Abstract
Postsynaptic density protein 95 (PSD-95) is essential for synaptic maturation and plasticity. Although its synaptic regulation is widely studied, the control of PSD-95 cellular expression is not understood. We find that Psd-95 is controlled post-transcriptionally during neural development. Psd-95 is transcribed early in mouse embryonic brain, but most of its product transcripts are degraded. The polypyrimidine tract binding proteins, PTBP1 and PTBP2, repress Psd-95 exon 18 splicing, leading to premature translation termination and nonsense-mediated mRNA decay (NMD). The loss first of PTBP1 and then of PTBP2 during embryonic development allows splicing of Exon 18 and expression of PSD-95 late in neuronal maturation. Re-expression of PTBP1 or PTBP2 in differentiated neurons inhibits PSD-95 expression and impairs development of glutamatergic synapses. Thus, expression of PSD-95 during early neural development is controlled at the RNA level by two PTB proteins whose sequential down-regulation is necessary for synapse maturation.
Keywords: PTBP1, PTBP2, Psd-95, alternative splicing, neural development, synapse maturation, nonsense-mediated mRNA decay
PSD-95 is an abundant scaffold protein of the excitatory postsynaptic density (PSD), where it functions to cluster proteins such as glutamate receptors on the postsynaptic membrane and couple them with downstream signalling molecules 1, 2. Psd-95−/− mice have severe learning defects and exhibit both facilitation of long-term potentiation (LTP) and disruption of long-term depression (LTD) 3, 4. The increase in PSD-95 expression during development plays a key role in maturation of excitatory synapses 5–9. However, although the synaptic expression and posttranslational modification of PSD-95 protein have been examined in relation to neuronal plasticity, there is little understanding of how its cellular expression is regulated during development.
Neuronal differentiation and maturation require an orchestrated series of complex genetic regulatory events. The roles of transcriptional and miRNA-mediated post-transcriptional control in this process are actively studied and are best understood. The contributions of other genetic regulatory mechanisms to neural development are not as well defined. Notably, the splicing of many transcripts is altered during neuronal differentiation. These regulated splicing events change the structure and activity of many proteins in a manner that is often highly conserved across species. But how these isoform changes affect the differentiating neuron is largely unknown. Many of these neuron-specific alternative splicing events are controlled by the polypyrimidine tract binding proteins, PTBP1 and PTBP2 10, 11. PTBP1 (PTB) is highly expressed in non-neuronal cells and neural progenitor cells. Its down-regulation in differentiating neurons alters the splicing of many exons to produce a neuron-specific repertoire of functional proteins. The down-regulation of PTBP1 also induces expression of its homolog PTBP2 (also known as brPTB or nPTB) 10–12. These two similar proteins equally affect some exons, whereas other exons are more responsive to PTBP1 and thus change their splicing when these two proteins are exchanged during differentiation.
In addition to altering protein structure and function, alternative splicing can alter reading frame to induce translation termination and subsequent nonsense-mediated mRNA decay (NMD) of the spliced isoform. The NMD pathway allows the degradation of nonsense and frame shift mutant mRNAs, preventing production of truncated protein products 13, 14. NMD also acts as a quality control process to eliminate aberrantly spliced mRNAs. In addition, many splicing regulators limit their own expression through the autoregulation of their splicing to produce an NMD-targeted mRNA (alternative splicing-induced NMD, or AS-NMD) 15, 16. Besides these splicing regulatory proteins, microarray studies found additional transcripts to be induced when NMD is inhibited in mammalian cells 16–18. Some of these transcripts integrate NMD into stress responses and nutrient homeostasis 19, 20.
To understand the functional significance of PTBP1-mediated regulation during neuronal differentiation, we examined the physiological consequences of its reintroduction into differentiated neurons. Ectopic expression of PTBP1 did not alter neuronal cell fate, but strongly decreased PSD-95 protein expression. In examining the mechanism of this PSD-95 repression, we find that Psd-95 mRNA is transcribed throughout embryonic development but is subject to intense post-transcriptional repression by the two PTB proteins and the NMD pathway.
Results
PTB proteins block PSD-95 expression in neurons
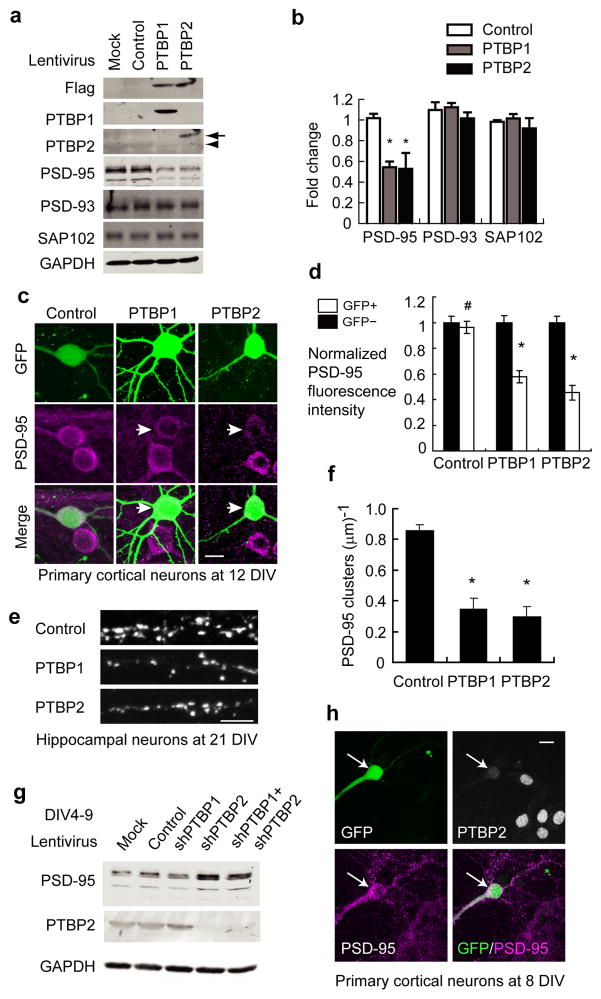
PTBP1 is expressed in neural progenitor cells but not differentiated neurons. To examine the effect of PTBP1 on mature neurons, we infected primary cortical cultures at 4 days in vitro (DIV) with lentivirus expressing flag-tagged PTBP1 and GFP. At 4 DIV, >95% of cells in the cultures expressed the neuronal marker Tuj1 and were committed to the neuronal cell lineage (Supplementary Fig. 1). We found that cultures infected with PTBP1 virus differentiated normally and appeared almost morphologically identical at 12 DIV from those infected with control virus expressing GFP only. Assessing several neuron specific markers in the PTBP1 expressing neurons, we found a significant reduction in PSD-95 protein. The repression was specific to PSD-95, as expression of the PSD-95 homologs PSD-93 and SAP102 was not affected (Fig. 1a–b).
Figure 1.
