
To assess potential functional implications of our findings, we used reverse transcriptase polymerase chain reaction (RT-PCR) to evaluate expression of DB129663 and BP432414 in human aortic samples and various cell lines. In light of the putative role of this region as a remote enhancer of BCL11B9 and relative proximity to VRK1, we also probed for expression of these genes (Supplementary Table S4). DB129663, BP432414, BCL11B and VRK1 were detected in whole aortic rings (Supplementary Table S4). VRK1 and DB129663 were expressed in cultured aortic smooth muscle cells, human umbilical vein endothelial cells (HUVECs) and adult cardiac fibroblasts. BCL11B was expressed in the same samples except cardiac fibroblasts. All transcripts were expressed in CD3+ cells.
To assess the potential clinical relevance of our finding of a locus on Chr14 strongly associated with CFPWV, we performed a lookup of our top SNP for association with CFPWV in the meta-analysis of 9 cohorts (rs1381289, Table 2) in results from separate clinical endpoint GWAS meta-analyses (see Supplement). We found an association with increased risk for coronary artery disease (hazard ratio [HR]=1.05, confidence interval [CI]=1.02 to 1.08 per allele, P=0.0013) and heart failure (HR=1.10, CI=1.03 to 1.16 per allele, P=0.004). To aid interpretation of these CVD endpoint associations, we used data from FHS to estimate the expected HR for a first major CVD event associated with presence of each allele of rs1381289, assuming that excess CVD risk was mediated by effects of the allele on CFPWV.4 In FHS, after adjusting for age and sex, CFPWV was associated with an excess CVD risk corresponding to a HR=1.65 per SD. Based on this estimate, and given that each minor allele of rs1381289 was associated with a 0.073 SD increase in CFPWV, the expected HR=1.047 per allele, which is comparable to the GWAS lookup results. In a separate meta-analysis that included 74,011 participants, rs1381289 was associated with higher pulse pressure (beta=0.18±0.06 mm Hg/allele, P=0.002), indicating that the increase in aortic stiffness associated with this SNP is detectable as a modest but significant increase in pressure pulsatility. The SNP was not associated with stroke (P>0.7), glomerular filtration rate estimated by using serum creatinine (P>0.6) or cystatin (P>0.5) or prevalent chronic kidney disease (P>0.6).
We also sought to replicate a previously reported association between CFPWV and a SNP (rs3742207) in COL4A1.10 After excluding 2 cohorts involved in the original report (Sardinia, HAPI), we found modest evidence of association for this SNP (rs3742207, beta=−0.025±0.011 SD/allele, P=0.017) (Supplementary Figure S1).
Discussion
We performed a meta-analysis of GWAS results for CFPWV from 9 community-based cohorts involving 20,634 participants spanning a broad age range and identified a locus of genome-wide significant association in an apparent gene desert on 14q32.2. This finding was replicated in 2 additional cohorts involving 5,306 participants. We identified a conserved sequence within the region of significant association surrounded by a cluster of primate-specific, noncoding RNAs (ncRNAs). We evaluated 2 of these ncRNAs, which have at least one associated SNP within an exon, and demonstrated that they are expressed in relevant human cardiac and vascular tissues and cell lines, including full thickness aortic rings, aortic smooth muscle cells, cardiac fibroblasts and HUVECs. In light of the putative role of the region of significant association as a gene enhancer,9;11;12 we also assayed for and demonstrated expression of flanking known genes, BCL11B and VRK1, in the same tissues and cell lines. Our findings indicate that the VRK1-BCL11B gene desert harbors a regulatory locus that modulates aortic stiffness. The association was consistent in younger and older participants, suggesting that the effects on CFPWV of genetic variation at this locus manifest early in life, prior to the marked increase in CFPWV that occurs from midlife onward. In addition, we demonstrated that the locus is associated with increased risk for cardiovascular disease, consistent with the hypothesis that increased aortic stiffness, as assessed by CFPWV, plays a causal role in the pathogenesis of cardiovascular disease. Further elucidation of potential mechanisms of aortic stiffening mediated through this locus may provide novel insights into the pathogenesis of aortic stiffening and could potentially offer insights into currently unavailable targeted interventions that prevent or attenuate aortic stiffening with advancing age and reduce the associated excess risk for major CVD events.
