Abstract
5′-methylthioadenosine (MTA) is a natural purine that is metabolized by methylthioadenosine phosphorylase (MTAP, E.C 2.4.2.28) in Eukarya and Archaea but generally not in bacteria. In this work, Rv0535, which has been annotated as a probable MTAP in M. tuberculosis, was expressed in and purified from E. coli BL21 (DE3). The purified protein displayed properties of a phosphorylase and MTA was the preferred substrate. Adenosine and S-adenosyl-L-homocysteine were poor substrates and no activity was detected with 5′-methylthioinosine, the other natural purines or the natural pyrimidines. Kinetic analysis of M. tuberculosis MTAP showed that the Km value for MTA was 9.1 μM. Rv0535 was estimated as a 30 kDa protein on a denaturing SDS-PAGE gel, which agreed with the molecular mass predicted by its gene sequence. Using gel filtration chromatography, the native molecular mass of the enzyme was determined to be 60 ± 4 kDa, and thus indicates that M. tuberculosis MTAP is a dimer. Differences in active site between mycobacterial and human MTAPs were identified by homology modeling based on the crystal of the human enzyme. A complete structure activity relationship analysis could identify differences in substrate specificity between the two enzymes to aid in the development of purine-based, anti-tuberculosis drugs.
Keywords: 5′-methylthioadenosine phosphorylase, Rv0535, purine metabolism, Mycobacterium tuberculosis
Introduction
One third of the world’s population is infected with Mycobacterium tuberculosis, the causative pathogen of tuberculosis (TB). As the second leading infectious disease in the world, TB causes approximately 2 million deaths every year (6, 24). TB remains a global health emergency because of the rising number of infections with Multiple Drug Resistant (MDR) and Extensively Drug Resistant (XDR) strains of the mycobacterium, increasing HIV co-infections, the lack of an effective vaccine that protects both children and adults, and poor patient compliance with current drug regimens. To eradicate this deadly disease, there is an urgent need to develop a new class of drugs that has an alternate mechanism of action than current drugs to treat active, latent, and drug-resistant TB. Rationally designed nucleoside analogs could fulfill the above criteria because initial studies on nucleoside analogs have shown cytotoxicity against active and latent mycobacteria. While most nucleoside analogs are currently used to treat cancer and viral infections, they are not used against TB and therefore, resistance to nucleoside analogs is unlikely initially. Thus, to aid in the development of nucleoside analogs against TB, it is important to identify and understand differences between mycobacterial and human purine metabolic enzymes.
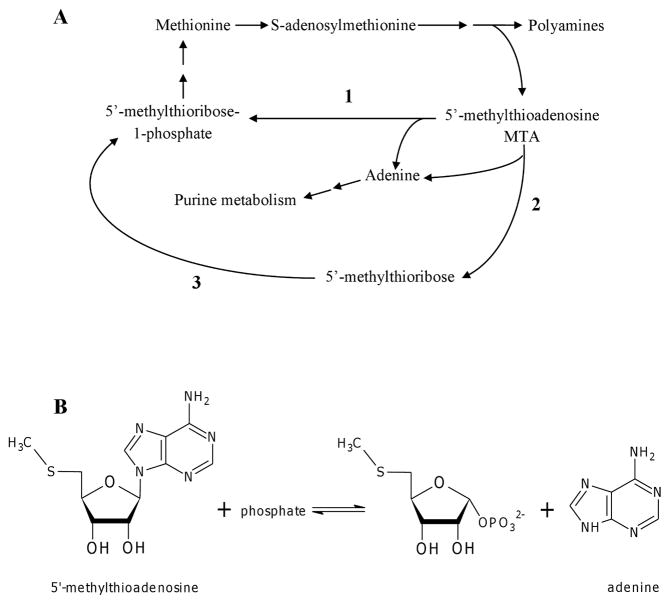
Methionine is an essential amino acid that acts as the initiator amino acid during protein synthesis. It can be converted to S-adenosylmethionine (SAM) which is important for transmethylation reactions (1, 13). Spermidine synthase uses decarboxylated SAM to produce polyamines (Figure 1A), and in the process releases 5′-methylthioadenosine (MTA), a natural purine that can inhibit spermine synthase, spermidine synthase, and mammalian S-adenosyl-L-homocysteine (SAH) hydrolase (14, 16, 9). In bacteria, MTA levels are kept low by MTA/S-adenosyl-L-homocysteine nucleosidase (MTAN, E.C 3.2.2.16), which hydrolyzes the glycosidic bond of MTA to produce adenine (Ade) and 5′-methylthioribose. Ade participates in the purine salvage pathway to produce Ade nucleotides while 5′-methylthioribose is phosphorylated by a kinase to 5′-methylthioribose-1-phosphate, which can be used in the methionine salvage pathway. In Eukarya, including trypanosomes, and Archaea, MTA phosphorylase (MTAP, E.C 2.4.2.28) cleaves MTA in the presence of phosphate to produce Ade and 5′-methylthioribose-1-phosphate (Figure 1B) (1, 8). Although MTAP had been described in Pseudomonas aeruginosa (1), a recent study showed that the enzyme was 5′-methylthioinosine phosphorylase (MTIP) instead of MTAP (8). Thus, it is believed that MTAP is not present in bacteria.
Fig 1.
(A) Metabolism of MTA. Reaction 1 is usually catalyzed by MTAP from Eukarya and Archaea, whereas bacteria recycle MTA via Reaction 2 (by MTAN) and Reaction 3 (by methylthioribose kinase). (B) MTAP catalyzes the reversible phosphorolysis of MTA to Ade and ribose-1-phosphate.
However, we have previously shown that unlike other bacteria, M. smegmatis expressed a phosphorylase that could cleave MTA, thereby marking the first report of a bacterial MTAP (3). According to Uniprot, M. smegmatis MTAP shares 196 amino acids (74% identity) with M. tuberculosis Rv0535 (GenBank accession no. O06401), which has been annotated as a probable MTAP (22). In order to prove the identity as Rv0535 as encoding MTAP and to investigate its biochemical properties, Rv0535 was expressed in and purified from E. coli BL21 (DE3) cells. Biochemical analysis of the recombinant gene product allowed the definitive assignment of Rv0535 as the MTAP gene in M. tuberculosis. Furthermore, our initial studies indicated differences between human and mycobacterial MTAPs that could be exploited to design a nucleoside analog that can be cleaved to a toxic product by the mycobacterial enzyme only, leading to selective toxicity against mycobacterial cells. A greater understanding of the differences between the two enzymes could aid in a TB drug discovery effort.
Materials and Methods
Reagents
The natural nucleosides, nucleobases, and ribose-1-phosphate were purchased from Sigma-Aldrich. 5′-Methylthioinosine and MT-DADMe-Immucillin A were gifts from Dr. Vern Schramm (Albert Einstein College of Medicine of Yeshiva University). Complete EDTA-free Protease inhibitor tablets were purchased from Roche. Sodium dodecyl sulfate – 12% polyacrylamide gel electrophoresis (SDS-12% PAGE) minigels, Tris/Glycine/SDS running buffer, Coomassie stain, Silver Stain Plus reagents, Precision Plus All Blue Protein™ standards, and Bradford dye reagent were purchased from Bio-Rad Laboratories. EZ-Run™ Pre-stained protein marker was purchased from Fisher Scientific.