PTB proteins repress PSD-95 expression cell-autonomously. (a) Western Blot of various proteins in the primary cortical cultures infected with PTBP1, PTBP2 or control lentivirus. The arrow and arrowhead point to the exogenous flag-PTBP2 and the endogenous PTBP2 respectively. (b) Protein levels of PSD-95, PSD-93 and SAP102 were quantified and normalized to mock infected cells. The plotted bars represent the mean values±s.e.m. (N=3). An asterisk indicates a p-value<0.01 determined by one-way ANOVA. (c) GFP plasmids were co-transfected with pcDNA3.1 control vector, flag-PTBP1 or flag-PTBP2 expression vector in primary cortical cultures. Arrows mark the transfected neurons expressing PTBP1 or PTBP2. Scale bar: 10 μm. The somatic PSD-95 immunofluorescent signals (magenta) of transfected (GFP+) and untransfected (GFP−) neurons were quantified in (d). The bar plot represents mean values±s.e.m. (N=23). * indicates p<1×10−5. # indicates p=0.35. The p-values were determined by two-tailed pair-wise t-test comparing GFP+ and GFP− cells. (e) Similar transfection was performed in dissociated hippocampal cultures. The PSD-95 cluster densities were reduced on dendrites from neurons expressing PTBP1 or PTBP2 and quantified in (f). The bar plot represents mean values±s.e.m. (N=11). * indicates p<10−8. (g) Western blots of PSD-95, PTBP2 and GAPDH from primary cortical cultures infected with control hairpin, shPTBP1 and shPTBP2 lentivirus at 4 DIV. Samples were harvested at 9 DIV. (h) Primary cortical neurons were co-transfected with shPTBP2 and GFP plasmids at 4 DIV and stained for PSD-95 and PTBP2 at 8 DIV. shPTBP2-transfected neurons (GFP+) enhanced PSD-95 expression compared to neighbouring untransfected neurons.
PTBP2 is induced as neural progenitors differentiate and PTBP1 is depleted. Re-expression of PTBP1 also repressed PTBP2. The repression of PSD-95 by PTBP1 could thus be due to a decrease in PTBP2. However, infecting cultures with a PTBP2 lentivirus had equivalent effects to PTBP1 in repressing PSD-95 proteins at 12 DIV (Fig. 1a–b).
The lentiviral infections were very efficient, with most of the neurons in the culture expressing PTBP1 or PTBP2. To observe the effect of the PTB proteins on single cell morphology and to confirm that the observed PSD-95 repression was autonomous to neurons expressing either PTB protein, we co-transfected primary cortical cultures with PTB expression plasmids and a GFP expression plasmid. This allowed flag-tagged PTBP1 or PTBP2 expression in sporadic cells of the primary cultures. The GFP positive cells all expressed exogenous PTB proteins as measured by anti-flag co-staining. PSD-95 protein levels in the GFP positive neurons at 12 DIV were compared to those of neighbouring GFP negative neurons by immunofluorescent staining and confocal microscopy (Fig. 1c). Both the total PSD-95 fluorescence in the soma (Fig. 1d) and the average PSD-95 voxel intensity relative to cell volume (Supplementary Fig. 2) were decreased by 50% in the PTBP1 expressing cells. A control vector had no effect on PSD-95 protein expression. Transfection with a flag-tagged PTBP2 expression plasmid had the similar effect in repressing PSD-95 protein (Fig. 1c–d). To examine whether PTBP1 or PTBP2 also impaired synaptic expression of PSD-95 proteins, we performed similar transfection experiments in low-density hippocampal cultures at DIV 9–11. Hippocampal neurons expressing ectopic PTBP1 or PTBP2 exhibited markedly reduced density of PSD-95 puncta along their dendrites at 21 DIV (Fig. 1e–f). These data together with the lentiviral experiments indicate that either re-expression of PTBP1 or over-expression of PTBP2 is sufficient to repress PSD-95 protein expression in differentiated neurons.
We next tested whether the loss of PTB proteins was sufficient to increase PSD-95 expression. Early knockdown of PTBP1 by shRNA lentivirus in primary cortical cultures during DIV 0–4 induced PTBP2 expression and the neurons appeared to differentiate normally. In contrast, depletion of PTBP2 caused a morphological deterioration and cell death at 5 DIV, indicating early requirement for PTBP2 in neuronal survival. If we infected the cultures with shRNA lentiviruses at 4 DIV instead of earlier, these neurons survived well to 9 DIV when we analyzed the protein expression. In these conditions, PTBP2 knockdown led to a nearly 2-fold increase in PSD-95 protein expression (Fig. 1g). PTBP1 expression was already very low by 4 DIV, so knockdown of PTBP1only marginally increased PTBP2 expression and did not affect PSD-95 expression. Double knockdown of PTBP1 and PTBP2 yielded similar results to the single PTBP2 knockdown (Fig. 1g). The specificity of this PTBP2 shRNA was confirmed by co-infection of neurons with the lentivirus expressing PTBP2 cDNA insensitive to the shPTBP2. PTBP2 transduction in these neurons reversed the effect of the PTBP2 shRNA (Supplementary Fig. 3). A different shRNA targeting different PTBP2 sequences also enhanced PSD-95 expression (Supplementary Fig. 4), indicating again that the change in PSD-95 was not due to an off-target effect of the shRNA. To confirm that the increase in PSD-95 is an autonomous effect in the cells depleted of PTBP2, primary cortical neurons were co-transfected with shPTBP2 and GFP plasmids at 4 DIV. Under these conditions, the PTBP2 deficient neurons exhibited significantly enhanced PSD-95 expression compared to neighbouring untransfected neurons (Fig. 1h). These data demonstrate that the PTB proteins are negative regulators of PSD-95 protein expression.
PTB proteins repress synapse maturation
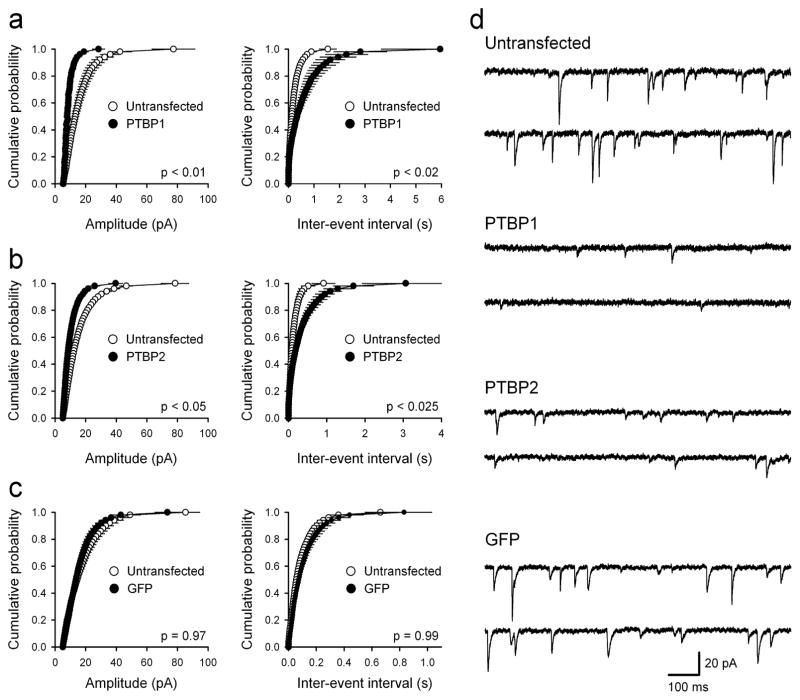
PSD-95 is involved in the maturation of excitatory synapses and has a crucial role in the synaptic targeting of AMPA-type glutamate receptors. Overexpression of PSD-95 in hippocampal cultures augments the amplitude and frequency of AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) and promotes synapse maturation 5. We thus examined whether PTBP1 and PTBP2 could regulate PSD-95-related neuronal physiology. In repressing PSD-95, re-expression of the PTB proteins in mature neurons should have potent, inhibitory effects on excitatory synaptic transmission. To examine this, we recorded mEPSCs from hippocampal neurons transfected with PTBP1, PTBP2 and control plasmids. Indeed, we find that co-transfection of PTBP1 and GFP led to an approximate two-fold decrease in both the amplitude and frequency of mEPSCs compared to non-transfected neurons from the same cultures (Fig. 2a,d). Over-expression of PTBP2 had a similar effect (Fig. 2b,d) while transfection of a pcDNA control plasmid with GFP had no effect on either mEPSC amplitude or frequency (Fig. 2c,d). Thus, down-regulation of PSD-95 protein expression following re-expression of the PTB proteins in mature neurons is associated with significant deficits in excitatory synaptic transmission.