CFPWV has emerged recently as an important risk factor for various afflictions of aging, including CVD, cognitive dysfunction and chronic kidney disease. CFPWV is easily and reproducibly measured in a few minutes using relatively inexpensive technology and is widely considered the present gold standard noninvasive measure of aortic stiffness. Aside from age, with which it is strongly related, CFPWV has consistent but relatively modest relations with standard CVD risk factors and is moderately heritable, rendering the phenotype optimal for genetic studies.
A number of prior studies have evaluated potential genetic correlates of various measures of arterial stiffness including CFPWV. Family-based studies have identified several regions of potential linkage for stiffness measures using a microsatellite-based whole genome approach.6 Genetic association studies have found relations between measures of arterial stiffness and polymorphisms in various candidate genes, including genes for the angiotensin-II type 1 receptor,13 fibrillin-1,14 angiotensin converting enzyme,15;16 alpha adducin,15 aldosterone synthase,15;17 beta adrenergic receptors,18 endothelin A and B receptors,19 matrix metalloproteinases 3 and 9,20;21 endothelial nitric oxide synthase,22 the large conductance calcium-activated potassium channel,23 estrogen genes24 and various inflammatory genes.25;26 However small sample sizes, ascertainment bias (hypertensive, known coronary artery disease, etc.) and relatively weak associations may have limited the generalizability and consistency of findings. Notably, none of the aforementioned candidate genes are in the top hits of the present analysis (Tables 2 and S3). Two prior CFPWV GWAS publications10;27 were similarly based on relatively small sample sizes and employed less dense genotyping, which limited power to detect associations. A relative paucity of genome-wide significant findings remains a concern with our present publication involving 9 discovery and 2 replication cohorts and an aggregate sample size of nearly 26,000 individuals. Additional work and larger samples will be required in order to determine whether any additional loci in the group of excess low P-values below ~1x10−4 are true positive associations.
Prior studies provide evidence that the region of association with CFPWV that we have identified on 14q32.2 lies in the vicinity of one or more gene enhancers. The region encompasses various regulatory features, including several DNAse-I hypersensitive sites and transcription factor binding sites and high levels of nuclear matrix attachment.9;11;12 Chromatin modifications in the region, including high levels of acetylation of histone 3 at lysine 27 (H3K27) and monomethylation at lysine 4 (H3K4) assessed in a lymphoblastoid cell line, are consistent with enhancer function (http://genome.ucsc.edu).28 Despite considerable genomic separation from the enhancer cluster (~1 MB telomeric), BCL11B is thought to be a target of one or more of the enhancers in this locus.9;11;12 BCL11B is located on the minus strand, positioning the enhancer cluster in the remote 3′ region of the gene. The closest known gene in the opposite direction, VRK1, is ~1.1 MB centromeric to the enhancer cluster and is on the plus strand, again positioning the enhancer in the remote 3′ region of VRK1, suggesting that one or both genes could potentially be targets of a remote 3′ enhancer in this region.
In support of BCL11B as a target, numerous translocations have been described that insert fragments of 5q35 at various positions in a breakpoint cluster region that falls between the enhancer region and 3′ BCL11B. These translocations interpose the homeobox genes TLX3 or NKX2-5 between the enhancer region and 3′ BCL11B and result in ectopic activation of the inserted homeobox gene, dysregulated T-cell proliferation and acute T-cell lymphoblastic leukemia.9;11;12 T-cell regulatory signals directed at BCL11B may interact with the enhancer to drive ectopic activation of the interposed homeobox gene, leading to cell-specific malignant transformation. Su et al. used translocation data to map enhancer function to a 58 kB segment of the genome that corresponds to the telomeric shoulder of our locus of significant association with CFPWV.12 Relative proximity of the enhancer to BCL11B, the important role that BCL11B plays in T-cell development and the T-cell specificity of malignant transformations involving translocations into the region support the hypothesis that BCL11B is a target of this enhancer.