Cloning of the Rv0535 gene from M. tuberculosis H37Rv genomic DNA
The Rv0535 genewas amplified by PCR from strain H37Rv genomic DNA, using the high fidelity DNA polymerase Dynazyme EXT (Finnzymes, Inc., MA), and NdeI primer (5′-AATTCATATGATGCACAACAATGGGCGCATG-3′) and BamH1 primer (5′-ATTAGGATCCTCATGGCAGCTCGAACGGCAA-3′). The PCR product was inserted into the Nde1/BamH1 site of the pET28a (+) expression vector (Novagen/EMD Chemicals Inc., CA). The entire coding sequence in the recombinant vector was verified by automated DNA sequencing to confirm the identity, integrity, and absence of PCR-introduced mutations in the cloned fragment.
Protein expression and purification
The resulting recombinant vector was transformed into E. coli BL21 (DE3) (Novagen) competent cells. A single colony of transformed cells was selected from an Luria-Bertani (LB) agar plate containing kanamycin (50 μg/ml), and grown aerobically in 10 ml of LB media supplemented with kanamycin (50 μg/ml) at 37 °C. The resulting culture was transferred into 1.2 L of the same media and grown at 24 °C until the A600 reached 0.6 – 0.8. Protein expression was induced by the addition of isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the culture was grown aerobically at 37 °C for 5 hours. The cells (6g) were then harvested by centrifuging for 20 min at 12,227 × g at 4 °C, and the cell pellet was rinsed twice with Buffer A (20 mM Tris-HCl, 300 mM NaCl, and 20 mM imidazole, pH 7.5). The pellet was resuspended in 20 ml Buffer A containing one Complete EDTA-free protease inhibitor tablet. The cell suspension was lysed using a French pressure cell and then centrifuged for 1 hour at 100,000 × g at 4 °C. The clarified supernatant was collected and applied to a pre-equilibrated nickel affinity column (HisTrap HP, GE Healthcare). The column was washed with 5 column volumes of Buffer A, and the proteins were eluted using a linear imidazole gradient of Buffer A to Buffer B (20 mM Tris-HCl, 300 mM NaCl, and 500 mM imidazole, pH 7.5). Fractions with the most MTA cleavage were pooled and dialyzed overnight against two changes of 1 L of Buffer C (50 mM Tris HCl, 150 mM NaCl, 1 mM dithiothreitol, and 20% glycerol, pH 7.6) at 4 C (Spectra/Por 4 Membrane Tubing, 12,000 to 14,000 Dalton MWCO, Fisher Scientific). Protein concentrations were obtained by the Bradford method (2) using bovine serum albumin as standard.
Activity Assays
Enzyme activity was followed by measuring the formation of product using reverse phase high performance liquid chromatography (HPLC) as described previously (3). The xanthine oxidase coupled spectrophotometric method of Savarese et al., (18) and Jensen and Nygaard (4) was modified to a 96-well plate format and was used to rapidly detect MTA cleavage in fractions eluting from the purification column as described previously (3).
Enzyme kinetics
Steady-state kinetic constants were determined by varying the concentration of one substrate at fixed saturating concentration of the co-substrate. The amount of product formed was measured by reverse-phase HPLC and the substrate conversions were maintained below 10%. The data was fitted using the nonlinear regression function of SigmaPlot 2004 (Systat Software, Inc).
NanoLC-tandem mass spectrometry
Mass spectrometry analysis was conducted at the Targeted Metabolomics and Proteomics Laboratory at the University of Alabama at Birmingham as described previously (15). Briefly, the protein band from an SDS-PAGE gel was excised and destained. Following trypsin digestion, peptides were applied to a C18 reverse-phase cartridge, and the eluted peptides were analyzed on an Applied Biosystems-MDS-Sciex (Concorde, Ontario, Canada) 4000 Qtrap mass spectrometer. The tandem mass spectrometry data thus obtained was processed to provide protein identification using an in-house MASCOT search engine (Matrix Science) with the M. tuberculosis NCBInr protein database. The search parameters accounted for the oxidation of methionine residues, and a fixed carbamidomethylation of cysteines. One missed cleavage site for trypsin was allowed.
Results
Expression and purification of recombinant Rv0535
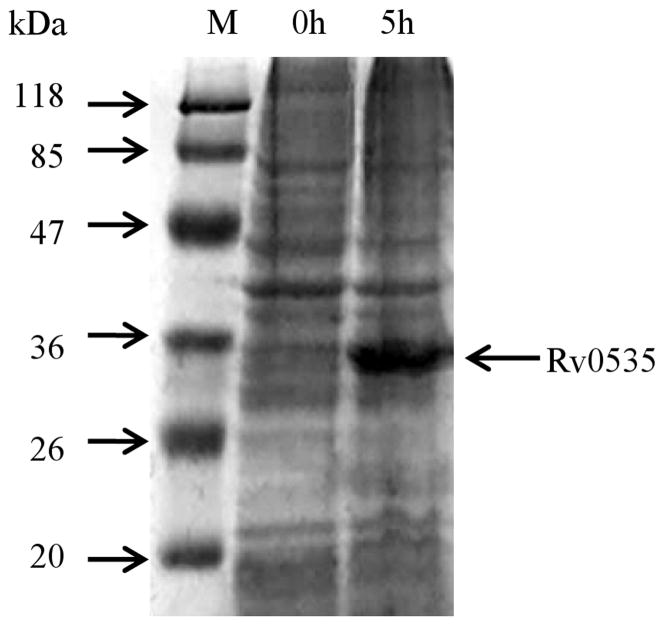
The Rv0535 sequence was amplified by PCR and the product was ligated into the pET28a (+) expression vector. The recombinant plasmid was then introduced into E. coli BL21 (DE3) competent cells. After induction with IPTG, the cells were harvested by centrifugation and lysed using a French pressure cell. The cytoplasmic fraction of the cells was loaded onto a denaturing SDS-PAGE gel and protein bands were visualized with Coomassie stain. As shown in Figure 2, after induction with IPTG, a new protein band that corresponded with the predicted monomer size of Rv0535 (30.276kDa, with histidine tags) was detected. The new protein band was excised from the gel and analyzed by NanoLC-tandem mass spectrometry, which identified the protein as Rv0535 (with 68% sequence coverage).
Fig. 2.
SDS-PAGE (12%, Coomassie stain) analysis of crude cell extracts from E. coli BL21 (DE3) cells before and after Rv0535 induction with IPTG. M, EZ-Run™ Pre-stained protein marker.