Figure 2.
PTB proteins repress AMPA receptor-mediated synaptic transmission in cultured hippocampal neurons. (a) Over-expression of PTBP1 (n = 8) reduces both mEPSC amplitude and frequency compared to untransfected neurons from the same cultures (n = 8). (b) Both mEPSC amplitude and frequency are also reduced in neurons expressing PTBP2 (n = 9) compared to untransfected controls (n = 9). (c) GFP expression alone (n = 9) has no effect on either mEPSC amplitude or frequency compared to untransfected neurons (n = 9). P values correspond to K-S test comparisons of cumulative distributions. (d) mEPSC sample traces from untransfected, PTBP1-, PTBP2-, and GFP alone expressing cells.
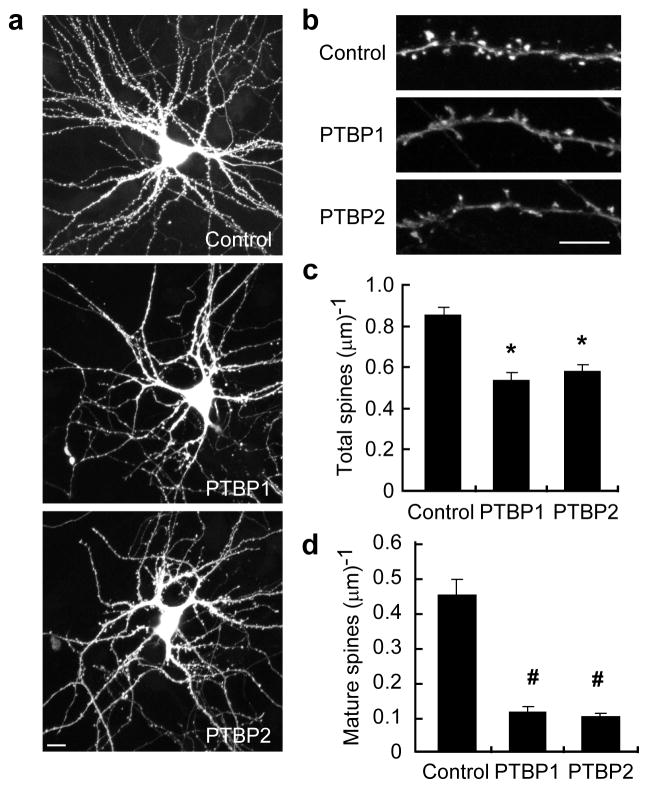
We also tested whether there were morphological changes that accompany PSD-95 reduction after PTBP1/PTBP2 expression in differentiated neurons. PSD-95 was previously shown to increase the number and size of dendritic spines 5. We again co-transfected dissociated hippocampal cultures with PTBP1/PTBP2 and GFP plasmids and then examined the spine morphology of transfected neurons via GFP fluorescence. Compared to control, PTBP1 or PTBP2 transfected neurons had a lower spine density (Fig. 3). These spines also did not appear as mature, with many having small or no spine heads. Indeed, PTBP1 or PTBP2 expression dramatically reduced the density of mushroom-shaped dendritic spines (Fig. 3b,d). These data indicate that the PTB proteins inhibit PSD-95 expression and PSD-95-mediated synapse maturation.
Figure 3.
PTB proteins inhibit dendritic spine maturation. Hippocampal neurons were co-transfected with a GFP plasmid and control, PTBP1, or PTBP2-expressing plasmids. (a, b) Spine morphology of transfected neurons at 21 DIV was visualized by GFP fluorescence. Scale bars, 10 μm (a), 5 μm (b). The density of total spines (c) and mature spines (d) were significantly reduced in PTBP1 or PTBP2 expressing neurons. The bar plot represents mean values±s.e.m. (N=22). * indicates p<10−4. # indicates p<10−5.
Psd-95 exon 18 splicing is regulated by the PTB proteins
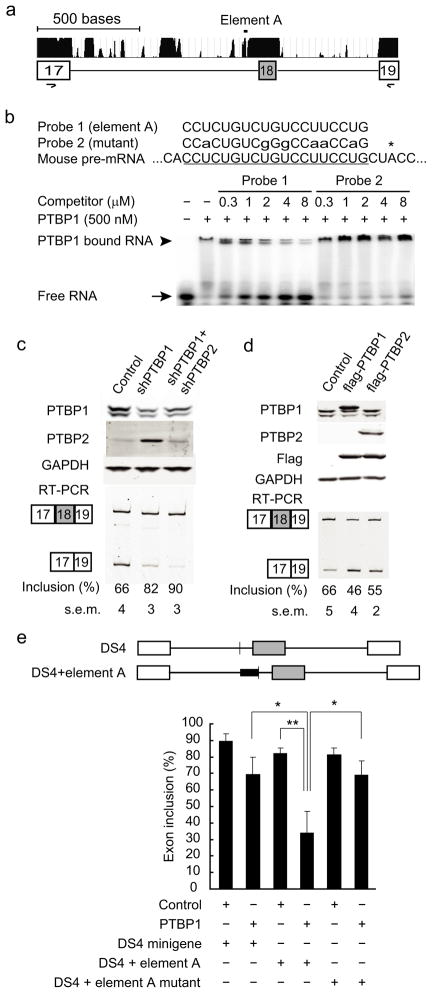
The PTB proteins are known to control many neuronally-regulated exons. Examining the Psd-95 gene locus, we found multiple blocks of intronic sequence flanking exon 18 that are conserved across mammalian species (Fig. 4a), a common feature of highly regulated exons 21–24. PSD-93 and SAP102, which are not repressed by the PTB proteins, do not have a paralogous exon and flanking introns. The 914 nucleotide intron (Intron 17) upstream of Psd-95 Exon 18 contains 20 non-overlapping tracts of 10 or more nucleotides that contain only C and U, with many of these sequences conserved across mammalian species. These polypyrimidine elements are a hallmark of exons regulated by PTBP1 25–27. To confirm the binding of PTBP1 to this region, we performed electrophoretic mobility shift assays (EMSA) with a 19 nucleotide pyrimidine-rich RNA of Intron 17 (Element A) found immediately upstream of the putative Exon 18 branch point (Fig. 4a–b). This is a common location for repressive PTB binding sites 28. Recombinant His-tagged PTBP1 protein efficiently bound to RNA probe 1, containing Element A, quantitatively shifting it into a slowly migrating complex. The sequence specificity of the interaction was confirmed in experiments showing that PTBP1 binding could be competed with cold wildtype Element A RNA (Probe 1) but not with a mutant RNA (Probe 2, Fig. 4b).