BCL11B codes for chicken ovalbumin upstream promoter transcription factor (COUP-TF) interacting protein 2 (CTIP2), which is a cofactor in the COUP-TF family of transcription factors29 and a direct transcriptional repressor.30 There are several potential mechanisms for an effect of BCL11B on aortic stiffness. COUP-TFII (NR2F2) modulates the angiopoietin-1 and vascular endothelial growth factor pathways and plays a critical role in the development of the heart and great vessels.31 In addition, BCL11B is a C2H2 zinc finger protein that can directly bind DNA in a sequence specific manner. Acting in part through an interaction with SIRT1, BCL11B can effect transcriptional repression of various genes that may be relevant to aortic stiffness.32–35 In addition to direct effects of BCL11B on aortic function, there are potential indirect effects mediated through the known role that BCL11B plays in T-cell function. T-cell specific deletion of BCL11B at the CD4+ single positive stage is associated with increased numbers of proinflammatory T-cells that could potentially infiltrate the aorta and promote inflammation, fibrosis and stiffening.36;37 Additional work will be required to establish the potential role that genetic variation in the chromosome 14 locus may play in BCL11B expression and aortic function.
We demonstrated that two overlapping ESTs that fall completely within the region of highly significant association with CFPWV are expressed in aortic tissue and cell lines. These primate-specific, potentially regulatory ncRNAs are expressed in cDNA extracts from full thickness human aortic rings and various human cell lines, including aortic smooth muscle cells, HUVECs and cardiac fibroblasts. One of the highly associated SNPs in the region (rs710285) is located in an exon of DB129663, suggesting a possible functional effect. The enhancer core region mapped by Su et al. corresponds to the putative promoter region of DB129663. Thus, enhancer function at our chromosome 14 locus may target DB129663, which appears to be a ncRNA of unknown function. Additional work will be required to test this hypothesis and further define the function of DB129663 and other ncRNAs in the region.
Several additional SNPs with suggestive associations to CFPWV (10−8 < P < 10−5) may merit further consideration and additional replication genotyping. The locus on chromosome 10 with the second lowest P-value in our GWAS meta-analysis lies in the vicinity of a putative protein coding gene that may represent a novel member of the low density lipoprotein receptor-related protein (LRP) family.38 We found moderate evidence for association at the LRP4 locus, which is also associated with stroke39 and bone mineral density.40;41 The LRP4 locus includes a nonsynonymous SNP (rs6485702) that has been related to bone mineral density,40 although a separate report involving several of our cohorts positioned the region of highest association with bone mineral density in the promoter region of ARHGAP1.41 Bone density and arterial stiffness are related phenotypes42 that may share many common pathways. The recently observed inhibitory role that LRP4 plays in Wnt signaling in bone43 coupled with the adverse effects of Wnt signaling in the aorta44 suggests that a mutation that impairs the ability of LRP4 to modulate the Wnt signaling cascade could simultaneously contribute to osteopenia and aortic stiffening. The chromosome 11 locus that encompasses LRP4 and additional potential candidates, including ARHGAP1 and F2, represents a long LD block that was also associated with stroke in a prior meta-analysis that included several of the cohorts in our study.39 The direction of effect in the prior study (higher risk for the minor allele) and ours (stiffer aorta with the minor allele) was consistent with the known association between increased CFPWV and increased risk for stroke. In addition, a prior family-based linkage analysis for myocardial infarction found a single significant linkage peak in the vicinity of our chromosome 14 locus.45 These regions of overlap with prior results involving separate but related phenotypes support the clinical relevance of our associations and suggest that several genetic variants that impact CFPWV may eventually manifest as age-related morbidity and major cardiovascular events.
We also attempted to replicate a previously reported association of CFPWV with a SNP in the COL4A1 locus in the only published GWAS that has evaluated CFPWV.10 The present results found modest evidence of association with some heterogeneity of effect, suggesting that additional work will be required to determine whether variation in LD patterns or other factors could potentially account for heterogeneous effects at this locus.
There are limitations of our study that should be considered. The cohorts comprised exclusively white participants of European descent. Thus, our findings may not generalize to other populations. Slightly different methods were used to assess CFPWV in the various cohorts. However, our use of standardized residuals generated within each cohort should have minimized the effects of these technical differences between studies. A major strength of our study is the use of data from 11 large community-based cohorts that routinely ascertained CFPWV, which should enhance generalizability of our findings.