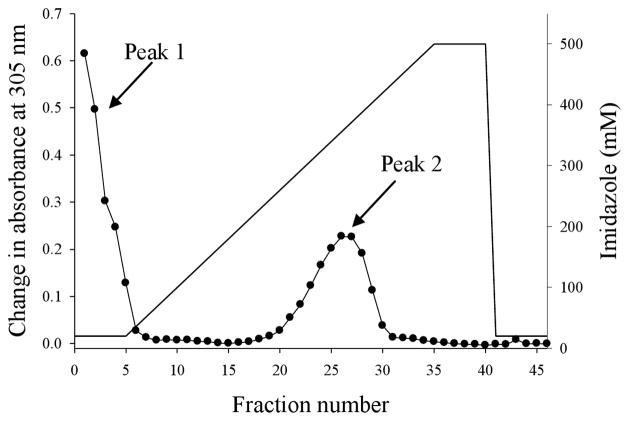
The pET28a (+) expression vector yielded recombinant Rv0535 that was fused to His-Tag at the N-terminus. Thus, a nickel affinity column (HisTrap HP) was used to purify the recombinant protein, after its expression had been induced with IPTG in E. coli BL21 (DE3) cells. The cytoplasmic fraction of the cells was applied to the purification column that was attached to an automated FPLC system. The column was washed with 5 column volumes of Buffer A, after which the concentration of imidazole was increased linearly, and one-ml fractions were collected. Fractions (including column flow-through and wash fractions) were tested for their ability to cleave MTA to Ade using the xanthine oxidase assay. Two peaks of MTA cleavage were seen. The first peak was in the flow-through and wash fractions and the second peak was in fractions 20 to 30 (Figure 3).
Fig. 3.
Elution profile of Rv0535 from the HisTrap HP column. Crude cell extract from E. coli BL21 (DE3) cells expressing Rv0535 was applied to the nickel affinity column and eluted with 20 – 500 mM imidazole gradient. Each fraction was tested for MTA cleavage, and the formation of Ade was measured by the change in absorbance at 305 nm after a 3-hour incubation with MTA.
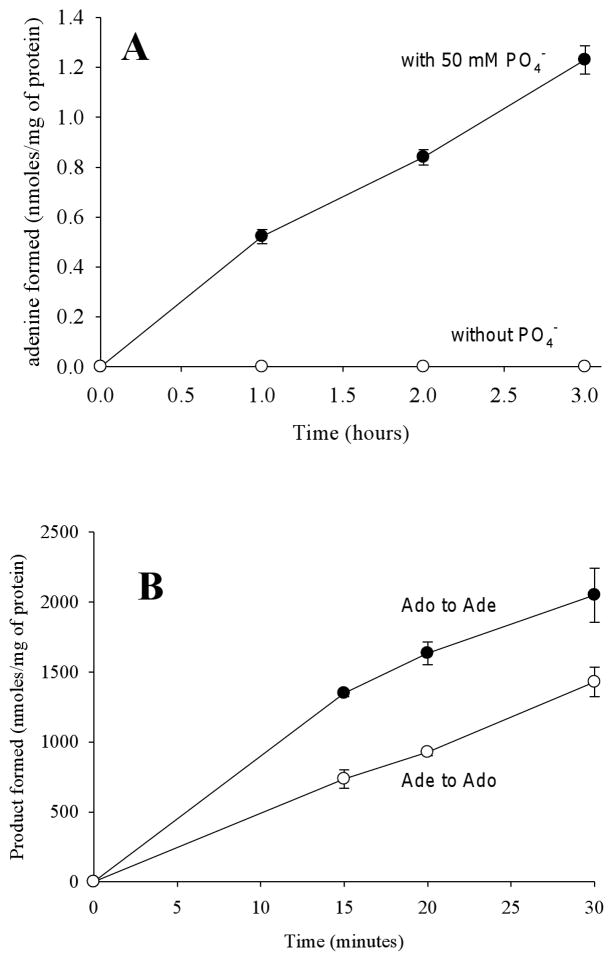
MTAP can only cleave MTA in the presence of phosphate, whereas MTAN can hydrolytically cleave MTA in the absence of phosphate. To determine whether peak 2 (fractions 20 to 30) contained MTAP, fractions 20 to 30 were pooled and dialyzed, after which MTA cleavage was assayed in the presence or absence of 50 mM phosphate. MTA to Ade conversion was measured by HPLC. As shown in Figure 4A, MTA cleavage was only seen in the presence of 50 mM phosphate. To further investigate whether fractions 20 to 30 contained a phosphorylase, the forward and reverse reactions were assayed. Since methylthioribose-1-phosphate is not commercially available, this experiment was performed with 1 mM Ado plus 50 mM phosphate or 1 mM Ade plus 1 mM ribose-1-phosphate. Both Ado phosphorolysis and synthesis were observed (Figure 4B), indicating that the cleavage of Ado is reversible. Because inosine (Ino) cleavage was not observed in the purified sample (see substrate specificity below), E. coli PNP cannot account for the phosphorolytic cleavage or synthesis of Ado observed in our experiments. Similar to the formation of Ado from Ade and ribose-1-phosphate, we anticipate that this sample (fractions 20 to 30) can catalyze the formation of MTA from Ade and methylthioribose-1-phosphate. Taken together, these results indicated that the MTA cleavage activity in peak 2 was due to a phosphorylase.
Fig. 4.
Ade formation requires phosphate and is reversible. (A) Purified Rv0535 was incubated with MTA in the presence or absence of 50 mM phosphate and Ade formation was monitored by HPLC. (B) The phosphorolysis and synthesis of Ado by Rv0535 was monitored by measuring the formation of Ade and Ado respectively by HPLC. Each data point represents the mean ± standard deviation for 3 determinations.
Peak 1 contained the column flow through and wash fractions, suggesting the presence of a second MTA cleavage enzyme that did not bind to the HisTrap column. When the phosphate requirement of these fractions was analyzed, MTA cleavage was observed in the absence of phosphate, indicating the presence of a hydrolase (data not shown). According to sequence homology, the BL21 (DE3) strain of E. coli is expected to express an MTAN, and therefore peak 1 is due to the elution of endogenous E. coli MTAN that did not bind to the HisTrap column.
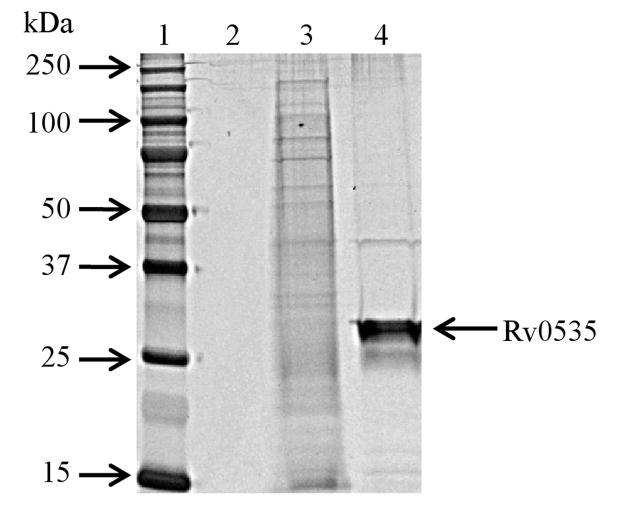
Purified Rv0535 (peak 2) was loaded and visualized on a silver stained, SDS-PAGE gel (Figure 5), which revealed one major band corresponding to the predicted monomer size of Rv0535. As shown in Table 1, our one-step purification scheme led to a 46-fold purification, with the removal of more than 99.9% of total protein. Table 1 also showed that only 5% of the total activity was recovered. While total activity represents MTA cleavage, the value in the crude cell extract does not discriminate between the contributions from endogenous E. coli MTAN or recombinant Rv0535. There was no difference in MTA cleavage in the crude cell extract in the presence or absence of phosphate (data not shown), suggesting that almost all of the activity detected in the crude was due to endogenous MTAN from the host cells. Since the purified Rv0535 fractions (Peak 2) could only cleave MTA in the presence of phosphate, it is likely that most of the Rv0535 was recovered during the purification step.