Figure 4.
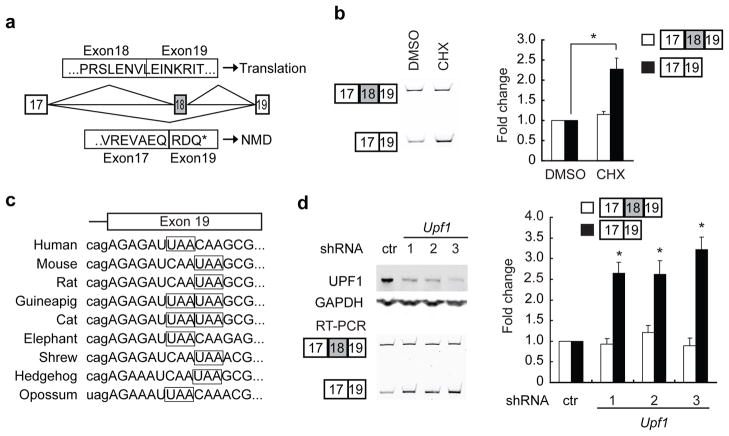
PTB proteins inhibit Psd-95 Exon 18 splicing. (a) Gene structure and the phastCons conservation scores of Psd-95 Exon 17 to Exon 19. (b) EMSA analysis of recombinant PTBP1 protein binding to an intronic RNA sequence (element A) upstream of Exon 18. Sequences of the wild-type (Probe 1) and mutant (Probe 2) RNA probes are aligned to element A (underlined), with the asterisk marking the putative branch. (c) Western blots (top panels) and Exon 18 splicing (bottom panels) in N2a cells after shRNA-mediated knockdown of PTBP1 and PTBP2. PCR primers are in exon 17 and exon 19 (a). The statistical significance of the splicing changes between each condition was determined, with p<0.0001 using One-way ANOVA test (N=6). (d) Western blots (top panels) and Exon 18 splicing (bottom panels) in N2a cells after transient transfection of flag-tagged PTBP1 and PTBP2. P values <0.02 for flag-PTBP1 or flag-PTBP2 transfected cells compared to control (N=3). (e) The structure of DS4 minigene and its variant (upper panel). The white boxes and grey box represent constitutive and alternative exons respectively, and horizontal lines represent the introns. The sequences of wild-type and mutant element A (solid thick bars) are added just upstream of the branch point (thin vertical line). Insertion of the element A significantly enhanced PTB repression of the DS4 alternative exon in N2A cells (lower panel). On the contrary, a mutant element (sequence of Probe 2) had no similar effect. The bar plot represents mean values±s.d. * indicates p<0.02, ** indicates p<0.005.
The binding of PTBP1 to this intron was also observed in a genome-wide mapping of PTBP1-binding sites by crosslinking and immunoprecipitation (CLIP) of HeLa cell RNA 29. Notably, HeLa cells express low levels of Psd-95 mRNA. To confirm the in vivo binding of the PTB proteins in brain, we performed CLIP followed by RT-PCR on E16 mouse brain. Immunoprecipitation with either PTBP1 or PTBP2 antibodies, but not Flag antibody, pulled down Psd-95 pre-mRNA containing Element A (Supplementary Fig. 5). In contrast, neither the related Sap102 transcript nor the Gapdh transcript was co-precipitated with the PTB proteins. These results indicate that Psd-95 Exon 18 is likely a direct target of the PTB proteins.
To demonstrate the regulation of Exon 18 by the PTB proteins, we performed RNAi-mediated knockdown of the protein in mouse neuroblastoma Neuro2-a (N2a) cells. Similar to HeLa cells, N2a and many other cell lines express Psd-95 mRNA although in smaller amounts than in neurons. Reducing PTBP1 protein levels by 75% increased Exon 18 splicing from 66% to 82% (Fig. 4c). A second shRNA targeting a different PTBP1 sequence also stimulated Exon 18, indicating that the change in splicing was due to PTBP1 depletion and not an off-target effect (Supplementary Fig. 6). As seen previously, PTBP1 depletion induced PTBP2 protein expression 10–12. Knocking down both PTB proteins further increased Exon 18 inclusion to 90% (Fig. 4c). We also performed the converse experiment of transiently expressing flag-tagged PTBP1 or PTBP2 in N2a cells. Consistent with PTBP depletion, overexpression of PTBP1 or PTBP2 repressed Exon 18 inclusion from 66% to 46% or to 55%, respectively (Fig. 4d). Ectopic expression of RNAi-resistant PTBP1 or PTBP2 cDNAs also rescued the effects of PTBP1 and PTBP2 knockdown on Exon 18 splicing (Supplementary Fig. 7). Thus, both PTBP1 and PTBP2 repress Exon 18 splicing, with PTBP1 possibly having a stronger effect in N2a cells.
To further confirm that Exon 18 repression was due to the direct binding of the PTB proteins, we inserted the sequence of Element A into a minigene construct (DS4) immediately upstream of the alternative exon branch point 25 and expressed this plasmid in cells. As seen previously, the splicing of DS4 itself was only weakly inhibited by coexpressed PTBP1. Addition of Element A significantly increased repression by PTBP1 (Fig. 4e). In contrast, insertion of a mutant element (Probe 2 in Fig. 4b) did not alter DS4’s response to PTBP1. Note that there are multiple potential PTB binding sites in this region in addition to Element A. These data show that the PTB proteins repress Exon 18 splicing through interaction with its upstream intronic sequences.
The Psd-95 ΔExon 18 isoform is a conserved NMD target
Skipping of Exon 18 shifts the Psd-95 reading frame and results in premature translation termination at a conserved stop codon in Exon 19 (Fig. 5a,c). In the absence of Exon 18, the reading frame terminates 80 nt upstream of the exon 19-exon 20 splice junction, making the transcript a candidate for degradation by the nonsense-mediated mRNA decay pathway 13, 14. Indeed, Exon 18 was previously identified in a large scale screen for NMD-inducing splicing events 16.
Figure 5.