In conclusion, we performed the first large scale GWAS of CFPWV, which is a moderately heritable measure of aortic stiffness and important risk factor for cardiovascular events. We identified a highly significant locus of association at 14q32.2 in the VRK1-BCL11B gene desert in an LD block that harbors one or more gene enhancers. We have also shown that genetic variation at this locus is associated with increased risk for major CVD events, providing strong support for the hypothesis that increased CFPWV contributes to the pathogenesis of CVD. We have shown that 2 potentially regulatory ncRNAs as well as flanking genes, BCL11B and VRK1, are expressed in human aorta. Further work will be required to define precise mechanisms mediating the association between CFPWV and genetic variation in the VRK1-BCL11B gene desert. Elucidation of pathways affected by this locus will provide new insights into the process of aortic stiffening in humans and could yield potential targets for specific interventions that reverse or attenuate aortic stiffening and prevent the associated morbidity and mortality.
Supplementary Material
Acknowledgments
Funding
The Age, Gene/Environment Susceptibility Reykjavik Study is funded by the National Institutes of Health, the National Institute on Aging Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The Baltimore Longitudinal Study of Aging (BLSA) is supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The ERF study was supported by grants from the Netherlands Organization for Scientific Research, Erasmus Medical Center and the Centre for Medical Systems Biology (CMSB) and the Netherlands Kidney Foundation. The Framingham Heart Study was partially supported by the National Heart, Lung, and Blood Institute’s Framingham Heart Study and its contract with Affymetrix, Inc for genotyping services. Additional support was provided by the National Heart, Lung, and Blood Institute, the National Institutes on Aging, and a grant from the Donald W. Reynolds Foundation. The Amish studies are supported by grants and contracts from the NIH National Institute on Aging, National Heart, Lung, and Blood Institute, National Center for Research Resources, and the National Institute of Diabetes, Digestive, and Kidney Disease. The Dynamics of Health, Aging and Body Composition (Health ABC) Study was supported by contracts from the National Institutes on Aging. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research and Research Institute for Diseases in the Elderly. This study was supported by the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The SardiNIA team is supported by contracts from the National Institute on Aging and, in part, by the Intramural Research Program of the NIH, National Institute on Aging and by research grants from the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute. The Anglo-Cardiff Study was supported by grants and fellowships from the British Heart Foundation, the Cambridge BioMedical Research Centre Award, and the Addenbrooke’s Charitable Trust. The Asklepios Study is supported by a Fonds voor Wetenschappelijk Onderzoek–Vlaanderen FWO research grant. A full list of funding sources appears in the online-only Data Supplement Note.
Footnotes
Conflict of Interest Disclosures: G.F.M. is owner of Cardiovascular Engineering, Inc, a company that designs and manufactures devices that measure vascular stiffness. The company uses these devices in clinical trials that evaluate the effects of diseases and interventions on vascular stiffness. The remaining authors report no conflicts.
References
- 1.Laurent S, Boutouyrie P, Asmar R, Gautier I, Laloux B, Guize L, et al. Aortic stiffness is an independent predictor of all-cause and cardiovascular mortality in hypertensive patients. Hypertension. 2001;37:1236–1241. doi: 10.1161/01.hyp.37.5.1236. [DOI] [PubMed] [Google Scholar]
- 2.Mattace-Raso FU, van der Cammen TJ, Hofman A, van Popele NM, Bos ML, Schalekamp MA, et al. Arterial stiffness and risk of coronary heart disease and stroke: the Rotterdam Study. Circulation. 2006;113:657–663. doi: 10.1161/CIRCULATIONAHA.105.555235. [DOI] [PubMed] [Google Scholar]