Fig. 5.
SDS-PAGE gel (12%; silver stain) of pooled Rv0535 fractions. Lane 1, molecular weight marker (Precision Plus All Blue Protein Standards); lane 2, empty; lane 3, crude cell extract (E. coli BL21 (DE3) expressing Rv0535); lane 4, pooled MTAP fractions from Peak 2 (Figure 3). Lanes 3 and 4 each contain 0.5 μg of protein.
Table 1.
Purification of M. tuberculosis Rv0535 from E. coli BL21 (DE3)
| Purification Step | Total protein (mg) | Total Activity (nmoles hr−1) | Specific Activity a (nmoles mg−1 hr−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude cell extract | 180 | 89,000 | 480 | 1 | 100 |
| HisTrap HP | 0.20 | 4,400 | 22,000 | 46 | 5 |
Activity determined with 100 μM MTA plus 50 mM phosphate
Substrate specificity of Rv0535
In our previous study, we have shown that endogenous M. smegmatis MTAP could cleave MTA and Ado but not the other natural purines or pyrimidines (3). Although M. smegmatis and M. tuberculosis MTAP share 196 amino acids (74% identity), it is possible that they have different substrate specificity. Therefore, we studied the substrate preference of Rv0535 by evaluating its ability to cleave the natural purines, including SAH, and the pyrimidines. As shown in Table 2, MTA was the preferred substrate of Rv0535. Ado cleavage was also seen at 2% of the MTA cleavage activity. This is in contrast to M. smegmatis MTAP, where Ado cleavage was 16% of the MTA cleavage activity (3). SAH cleavage was very low (0.8% of MTA cleavage), further indicating that Rv0535 does not encode for the bacterial MTAN, which can cleave both MTA and SAH. No activity was detected with the other purines (Ino, guanosine, or xanthosine) or the pyrimidines (cytidine, thymidine, or uridine). Similar to human and M. smegmatis MTAP, cleavage of 2′-deoxyadenosine was not observed with Rv0535 (7, 3).
Table 2.
Substrate specificity of Rv0535
| Nucleosides (100 μM) | Enzyme Activity (nmoles mg−1 hr−1)a | Relative activity (%) |
|---|---|---|
| Purines | ||
| 5′-Methylthioadenosine | 99,000 ± 3,000 | 100 |
| Adenosine | 2,100 ± 200 | 2.1 |
| S-Adenosyl-L-homocysteine | 800 ± 100 | 0.81 |
| 2′-Deoxyadenosine | -- | -- |
| Inosine | -- | -- |
| Guanosine | -- | -- |
| Xanthosine | -- | -- |
| 5′-Methylthioinosine | -- | -- |
| Pyrimidines | ||
| Cytidine | -- | -- |
| Thymidine | -- | -- |
| Uridine | -- | -- |
values represent mean ± standard deviation for 3 determinations
-- less than 700 nmoles mg−1 hr−1
MTA can be deaminated to 5′-methylthioinosine (MTI), a reaction that led to the misidentification of an MTIP as an MTAP in P. aeruginosa (8). To investigate whether the enzyme activity seen in our purified sample was due to an MTIP, the ability of Rv0535 to cleave MTI was assessed by HPLC. No activity was seen with MTI, indicating that Rv0535 was not MTIP. Further, P. aeruginosa MTIP can cleave Ino and Ado but not MTA (8), which is in contrast to our results above. Taken together, our substrate specificity results support our assessment of Rv0535 as an MTAP instead of an MTIP.
Kinetic properties of Rv0535
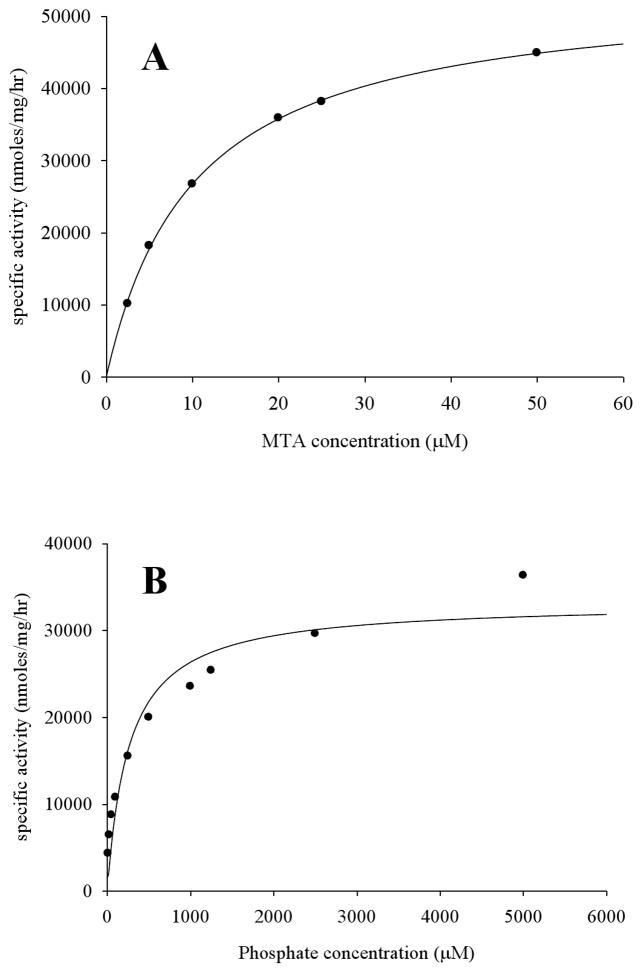
The steady-state kinetic constants of Rv0535 were determined for MTA and phosphate (Table 2). The initial rate of reaction at different substrate concentrations was used to generate saturation curves, which were described by a hyperbolic function and thus displayed Michaelis-Menten kinetics (Figure 6). The Km values for MTA and phosphate were 9 ± 3 μM (n=3) and 260 ± 80 μM (n=3), respectively. The Km value for Ado cleavage was 1700 ± 200 μM (n=3), with 16% of the relative Vmax with MTA, and more than a 6-fold decrease in Kcat, thereby indicating that Ado was a poor substrate for Rv0535.
Fig. 6.
Determination of steady-state kinetic constants. Representative saturation curves for (A) MTA as the variable substrate in the presence of 50 mM phosphate, and (B) phosphate as the variable substrate in the presence of 200 μM MTA.
Inhibitors of Rv0535
Fabianowska-Majewska et al., have shown a 92 % decrease in MTA cleavage activity in the presence of 1 mM Ado (500 μM MTA) and therefore, even though Ado is a poor substrate for human MTAP, it could inhibit the human enzyme (7). To see whether Ado could inhibit Rv0535, Ade formation was monitored in the presence of 100 μM MTA and either 1 mM Ado or 3 mM Ado. No inhibition was seen at either concentration of Ado (data not shown). Thus, unlike the human enzyme, Rv0535 was not inhibited by Ado.