Nonsense-mediated decay of Psd-95 ΔExon 18 isoforms. (a) Codon translation of Psd-95 full length isoform and Psd-95 ΔExon 18 isoform. Exons are numbered in the boxes and connected by horizontal lines representing the introns. Above are the encoded amino acids of the Exon 18-included isoform. Below are those of the Exon-18-skipped isoform. The ΔExon 18 isoform contains a premature termination codon (PTC, *) in Exon 19. (b) Conservation of the PTC (in boxes) among various mammals. Intron sequence is in lowercase and exon in uppercase. (c) RT-PCR detection of Exon 18 splicing in total RNA from N2a cells treated with DMSO or cycloheximide (CHX). CHX treatment increases the Exon 18-skipped isoforms. The relative isoform amount was normalized to the corresponding isoform in DMSO-treated cells and plotted. The plotted bars represent the mean values±s.e.m. (N=3). An asterisk indicates a p-value<0.01 using the corresponding isoform in DMSO-treated cells for comparison. (d) Immunoblot confirming the knockdown of UPF1 in N2a cells after transient transfection of three different shRNAs specifically targeting Upf1. Total RNA samples were extracted and assayed via RT-PCR for the Psd-95 Exon 18+ and Exon 18− transcripts. Isoform levels were normalized to the corresponding isoform in control shRNA-transfected cells. The plotted bars represent the mean values±s.e.m. (N=3). An asterisk indicates a p-value<0.01 in comparison to the exon-skipped isoform in control hairpin-transfected cells.
Due to its loss to NMD, the ΔExon 18 isoform should be produced at higher levels than are observed in steady-state mRNA. Transcripts subject to NMD can be stabilized by blocking translation because initial translation is required to trigger NMD 30. To confirm that the ΔExon 18 transcript was targeted by NMD, we treated N2a cells with the translation elongation inhibitor, cycloheximide. This treatment led to an increase in the ΔExon 18 isoform without affecting the full length Exon 18-plus isoform (Fig. 5b). We also examined ΔExon 18 isoform levels after knocking down the essential NMD factor Upf1. Depletion of UPF1 protein expression by 52 to 70% using several different shRNAs led to a 2.6 to 3.2 fold increase in the ΔExon 18 isoform, while having minimal effects on the full length isoform (Fig. 5d). Thus, the Psd-95 ΔExon 18 isoform is produced at significant levels in cells but is then degraded by NMD in N2a cells.
Psd-95 mRNA is actively transcribed but lost to AS-NMD
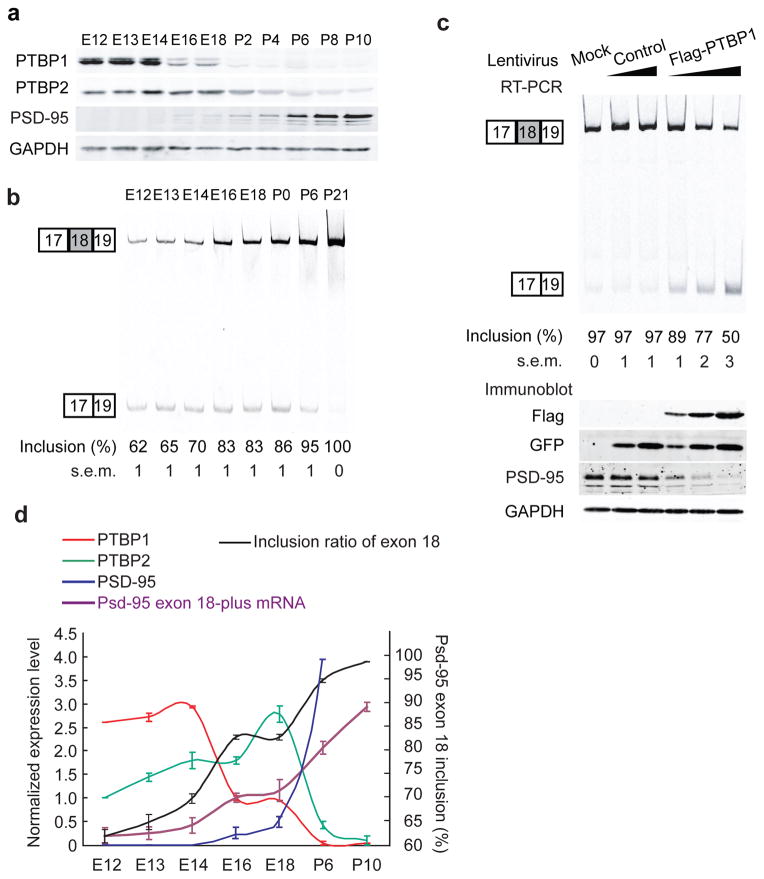
PSD-95 protein is induced during neuronal maturation 31, but its mRNA expression and splicing during early development have not been well examined. To track Psd-95 and PTB expression in the developing brain, we isolated protein and RNA from dissected cerebral cortices at various developmental stages. PTBP1 protein was most abundant from embryonic day 12 to embryonic day 14 (E12–E14) and then decreased significantly from E14 to E16, with a second drop in expression after birth (Fig. 6a,d). PTBP2 protein was present at E12 and increased through E18 as PTBP1 decreased (Fig. 6a,d). Although PTBP2 was expressed in the adult brain, we found that its expression was highest at E18 and then declined after birth. Psd-95 steady-state mRNA was readily detectable at E12 when 38% was the ΔExon 18 isoform, although its real splicing ratio is presumably underestimated due to NMD (Fig. 6b,d). There was a notable increase in the Exon 18-plus transcript between E14 and E16, concurrent with the drop in PTBP1 protein (Fig. 6d). An additional postnatal increase in the Exon 18-plus transcript coincided with the down-regulation of both PTB proteins after birth (Fig. 6d). Thus, Psd-95 is transcribed in embryonic brain, but Exon 18 is repressed when the PTB proteins are present. PSD-95 protein becomes detectable from E16 and then exhibits a steady increase in expression after birth in parallel with the increase in Exon 18-plus mRNA.
Figure 6.
Sequential downregulation of PTBP1 and PTBP2 induce Exon 18 splicing during neural development. (a) Western blot of PTBP1, PTBP2, PSD-95 and GAPDH in cerebral cortices from E12 to P10. Each lane was loaded with equal amounts of total proteins. (b) RT-PCR assay of Psd-95 Exon 18 splicing in cerebral cortices from E12 to P21. Each lane had equal amount inputs of total RNA. N=3. (c) Temporal expression of PTBP1 (red), PTBP2 (green), PSD-95 (blue), Psd-95 Exon-18 plus mRNA transcript (purple) and the inclusion ratio of Exon 18 (black) in mouse cerebral cortex from E12 to P10. PTBP1, PTBP2 and PSD-95 protein expression at E16, E12 and P6 are normalized to 1, 1, and 4 respectively. Psd-95 Exon 18-plus transcript is normalized to its level at E16. The up-regulation of Exon 18 splicing, Exon 18-plus transcripts and PSD-95 proteins are correlated with sequential down-regulation of both PTBP1 and PTBP2 proteins during this period. The plots represent the mean values±s.e.m. (N=3) (d) Primary cortical cultures were infected with increasing doses of control lentivirus or PTBP1 lentivirus. RT-PCR of Psd-95 Exon 18 alternative splicing (top panel) and protein levels of flag-PTBP1, GFP, PSD-95 and GAPDH (bottom panel) are shown. P<0.001 when comparing PTBP1-infected neurons to control neurons (N=3).