- 3.Willum-Hansen T, Staessen JA, Torp-Pedersen C, Rasmussen S, Thijs L, Ibsen H, et al. Prognostic value of aortic pulse wave velocity as index of arterial stiffness in the general population. Circulation. 2006;113:664–670. doi: 10.1161/CIRCULATIONAHA.105.579342. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell GF, Hwang SJ, Vasan RS, Larson MG, Pencina MJ, Hamburg NM, et al. Arterial stiffness and cardiovascular events: the Framingham Heart Study. Circulation. 2010;121:505–511. doi: 10.1161/CIRCULATIONAHA.109.886655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell GF, Guo CY, Benjamin EJ, Larson MG, Keyes MJ, Vita JA, et al. Cross-sectional correlates of increased aortic stiffness in the community: the Framingham Heart Study. Circulation. 2007;115:2628–2636. doi: 10.1161/CIRCULATIONAHA.106.667733. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell GF, DeStefano AL, Larson MG, Benjamin EJ, Chen MH, Vasan RS, et al. Heritability and a genome-wide linkage scan for arterial stiffness, wave reflection, and mean arterial pressure: the Framingham Heart Study. Circulation. 2005;112:194–199. doi: 10.1161/CIRCULATIONAHA.104.530675. [DOI] [PubMed] [Google Scholar]
- 7.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutouyrie P, Tropeano AI, Asmar R, Gautier I, Benetos A, Lacolley P, et al. Aortic stiffness is an independent predictor of primary coronary events in hypertensive patients: a longitudinal study. Hypertension. 2002;39:10–15. doi: 10.1161/hy0102.099031. [DOI] [PubMed] [Google Scholar]
- 9.Nagel S, Scherr M, Kel A, Hornischer K, Crawford GE, Kaufmann M, et al. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67:1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- 10.Tarasov KV, Sanna S, Scuteri A, Strait JB, Orru M, Parsa A, et al. COL4A1 is associated with arterial stiffness by genome-wide association scan. Circ Cardiovasc Genet. 2009;2:151–158. doi: 10.1161/CIRCGENETICS.108.823245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel S, Kaufmann M, Drexler HG, MacLeod RA. The cardiac homeobox gene NKX2-5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t(5;14)(q35.1;q32.2) Cancer Res. 2003;63:5329–5334. [PubMed] [Google Scholar]
- 12.Su XY, Della-Valle V, Andre-Schmutz I, Lemercier C, Radford-Weiss I, Ballerini P, et al. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t(5;14)(q35;q32) Blood. 2006;108:4198–4201. doi: 10.1182/blood-2006-07-032953. [DOI] [PubMed] [Google Scholar]
- 13.Benetos A, Topouchian J, Ricard S, Gautier S, Bonnardeaux A, Asmar R, et al. Influence of angiotensin II type 1 receptor polymorphism on aortic stiffness in never-treated hypertensive patients. Hypertension. 1995;26:44–47. doi: 10.1161/01.hyp.26.1.44. [DOI] [PubMed] [Google Scholar]
- 14.Medley TL, Cole TJ, Gatzka CD, Wang WY, Dart AM, Kingwell BA. Fibrillin-1 genotype is associated with aortic stiffness and disease severity in patients with coronary artery disease. Circulation. 2002;105:810–815. doi: 10.1161/hc0702.104129. [DOI] [PubMed] [Google Scholar]
- 15.Balkestein EJ, Staessen JA, Wang JG, van der Heijden-Spek JJ, Van Bortel LM, Barlassina C, et al. Carotid and femoral artery stiffness in relation to three candidate genes in a white population. Hypertension. 2001;38:1190–1197. doi: 10.1161/hy1101.095992. [DOI] [PubMed] [Google Scholar]
- 16.Benetos A, Gautier S, Ricard S, Topouchian J, Asmar R, Poirier O, et al. Influence of angiotensin-converting enzyme and angiotensin II type 1 receptor gene polymorphisms on aortic stiffness in normotensive and hypertensive patients. Circulation. 1996;94:698–703. doi: 10.1161/01.cir.94.4.698. [DOI] [PubMed] [Google Scholar]
- 17.Pojoga L, Gautier S, Blanc H, Guyene TT, Poirier O, Cambien F, et al. Genetic determination of plasma aldosterone levels in essential hypertension. Am J Hypertens. 1998;11:856–860. doi: 10.1016/s0895-7061(98)00048-x. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Srinivasan SR, Boerwinkle E, Berenson GS. Beta-adrenergic receptor genes are associated with arterial stiffness in black and white adults: the Bogalusa Heart Study. Am J Hypertens. 2007;20:1251–1257. doi: 10.1016/j.amjhyper.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Lajemi M, Gautier S, Poirier O, Baguet JP, Mimran A, Gosse P, et al. Endothelin gene variants and aortic and cardiac structure in never-treated hypertensives. Am J Hypertens. 2001;14:755–760. doi: 10.1016/s0895-7061(01)02162-8. [DOI] [PubMed] [Google Scholar]
- 20.Medley TL, Kingwell BA, Gatzka CD, Pillay P, Cole TJ. Matrix metalloproteinase-3 genotype contributes to age-related aortic stiffening through modulation of gene and protein expression. Circ Res. 2003;92:1254–1261. doi: 10.1161/01.RES.0000076891.24317.CA. [DOI] [PubMed] [Google Scholar]