Transition state analogs for bacterial purine enzymes with inhibitory constants in the femtomolar and picomolar range have been synthesized (11, 20), some of which are being investigated as antimicrobials (17). MT-DADMe immucillin is one of the best transition state analogs, and is a potent inhibitor of human MTAP. We investigated the effect of this transition state analog on MTA cleavage by Rv0535. The IC50 with MT-DADMe immucillin was 25 ± 6 nM or 70 ± 6 nM nM in the presence of 10 μM or 100 μM MTA, respectively (data not shown), which indicated that MT-DADMe immucillin was a potent inhibitor of Rv0535. Since transition state analogs have been described as slow-onset inhibitors, it is likely that the IC50 values obtained in our studies are greater than the true overall inhibition constant values of this inhibitor.
Other characteristics of Rv0535
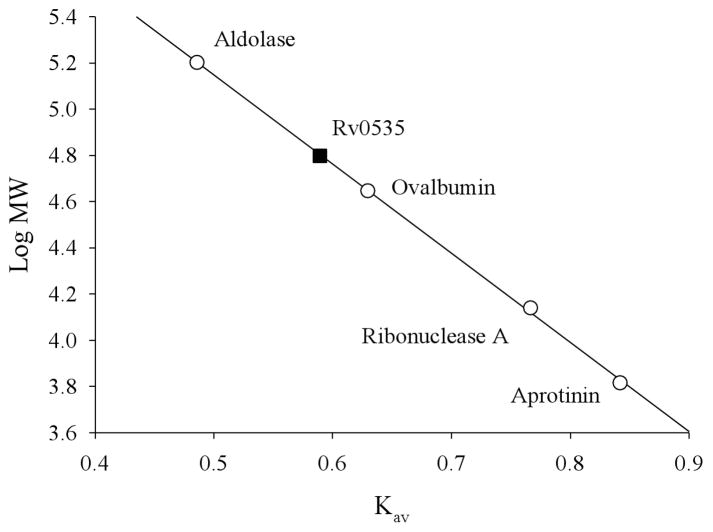
The native molecular weight of Rv0535 was estimated by gel filtration column chromatography (Superdex™ 200 PG, GE Lifesciences). Briefly, the column was calibrated using the UV absorbance spectra of the molecular weight protein standards (ferritin, conalbumin, carbonic anhydrase, and ribonuclease A). Purified Rv0535 (200 μl) was applied to the column and eluted with an isocratic run of Buffer C. Fractions eluting from the filtration column were tested for MTA cleavage activity. The purified protein eluted as a single peak and based on its elution volume, the molecular weight of the native form of the enzyme was calculated as 60 ± 4 kDa (n=3) (Figure 7). Since the monomeric form of Rv0535 is approximately 30 kDa, our size exclusion data indicates that Rv0535 is a functional dimer. Although dimeric MTAPs are uncommon, our data agrees with the molecular weight of the M. smegmatis MTAP (64 ± 5 kDa, with a monomer of 27 kDa) (3).
Fig. 7.
Estimation of the molecular weight of native Rv0535. Purified Rv0535 was applied to a Superdex 200 PG size exclusion column, and MTA cleavage activity was measured in fractions eluting from the column. The fraction with the most activity was used to calculate Kav according to the following equation: Kav=(Ve−Vo)/(Vc−Vo), where Vo is the column void volume, Ve is the elution volume, and Vc is the geometric column volume. Vo was determined based on the Ve of Blue Dextran 2000. MW, molecular weight.
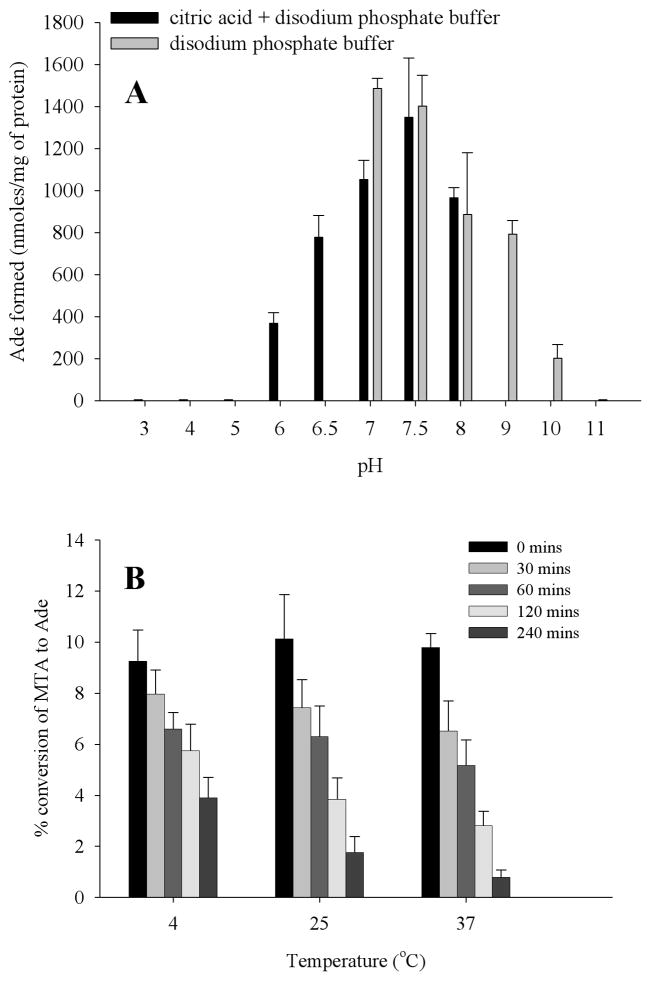
To investigate the effect of pH on enzyme activity, Rv0535 was incubated with MTA at different pH as described previously (3). The formation of Ade at each pH was measured by HPLC and the optimal pH of the reaction was between pH 7 and 7.5 (Figure 8A). The enzyme was inactive above pH 10 and below pH 6, indicating that the enzyme is inactive at its theoretical pI of 5.75.
Fig. 8.
Effect of pH (A) and temperature (B) on Rv0535 activity. The amount of Ade formed was measured by HPLC, after the enzyme had been incubated at different pH (A) or at 4 °C, 25 °C, or 37 °C for different time periods (B). Error bars indicate mean ± standard deviation for 3 determinations.
The stability of Rv0535 at various temperatures was also studied. The purified protein was incubated at 4 °C, 25 °C, and 37 °C for specific amounts of time, after which MTA cleavage (at 37 °C) was assayed by HPLC. As shown in Figure 8B, there was a 12% or 40% decrease in MTA cleavage activity after the enzyme had been incubated at 4 °C for 30 minutes or 4 hours respectively. In contrast, there was a 20% or 90% decrease in MTA cleavage activity after being incubated at 37 °C for 30 minutes or 4 hours respectively. While there was no difference between the stability of the enzyme at 25 °C or 37 °C after a 30 minute incubation at either temperatures, there was a considerable decrease in the rate of reaction when the assay was performed at 25 °C instead of 37 °C (data not shown). Therefore, enzyme assays were carried out at 37 °C. Additionally, there was no change in activity in aliquots stored at −20 °C (data not shown). Therefore, our data indicated that the recombinant protein was not stable at 4 °C, once the aliquot had been thawed. Because of the instability of the protein at 4 °C, necessary precautions were taken when handling the enzyme and to limit its loss of activity, the enzyme was incubated no longer than 15 minutes at 37 °C for the HPLC enzyme assay. Although the reason for the instability is not known, it could be due to the histidine tags that are on the N-terminus of the recombinant protein. It is possible that the removal of the polyhistidine tags by peptidases, using a different tag, or purifying endogenous M. tuberculosis MTAP could resolve the stability issues.