To examine whether the PTB-mediated repression of PSD-95 protein expression was due to changes in Psd-95 splicing, we re-examined the PTB lentivirus-infected cells. We infected primary cortical neurons with increasing doses of PTBP1 expressing lentivirus to counteract the loss of the PTB proteins during development. At 12 DIV, control cultures expressing GFP alone exhibited nearly 100% Exon 18 inclusion and expressed significant PSD-95 protein (Fig. 6c). In cultures infected with PTBP1 virus, Exon 18 inclusion was reduced to 89%, 77% and 50% with increasing dose of PTBP1 (Fig. 6c). This was accompanied by parallel decreases in Exon-18 plus transcript and PSD-95 protein, similar to the pattern of expression during embryonic development (Fig. 6a,c). These data indicate that the PTBP1 and PTBP2 proteins prevent accumulation of the productive Psd-95 Exon 18-plus transcript early in development. The loss of the PTB proteins during neural maturation is necessary for Exon 18 splicing and accumulation of the full-length transcript.
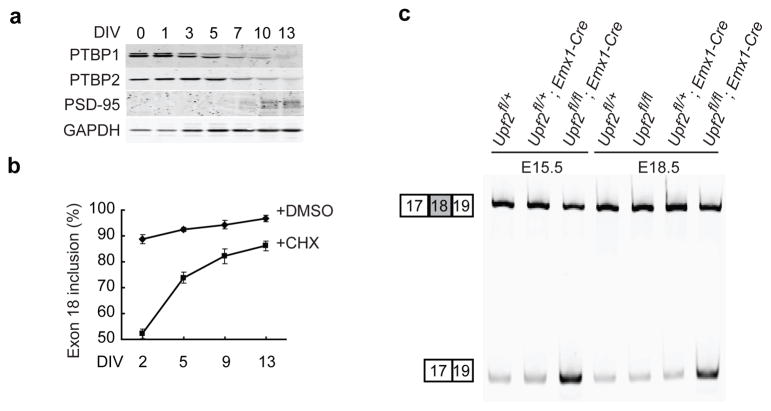
To further confirm the role of the PTB proteins and NMD in controlling Psd-95 expression, we examined NMD activity in primary cortical cultures. PTBP1, PTBP2 and PSD-95 protein expression were assayed as the cultures differentiated and matured, and agreed well with the levels observed in vivo. PTBP1 protein was high at 0 DIV when neurons were dissected from mouse E15 cerebral cortex, but began to decrease by 3 DIV, and declined to negligible levels by 13 DIV (Fig. 7a). As expected, the PTBP2 protein increased from 0 DIV to 5 DIV, as the PTBP1 level dropped, and then declined with further maturation. PSD-95 protein significantly increased when both PTBP1 and PTBP2 proteins decreased. We then measured the splicing of Psd-95 Exon 18 in these cultures with and without cycloheximide treatment to block NMD. Without cycloheximide, the ΔExon 18 isoform decreased from about 12% of the Psd-95 mRNA at 2 DIV to approximately 2% at 13 DIV (Fig. 7b). Cycloheximide treatment markedly stabilized the ΔExon 18 mRNA isoform without affecting the full-length isoform. Under these conditions, Exon 18 was skipped in nearly 50% of the mRNA at 2 DIV (corresponding to E17 in vivo). Thus, the true percentage of Exon 18 skipping in these cultures is underestimated due to the loss of the isoform to NMD. Importantly, the amount of ΔExon 18 mRNA stabilized by cycloheximide decreased as the cells mature. Accounting for NMD, the splicing of Exon 18 strongly increases over time, in parallel with the loss of the PTBP1 and PTBP2 proteins.
Figure 7.
Most of embryonic Psd-95 transcripts are actively degraded via the NMD pathway (a) Western blot of PTBP1, PTBP2, PSD-95 and GAPDH in primary cortical cultures at different days in vitro. Each lane was loaded with proteins from the same number of neuronal cells. GAPDH is normally not expressed as highly at 0 and 1 days after neurons are plated. (b) Splicing of Psd-95 Exon 18 in primary cortical cultures at different DIV after treatment with DMSO or cycloheximide (CHX) for 5 hours. The plots represent the mean values±s.e.m. (N=3) (c) RT-PCR assay of Psd-95 Exon 18 splicing in cerebral cortices of Upf2fl/fl; EMX1-Cre conditional knockout mice and their control littermates at E15.5 and E18.5.
To confirm that the ΔExon 18 isoform lost to NMD early in differentiation was being generated through the action of PTBP2, we transduced a PTBP2 targeted shRNA into differentiating neurons and treated with cycloheximide. As expected, depletion of PTBP2 protein from primary cortical neurons at DIV 4 increased Exon 18 splicing (Supplementary Fig. 8). This increase was also observed and more robust (from 63% to 84%) when NMD was blocked by cycloheximide treatment. These PTBP2 deficient neurons at DIV 4 skipped Exon 18 to the same degree as wild type neurons at DIV 10 when the PTBP2 level is much lower. Thus at early times, the ΔExon 18 isoform lost to NMD is generated by PTBP2. As the PTB proteins are depleted during neural development, Exon 18 splicing increases and less Psd-95 mRNA is targeted for decay.
To examine nonsense-mediated mRNA decay of ΔExon 18 mRNA isoform in vivo, we dissected cortices from the Upf2 knockout mice. Upf2 is an essential factor in most nonsense-mediated decay, and like Upf1, Upf2 null mice die embryonically 18, 32. To generate mice with a Upf2 null mutation in the nervous system, we crossed mice carrying a conditional floxed Upf2 allele (Upf2fl/fl) 18 with the Emx1-Cre mouse. The Emx1-Cre gene expresses Cre recombinase specifically in excitatory neurons of cerebral cortex leading to gene deletion only in these cells 33. Ablation of Upf2 in these neurons significantly stabilized the ΔExon 18 mRNA isoform in both E15.5 and E18.5 cerebral cortices (Fig. 7c). In cortex lacking UPF2, the ΔExon 18 isoform accounted for 60% and 46% of total Psd-95 transcript at E15.5 and E18.5 respectively, whereas in wild-type animals this isoform was only 17% and 13%. Similar to in vitro, younger embryonic neurons exclude Exon 18 to a higher degree. Thus, Psd-95 mRNA is highly transcribed during embryonic development but is actively degraded due to alternative-splicing-induced NMD.