- 21.Medley TL, Cole TJ, Dart AM, Gatzka CD, Kingwell BA. Matrix metalloproteinase-9 genotype influences large artery stiffness through effects on aortic gene and protein expression. Arterioscler Thromb Vasc Biol. 2004;24:1479–1484. doi: 10.1161/01.ATV.0000135656.49158.95. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell GF, Guo CY, Kathiresan S, Vasan RS, Larson MG, Vita JA, et al. Vascular stiffness and genetic variation at the endothelial nitric oxide synthase locus: the Framingham Heart study. Hypertension. 2007;49:1285–1290. doi: 10.1161/HYPERTENSIONAHA.106.085266. [DOI] [PubMed] [Google Scholar]
- 23.Kelley-Hedgepeth A, Peter I, Montefusco MC, Levy D, Benjamin EJ, Vasan RS, et al. The KCNMB1 E65K variant is associated with reduced central pulse pressure in the community-based Framingham Offspring Cohort. J Hypertens. 2009;27:55–60. doi: 10.1097/HJH.0b013e328317c8ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter I, Kelley-Hedgepeth A, Huggins GS, Housman DE, Mendelsohn ME, Vita JA, et al. Association between arterial stiffness and variations in oestrogen-related genes. J Hum Hypertens. 2009;23:636–644. doi: 10.1038/jhh.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnabel R, Larson MG, Dupuis J, Lunetta KL, Lipinska I, Meigs JB, et al. Relations of inflammatory biomarkers and common genetic variants with arterial stiffness and wave reflection. Hypertension. 2008;51:1651–1657. doi: 10.1161/HYPERTENSIONAHA.107.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita A, Nakayama T, Doba N, Hinohara S, Soma M. Polymorphism of the C-reactive protein (CRP) gene is related to serum CRP Level and arterial pulse wave velocity in healthy elderly Japanese. Hypertens Res. 2006;29:323–331. doi: 10.1291/hypres.29.323. [DOI] [PubMed] [Google Scholar]
- 27.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 (Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Avram D, Fields A, Nevrivy DJ, Ishmael JE, Leid M. Pretty On Top. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 33.Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B, et al. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28:3380–3389. doi: 10.1038/onc.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B, Jr, Martinez B, Crofoot K, et al. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281:32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu XM, Chapman GB, Wang H, Durante W. Adenovirus-mediated heme oxygenase-1 gene expression stimulates apoptosis in vascular smooth muscle cells. Circulation. 2002;105:79–84. doi: 10.1161/hc0102.101369. [DOI] [PubMed] [Google Scholar]
- 36.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, et al. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzik TJ, Hoch NE, Brown KA, McCann LA, Rahman A, Dikalov S, et al. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J Exp Med. 2007;204:2449–2460. doi: 10.1084/jem.20070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zubenko GS, Hughes HB, III, Zubenko WN. D10S1423 identifies a susceptibility locus for Alzheimer’s disease (AD7) in a prospective, longitudinal, double-blind study of asymptomatic individuals: Results at 14 years. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:359–364. doi: 10.1002/ajmg.b.31017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 41.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabit R, Bolton CE, Edwards PH, Pettit RJ, Evans WD, McEniery CM, et al. Arterial stiffness and osteoporosis in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:1259–1265. doi: 10.1164/rccm.200701-067OC. [DOI] [PubMed] [Google Scholar]
- 43.Choi HY, Dieckmann M, Herz J, Niemeier A. Lrp4, a novel receptor for Dickkopf 1 and sclerostin, is expressed by osteoblasts and regulates bone growth and turnover in vivo. PLoS One. 2009;4:e7930. doi: 10.1371/journal.pone.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al Aly Z, Shao JS, Lai CF, Huang E, Cai J, Behrmann A, et al. Aortic Msx2-Wnt calcification cascade is regulated by TNF-alpha-dependent signals in diabetic Ldlr−/− mice. Arterioscler Thromb Vasc Biol. 2007;27:2589–2596. doi: 10.1161/ATVBAHA.107.153668. [DOI] [PubMed] [Google Scholar]
- 45.Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, et al. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–214. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.