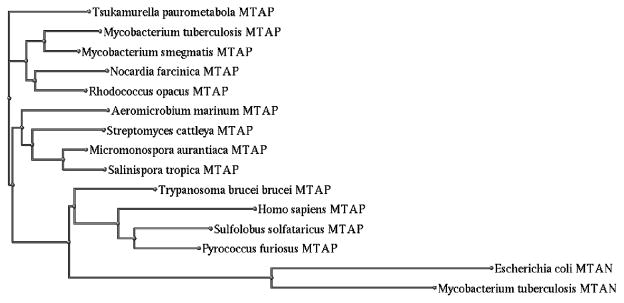
To evaluate whether other bacteria express MTAP, the Basic Local Alignment Search Tool (BLAST) function of Uniprot (22) was used to compare the amino acid sequence of M. tuberculosis MTAP (Rv0535) to other sequence databases. The program produced 250 hits with sequence similarity ranging from 50% to 100%. More than 95% of these hits correspond to predicted or probable MTAPs, as opposed to MTAPs that have been purified and characterized. From the 250 hits, one sequence from each parent species, organism or strain that also had more than 55% similarity with Rv0535 was selected for a phylogenetic analysis. The resulting 7 sequences were all probable bacterial MTAPs. Therefore, while mycobacterial MTAPs were the first bacterial MTAPs to be identified, it is possible that other bacteria also express MTAP.
Given that there are a few differences in substrate specificity between M. tuberculosis and M. smegmatis MTAP, M. smegmatis MTAP (75% identity) was also included in the analysis. To represent MTAP from Archaea, MTAP sequences from Sulfolobus Solfataricus (43% identity) and Pyrococcus furiosus (46% identity) were added. MTAPs that have been well studied were also included, namely human MTAP (to represent mammalian MTAP, 38% identity), and Trypanosoma brucei brucei MTAP (to represent trypanosome MTAP, 40% identity). Since MTAP and MTAN can both cleave MTA, M. tuberculosis MTAN (15% identity) and E. coli MTAN (14% identity) were also included in the analysis. The selected sequences were aligned using MUltiple Sequence Comparison by Log- Expectation (MUSCLE) and a phylogenetic tree was created using the NCBI Genome Workbench (version 2.4.0 (12)).
As shown in Figure 9, the sequences that had been selected from the BLAST hits are closely grouped together. M. tuberculosis MTAP is in closer proximity to pathogenic bacteria than to non-pathogenic bacteria, with the exception of M. smegmatis. Further, MTAPs that have been characterized were grouped separately from the predicted MTAPs. The Archaea and trypanosome MTAPs are closer to human MTAP as compared to bacterial MTAPs. While human and M. tuberculosis MTAP share 38% sequence identity, human and M. tuberculosis MTAN only share 18% identity. Similarly, M. tuberculosis MTAP and MTAN share 15% sequence identity. As suggested by the phylogenetic tree, despite having similar functions, MTAP sequences are different from MTAN sequences.
Fig. 9.
Phylogenetic analysis of MTA catabolizing enzymes from mycobacteria and other organisms. Amino acid sequences of MTAPs and MTANs from various sources were aligned with M. tuberculosis MTAP (Rv0535) and a phylogram was produced to represent the alignment.
Molecular model of Rv0535
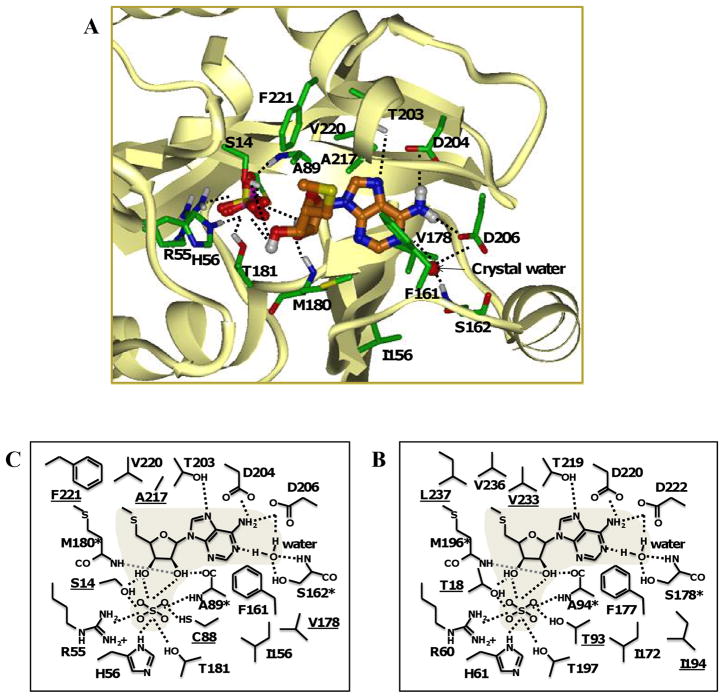
The crystal structure of human MTAP has been determined and was used to generate a homology model of M. tuberculosis MTAP. This approach could help explain the substrate specificity of M. tuberculosis MTAP, as well as predict modifications to nucleoside analogs that would lead to their increased or decreased cleavage by the enzyme. To this end, anM. tuberculosis MTAP homology model was built using human MTAP (PDB no. 1CG6) as a template. The human MTAP sequence as present in the crystal structure was aligned with the M. tuberculosis MTAP sequence and short loop structures missing in the crystal structures were modeled with MODELER. The sequence identity in the modeled region was 40.5%, and residues that interact with the substrate were identified.
Similar to other MTAPs, we predict that each subunit of M. tuberculosis MTAP contains one active site. Our size exclusion data indicated that M. tuberculosis MTAP was a dimer, whereas human MTAP is known to be a trimer. Therefore, our modeling data is limited to comparisons of a single subunit from the M. tuberculosis MTAP structure to one subunit of the human MTAP structure. Residues that form interactions with the substrate were identified, and all but one residue, were components of one subunit. The M. tuberculosis model predicted that one amino acid residue (His126) from a neighboring subunit could contribute to weak steric interactions with the substrate. This could indicate that the active site of M. tuberculosis MTAP is not buried and is located at the subunit interface.
As shown in Figure 10, the active site of M. tuberculosis MTAP consists of about 17 amino acid residues. In general, the residues closer to the N-terminal domain are likely to participate in phosphate binding (Ser14, Arg55, His56, Cys88, Ala89, Met180, and Thr181) whereas the C-terminal domain residues could participate in nucleoside binding (Ser162, Thr203, Asp204, and Asp206). Ser14 could participate in both phosphate binding and interaction with the 3′-hydroxy group on the ribose moiety. Thus, it is possible that the active site of M. tuberculosis MTAP could be divided into a phosphate-binding site, a purine-binding site, and a ribose-binding site.