Discussion
PSD-95 expression is enhanced late in neural development to promote spine maturation, stabilize excitatory synapses and enhance AMPA receptor insertion in the synapse 5–8, 34. We find that in immature neurons Psd-95 is under stringent post-transcriptional repression mediated by the two RNA binding proteins PTBP1 and PTBP2. These proteins target the Psd-95 transcript to the nonsense-mediated mRNA decay pathway by altering the splicing of Exon 18. The PTB proteins limit PSD-95 expression in immature neurons where abundant expression of PSD-95 could be deleterious. For example, overly abundant PSD-95 might result in precocious stabilization of early-generated synapses, and limit the fine-tuning of connections during synaptic pruning. We find that the increase in PSD-95 expression during development requires the sequential down-regulation of PTBP1 and PTBP2. Consistent with the idea that PTB inhibition of PSD-95 expression has a key role in synapse maturation, re-expression of PTBP1 or PTBP2 strongly represses AMPA receptor-mediated mEPSCs and reduces mature spine density in hippocampal neurons. Thus, the down-regulation of PTBP1 and PTBP2 during development is necessary for PSD-95 expression and synapse maturation.
Tissue specific expression enforced by AS-NMD
Alternative splicing of Exon 18 may have been acquired as a means of restricting expression of the mammalian Psd-95 gene to neurons. The ancestral homolog of Psd-95 in invertebrates, discs large 1, is expressed in epithelial cells and other non-neuronal cell types of Caenorhabditis elegans and Drosophila 35,36. These invertebrate genes do not have an equivalent exon to mouse Exon 18. All mammals have an exon 18 equivalent with its conserved flanking introns, although there is not a clearly homologous exon in other vertebrates. Since PTBP1 is widely expressed in non-neuronal cells in mammals, Psd-95 is largely repressed outside of neurons.
During development, the abundance of the productive Psd-95 transcript increases as the PTB proteins are depleted (Fig. 6d). Rather than modifying its activity through changes in protein sequence, the alternative splicing of Exon 18 is determining the overall level of Psd-95 mRNA, similar to the output of transcriptional control. This process of alternative splicing coupled to NMD is known to maintain the homeostatic levels of splicing factors in cultured cells, but for Psd-95 this mechanism modulates its expression during development. The NMD pathway itself may be subject to developmental regulation, as miR-128 represses Upf1 mRNA expression during neuronal differentiation leading to increased levels of many NMD-targeted transcripts 37. We find that the loss of UPF1 occurs later in development than the changes in PSD-95 observed here (data not shown). We have not observed an increase in the ΔExon 18 transcript during development that might result from a depletion of NMD activity (Fig. 6b). Presumably, the PTB proteins are depleted and there is little ΔExon 18 transcript to be stabilized by the time UPF1 declines.
It is possible that the remaining PTBP2 present in adult neurons could affect activity-dependent PSD-95 expression. However, we have not yet seen evidence for this. Cortical cultures stimulated with 50 μM glutamate for 5 minutes exhibit reduced PSD-95 protein within an hour 38, however these cells do not show changes in PTBP2 protein or ΔExon 18 transcript (data not shown). Similarly, mice that have undergone kainite-induced seizure have reduced PSD-95 in their hippocampi 39. Under these conditions, we do not observe increased levels of PTBP2. Thus, the role of PTBP1 and PTBP2 may be restricted to the developmental regulation of PSD-95 and not its subsequent control in the adult.
The splicing of a pre-mRNA has been shown to affect its subsequent localization and translation 40–42. In mature neurons where Exon 18 is fully included, some Psd-95 mRNA is localized at synapses where it is translationally controlled 43, 44. Most of the ΔExon 18 transcript is expressed at an early stage of synapse formation and it is unclear whether it can be targeted to synapses. However, it appears unlikely that the ΔExon 18 transcript is translated into stable protein. We have not been able to detect a putative ΔExon 18 protein in the E15.5 or E18.5 neocortices of Upf2 conditional knockout mice where the ΔExon 18 transcript is significantly stabilized. Similarly in mature neurons, when the ΔExon 18 transcript is increased via re-expression of PTBP1, its protein product remains undetectable (Fig. 6c). The ΔExon 18 protein could have a high turnover rate, be inefficiently translated or both.
Other neuronal mRNAs have structures that implicate splicing and NMD in their posttranscriptional control. The Arc mRNA contains introns within its 3′ UTR, making it a “constitutive” NMD target that always carries the eIF4A3 protein needed for inducing decay. This property is thought to limit Arc translation to precise times and locations in mature neurons 45. Earlier in development, Robo3, a Slit receptor, has two spliced isoforms (Robo3.1 and Robo3.2) that control the guidance of spinal commissural axons as they cross the midline 46. Robo3.1 protein mediates Slit attraction, whereas Robo3.2 mediates repulsion from Slit protein expressed at the midline. Robo3.2 arises from an mRNA whose translation terminates within a retained intron 46, 47, making it a potential NMD target. Factors controlling the splicing of Arc and Robo3 have not yet been identified, but the similar combination of posttranscriptional controls seen for PSD-95, Arc and Robo3 imply that this mechanism of neuronal gene regulation may be widespread.
PTBPs define three phases of neuronal differentiation
The changes in PTBP1 and PTBP2 protein expression define three phases of splicing regulation during neuronal differentiation. PTBP1 target exons are repressed in neural progenitor cells. Some exons that are more sensitive to PTBP1 than PTBP2 are induced early in development as PTBP1 levels drop 10, 11. Other exons that are affected by both PTBP1 and PTBP2 proteins, such as Exon 18, are not spliced strongly until later in development when the PTBP2 level is reduced. Exons that are repressed by both proteins have also been identified in RNAi knockdown experiments in tissue culture cells 10, 12. The ordered expression of these related splicing regulators allows neuronal stem cells, early differentiating neurons, and cells undergoing synaptic maturation to each be defined by a different splicing program.
Except for its timing, the control of PSD-95 is similar to the induction of PTBP2 expression during neuronal differentiation. PTBP2 transcripts are expressed in neuronal progenitor cells but PTBP2 exon 10 is repressed in these cells by the PTBP1 protein 10–12. The absence of exon 10 introduces a frameshift in the mRNA targeting it to the NMD pathway. During differentiation, the downregulation of PTBP1 induces the splicing of PTBP2 exon 10 and expression of PTBP2 protein 10. This same process apparently acts on Psd-95, except that both PTBP1 and PTBP2 proteins affect Psd-95 transcripts. This results in continued repression of Psd-95 through an intermediate period when PTBP1 protein is depleted but PTBP2 protein is still high. Induction of PSD-95 then occurs when the PTBP2 protein is reduced after birth (Supplementary Fig. 9).
PTBP2 exon 10 splicing is also promoted by the increase in another splicing regulator NSR100 during neural development and is likely affected by multiple additional factors 48. It will be interesting to determine which other splicing regulators can affect PSD-95 Exon 18 splicing, including NSR100 and factors known to be expressed in mature neurons such as the Nova or Fox family members.