Fig. 10.
M. tuberculosis MTAP homology model built using human MTAP (PDB no. 1CG6) as a template. (A) Ribbon structure of a single subunit of M. tuberculosis MTAP with MTA and sulfate in the active site. Schematic representation of the interactions formed between MTA and the M. tuberculosis MTAP (B) and human MTAP (C). Amino acids differences between M. tuberculosis and the human sequence are underlined. Residues with backbone atoms involved in interactions with the substrate are marked (*). Polar interactions are shown as dashed lines.
Of the 17 active site amino acids, 5 residues (Ser14, Cys88, Val178, Ala217, and Phe221) were different from human MTAP (Figure 10B and C). Ser14 and its human counterpart (Thr18) are both amino acids with polar, uncharged side chains and are both capable of forming analogous hydrogen bonding interactions with the substrate. However, the additional methyl group in Thr18 may have steric effects on the 5′-methylthio-group in MTA. Thr93 (from the human enzyme) forms more favorable interactions with the co-crystallized sulfate compared to Cys88. Val178 and its human counterpart (Ile194) are in close proximity to the purine ring, and although Ile194 has an additional methyl group and is therefore bulkier than Val178, steric/non-polar interactions are similar between the two species. The side-chain of Ala217 is in close proximity to the 5′-methylthio-group of the substrate and results in distinct non-polar interactions. The human counterpart of Ala217 (Val233) is significantly bulkier than Ala217 but Val233 forms more favorable non-polar interactions with the substrate. Thus, the substitution of Val233 with Ala217 in M. tuberculosis MTAP leads to a net loss in favorable non-polar interactions. Phe221 has a phenyl ring in close proximity to the 5′-methylthio- group of MTA and adds considerable bulk to that region as compared to its human counterpart (Leu237). The non-polar interactions formed between the human MTAP residue Leu237 and the 5′-methylthio-group of MTA are more favorable than the analogous steric interaction involving Phe221, the corresponding residue in M. tuberculosis MTAP.
Given that the molecular model of M. tuberculosis MTAP identified amino acids that participate in substrate interactions, we compared the aligned sequences of other bacterial MTAPs (from M. smegmatis, Nocardia farcinica, Rhodococcus opacus, and Tsukamurella paurometabola) to determine whether these amino acids were conserved among bacterial MTAPs. Of the 17 residues involved in the active site of M. tuberculosis MTAP, 12 were conserved among these bacteria and human MTAP. Most active site amino acids were conserved among bacterial MTAPs: there were only two active site residues that were different between the various bacterial enzymes. M. tuberculosis Ser14, which we suggest participates in both phosphate and ribose binding pockets, is conserved in M. smegmatis and R. opacus MTAPs, whereas N. farcinica and T. paurometabola MTAPs have a Thr at that position, similar to human MTAP. M. tuberculosis Phe221 is conserved in M. smegmatis, N. farcinica, and R. opacus MTAPs, but not in T. paurometabola MTAP, which has a Leu at that position, similar to human MTAP. In general, MTAPs can fall in two categories, with amino acids resembling either M. tuberculosis or human MTAP. Given the differences that exist between bacterial MTAPs, it is possible that they have different substrate specificity.
Discussion
MTAP is primarily expressed in Eukarya, including trypanosomes, and Archaea, while MTAN is expressed in bacteria. In our previous work in M. smegmatis, we had noted the phosphorolytic cleavage of MTA and presented the first evidence of an MTAP activity in mycobacteria. Based on sequence homology, both MTAP and MTAN are predicted to exist in M. smegmatis and M. tuberculosis. Rv0535 was expressed in E. coli BL21 (DE3) cells and the purified protein was biochemically characterized. The natural purines and pyrimidines were tested as substrates, and MTA was cleaved with maximal activity, thereby indicating that MTA was the preferred substrate. MTI was also investigated since MTIP from P. aeruginosa had been incorrectly defined as MTAP. In our work, we showed that MTI was not a substrate for this enzyme, and therefore Rv0535 does not encode for MTIP. Thus, using a recombinant protein strategy combined with biochemical analysis, we confirm the correct annotation of Rv0535 as MTAP in M. tuberculosis.
In this work, we report that M. tuberculosis MTAP had a Km of 9 μM with MTA, which is higher than the Km of human MTAP (1.5 ± 0.2 μM) (23), but lower than that of Sulfolobus solfataricus MTAP (24 μM) (4). Further, the Kcat of M. tuberculosis MTAP with MTA was 0.4 ± 0.2 s−1, which is low in comparison to other bacterial purine metabolic enzymes, especially E. coli MTAN (MTA, 4.0 ± 0.1 s−1) (21) and M. tuberculosis PNP ( inosine, 5.4 ± 0.1 s−1)(5). However, S. pneumonia MTAN has a comparable Kcat (MTA, 0.25 ± 0.04 s−1; SAH, 0.37 ± 0.05 s−1) (21). Variability in the specific activity of the recombinant enzyme was observed (in the range of 20,000 – 100,000 nmoles/mg/hr) and could be due to different purification efficiency and the instability of the protein at 4 °C. It is possible that the specific activity and Vmax (and thus Kcat) reported in this study are an underestimation of the actual values. On the other hand, a high turnover may not be required because of the presence of another enzyme (MTAN) that can also cleave MTA.
There are no known physiological explanations for the expression of both MTAP and MTAN in one organism. Until M. tuberculosis MTAN (Rv0091, Genbank Accession no. CAA9827) is expressed and its kinetic parameters are studied, the relationship between MTAP and MTAN will remain elusive. The concerted action of MTAN and methylthioribose kinase produces 5′-methylthioribose-1-phoshate, which can then be salvaged to methionine. To date, methylthioribose kinase has not been identified in the M. tuberculosis genome. Some bacteria such as E. coli do not possess methylthioribose kinase and therefore cannot recycle 5′-methylthioribose, which is excreted from the cell. If methionine salvage is important to the mycobacterium, it is possible that M. tuberculosis expresses both MTAP and MTAN to improve the efficiency of MTA cleavage, and MTAP produces 5′-methylthioribose-1-phosphate to rescue the methionine salvage pathway. It is also possible that because human cells express MTAP, mycobacterial MTAP is not expressed when the mycobacterium is found in its host. Further, it has been shown that methylthioribose kinase is expressed under starvation conditions in Bacillus subtilis (19). Thus, the expression of MTAP and MTAN under different growth conditions needs to be investigated.
The fact that human cells also express MTAP does not preclude M. tuberculosis target as a drug target. For example, parasites such as Trypanosome brucei brucei also express MTAP but the substrate specificity of the parasitic MTAP differs from that of the human form and these differences have been investigated for anti-parasitic drug development (10). The M. tuberculosis model indicated that the 5 different amino acid residues are in close proximity to or participate in the phosphate-binding site (Ser14 and Cys88), the purine-binding site (Val178), or the 5′-methylthioribose-binding site (Ser 14, Ala217, and Phe221). Thus, it is possible that modifications to the Ade moiety or ribose moiety of the substrate could result in different substrate specificity between the mycobacterial and human MTAP. A complete structure-activity relationship study of Rv0535 using Ado analogs could help identify key differences between human and M. tuberculosis MTAP, which could be exploited in a drug discovery effort.
Table 3.
Steady-state kinetic parameters of Rv0535 in the direction of MTA phosphorolysis.
| Substrate | Co-substrate | Km (μM) | Kcat (s−1) |
|---|---|---|---|
| MTA | 50 mM phosphate | 9 ± 3 | 0.4 ± 0.2 |
| Phosphate | 200 μM MTA | 260 ± 80 | 0.26 ± 0.02 |
Values represent mean ± standard deviation for 3 determinations
Acknowledgments
This work was supported by a grant from the National Institutes of Health, AI43241.
We thank Dr. Stephen Barnes and Dr. Mahmoud el Kouni, Department of Pharmacology and Toxicology, University of Alabama at Birmingham, for valuable advice, and Paula Allan, Southern Research Institute, for technical assistance. We also thank Dr. Vern Schramm, Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University for his contribution of 5′-methylthioinosine and MT-DADMe-Immucillin A.
Operation of the Targeted Metabolomics and Proteomics Laboratory is supported by grants from the National Center for Complementary and Alternative Medicine and the NIH Office of Dietary Supplements to the Purdue-UAB Botanicals Center for Age-Related Disease (P50 AT00477, C. Weaver, PI), from the National Cancer Institute to the UAB Center for Nutrient-Gene Interaction (U54 CA100949, S. Barnes, PI), from the National Institute of Digestive, Diabetes and Kidney Disease to the UAB O’Brien Acute Kidney Injury Center (P30 DK079337, A. Agarwal, PI) and the UAB Diabetes Research and Training Center (P60 DK079626, T. Garvey, PI), from the National Institute of Arthritis and Musculoskeletal Disease to the UAB Skin Disease Research Center (P30 AR050948, C. Elmets, PI), and from the UAB Center for Free Radical Biology and the UAB Lung Health Center. Support for the mass spectrometers used in the study came from NCRR Shared Instrumentation grants S10 RR19231 and RR027822.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kajal Buckoreelall, Email: kbuck@uab.edu.
Yanjie Sun, Email: yanjie_sunny@yahoo.com.
Judith V. Hobrath, Email: hobrath@southernresearch.org.
Landon Wilson, Email: landon.wilson@ccc.uab.edu.
William B. Parker, Email: parker@southernresearch.org.
References
- 1.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life. 2009;61:1132–1142. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- 2.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 3.Buckoreelall K, Wilson L, Parker WB. Identification and characterization of two adenosine phosphorylase activities in Mycobacterium smegmatis. J Bacteriol. 2011;193:5668–5674. doi: 10.1128/JB.05394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cacciapuoti G, Porcelli M, Bertoldo C, De Rosa M, Zappia V. Purification and characterization of extremely thermophilic and thermostable 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus. Purine nucleoside phosphorylase activity and evidence for intersubunit disulfide bonds. J Biol Chem. 1994;269:24762–24769. [PubMed] [Google Scholar]
- 5.Ducati RG, Santos DS, Basso LA. Substrate specificity and kinetic mechanism of purine nucleoside phosphorylase from Mycobacterium tuberculosis. Arch Biochem Biophys. 2009;486:155–164. doi: 10.1016/j.abb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Dye C, Lönnroth K, Jaramillo E, Williams BG, Raviglione M. Trends in tuberculosis incidence and their determinants in 134 countries. Bull World Health Organ. 2009;87:683–691. doi: 10.2471/BLT.08.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabianowska-Majewska K, Duley J, Fairbanks L, Simmonds A, Wasiak T. Substrate specificity of methylthioadenosine phosphorylase from human liver. Acta Biochim Pol. 1994;41:391–395. [PubMed] [Google Scholar]
- 8.Guan R, Ho MC, Almo SC, Schramm VL. Methylthioinosine Phosphorylase from Pseudomonas aeruginosa. Structure and Annotation of a Novel Enzyme in Quorum Sensing. Biochemistry. 2011;50:1247–1254. doi: 10.1021/bi101642d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershfield MS. Apparent suicide inactivation of human lymphoblast S-adenosylhomocysteine hydrolase by 2′-deoxyadenosine and adenine arabinoside. A basis for direct toxic effects of analogs of adenosine. Journal of Biological Chemistry. 1979;254:22–25. [PubMed] [Google Scholar]
- 10.el Kouni MH. Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacology & Therapeutics. 2003;99:283–309. doi: 10.1016/s0163-7258(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 11.Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL. Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Biol Chem. 2005;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- 12.National Center for Biotechnology Information. Genome Workbench Home Page. [Google Scholar]
- 13.Parveen Nikhat, Cornell KA. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol. 2011;79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pajula R, Raina A. Methylthioadenosine, a potent inhibitor of spermine synthase from bovine brain. FEBS Letters. 1979;99:343–345. doi: 10.1016/0014-5793(79)80988-6. [DOI] [PubMed] [Google Scholar]
- 15.Pressey JG, Pressey CS, Robinson G, Herring R, Wilson L, Kelly DR, Kim H. 2D-difference gel electrophoretic proteomic analysis of a cell culture model of alveolar rhabdomyosarcoma. J Proteome Res. 2011;10:624–636. doi: 10.1021/pr1008493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina A, Tuomi K, Pajula RL. Inhibition of the synthesis of polyamines and macromolecules by 5′-methylthioadenosine and 5′-alkylthiotubercidins in BHK21 cells. Biochem J. 1982;204:697–703. doi: 10.1042/bj2040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson JG. Enzymes as a special class of therapeutic target: clinical drugs and modes of action. Curr Opin Struct Biol. 2007;17:674–679. doi: 10.1016/j.sbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Savarese TM, Crabtree GW, Parks RE., Jr 5′-methylthioadenosine phosphorylase--I: Substrate activity of 5′-deoxyadenosine with the enzyme from Sarcoma 180 cells. Biochemical Pharmacology. 1981;30:189–199. doi: 10.1016/0006-2952(81)90077-0. [DOI] [PubMed] [Google Scholar]
- 19.Sekowska A, Mulard L, Krogh S, Tse JK, Danchin A. MtnK, methylthioribose kinase, is a starvation-induced protein in Bacillus subtilis. BMC Microbiol. 1:15–15. doi: 10.1186/1471-2180-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 21.Singh V, Shi W, Almo SC, Evans GB, Furneaux RH, Tyler PC, Painter GF, Lenz DH, Mee S, Zheng R, Schramm VL. Structure and Inhibition of a Quorum Sensing Target from Streptococcus pneumoniae. Biochemistry. 2006;45:12929–12941. doi: 10.1021/bi061184i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Research. 2010;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toorchen D, Miller RL. Purification and characterization of 5′-deoxy-5′-methylthioadenosine (MTA) phosphorylase from human liver. Biochem Pharmacol. 1991;41:2023–2030. doi: 10.1016/0006-2952(91)90145-u. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. Global tuberculosis control - epidemiology, strategy, financing WHO Report 2009. 2009. [Google Scholar]