Methods
RNA preparation and quantitative splicing analysis
Total RNA from brain tissues, cultured neurons and cell lines were prepared using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA were synthesized by Superscript III reverse transcriptase with random hexamer. PCR reactions contained 200,000–500,000 counts per minute of 32P-labeled or 0.2 μM FAM-labeled reverse primer. PCR reactions were run for 22 cycles for N2a cells and 20 cycles for neurons or cortices with an annealing temperature of 60°C PCR reactions were mixed 1:3 with 95% formamide and loaded onto 8% polyacrylamide, 7.5 M urea gels and electrophoresed. Subsequently, gels were dried and imaged on a Typhoon Imager (GE Healthcare). After scanning, the specific signals were quantified via ImageQuant TL. Primer sequences are as follows: PCR primer sequences for endogenous Psd-95: 5′-TCTGTGCGAGAGGTAGCAGA-3 ′ and 5 ′-AAGCACTCCGTGAACTCCTG-3′. I17_19ntWt and I17_19ntMut were cloned into the DS4 minigene at the ApaI site just upstream of the branch point of the alternative exon. RT-PCR primers for the minigenes are 5′-GACACCATGCATGGTGCACC-3′ and 5′-AACAGCATCAGGAGTGGACAGATCCC-3′.
Tissue culture and gene expression
Mouse primary cortical cultures and hippocampal cultures were prepared from gestational day 15 and 18 CD-1 mice respectively as described 49. Briefly, the tissues were dissected and the cells were dissociated after a 10 min digestion in Trypsin (Invitrogen). Cells were plated on plastic plates or cover slips coated with 0.1 mg/ml poly-L-lysine and 5 μg/ml Laminin. Four days after plating proliferation of non-neuronal cells was inhibited by exposure to 30 μM 5-fluoro-2-dexoyuridine. The N2a neuroblastoma cell line was grown following guidelines provided by the American Type Culture Collection. Transient gene expression and knockdown in N2a cells and primary neuronal cultures were carried out with Lipofetamine 2000 (Invitrogen) according to the manufacturer’s instructions. 200–250 thousand N2a cells were used for transfection in each well of a 12-well plate. Short hairpin RNAs were designed and cloned into the template vector psiRNA-W[H1.4] and lentiviral vectors. shPTBP1: 5′-GCAGTTGGAGTGACCTTACTTCAAGAGAGTAAGGTCACTTCAGCTGC-3′. shPTBP1 #2: 5′-GGCCTGTATCGGCTTCCACTTCAAGAGAGTGGAAGCTGATGCAGGCC-3′. shPTBP2: 5′-ATGTCAGTACTGAGTAATGTCTTCAAGAGAGACATTACTTAGTGCTG-3′. shPTBP2 #2: 5′-CAACTTAGTTGATATATGTGCTTCAAGAGAGCACGTATGTCAACTAAGTTG -3′. shUPF 1 # 1: 5′-CTGAAGCATCGCTGTGTGAGCTTCAAGAGAGCTCACACAGTGGTGCTTCAG -3′. shUPF1 #2: 5′-CAGCGTCTTGTTCTGGGTAATTCAAGAGATTACCCAGAATAAGATGCTG-3′. shUPF1 #3: 5′-CCTGTGTGGTTTGCTGTAATTTCAAGAGAATTACAGTAAACCACGCAGG-3′. The Lentiviruses were generated essentially as described 50. High titer of PTBP1 virus requires two rounds of concentration via 20,000 rpm centrifugation for 2 hours in a SW28 rotor. The lentivirus was incubated in primary cortical cultures over night and washed away in the second day to induce gene expression or gene knockdown. Cycloheximide was applied at the concentration of 0.5 mg/ml for 5 hours before cells were harvested for NMD analysis.
Quantitative immunoblotting and quantitative imaging analysis
Primary antibodies used were: PTBP1-NT 1:3000, PTBP2-IS2 1:1000, UPF1 1:500–1:1000 (Bethyl Laboratories, A300-036A), PSD-95 1:1000 (Millipore, MAB1598), PSD-93 1:1000 (Neuromab, 75-057), SAP102 1:1000 (Neuromab, 75-058), GAPDH 1:50,000 (Ambion), Flag M2 1: 3000 (Sigma), GFP 1:2000 (Abcam). After incubation with fluorescent-conjugated 2nd antibodies, the blots were scanned using Typhoon Imager. Specific Western blot band intensities were quantified using ImageQuant TL. After immunocytochemistry, Z-stack images were acquired with a 40x or a 100x objective of the CarlZeiss Laser Scanning System LSM 510 META confocal microscope. ImageJ were used to process stack images, select regions of interest and quantify fluorescent signals of somatic PSD-95. PSD-95 immunofluorescence of GFP neurons in one image were averaged and counted as one data point to compare with the GFP+ neuron in the same image. PSD-95 cluster density was quantified after 3D reconstruction using Imaris software (Bitplane). Quantitative analysis of dendrtitic spine was performed with FilamentTracer of Imaris. For either puncta analysis or spine analysis, at least 12 neurons of each group were examined. 1–3 dendrites of each neuron beyond 75 μm of the soma with a minimum section length of 20 μm were measured. Mushroom-type spines were identified using the default setting of the Matlab Spine Classifier in Imaris.
Electrophoretic mobility shift assay
RNA were synthesized by T7 RNA polymerase as previously described 25. 500 nM recombinant PTBP1 proteins were incubated with 50 nM [α-32P] RNA in DG buffer (20 mM HEPES-KOH pH 7.9, 80 mM K. glutamate, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 20% glycerol) and 2.2 mM MgCl2 for 30 min at room temperature. For the competition assay, 0.3, 1, 2, 4 and 8 μM cold RNA were incubated with PTBP1 proteins first for 30 min at room temperature before adding 50 nM [α-32P] Wt RNA and incubating for another 30 min. The reaction mixes were loaded onto 8% native PAGE gels and electrophoresed. Gels were dried and imaged on a Typhoon Imager (GE Healthcare).
Animals
Mice were maintained under the guidelines of University of California Los Angeles DLAM. Upf2fl/fl mice were bred to Emx1-Cre mice to generate Upf2fl/+; EMX1-Cre mice. Upf2fl/+; EMX1-Cre mice were then bred to Upf2fl/fl mice to generate Upf2 conditional knockout mice in the Emx1-Cre background (i.e. Upf2fl/fl; EMX1-Cre).
Electrophysiology
Primary hippocampal cultures were plated on coated glass coverslips at a density of 25,000–38,000 cells/cm2 and transfected at 6 DIV using lipofectamine. Electrophysiological experiments were performed at 17–21 DIV, and recordings from transfected neurons were interleaved with those of non-transfected controls from the same plating. Cultured neurons were maintained at 22 – 23 °C and perfused with an external solution containing (in mM): 134 NaCl, 2.5 KCl, 3 CaCl2, 1 MgCl2, 0.34 NaHPO4, 1 NaHCO3, 20 glucose, 10 HEPES, 1 μM tetrodotoxin, 100 μM pictrotoxin; pH 7.3, 315 mOsm. Neurons were whole-cell voltage-clamped at −60 mV using patch electrodes (7–9 MΩ) filled with an internal solution containing (in mM): 100 cesium methanesulfonate, 4 Mg-ATP, 0.3 GTP, 10 Na-phosphocreatine, 5 MgCl2, 0.6 EGTA, 30 HEPES; pH 7.3, 295 mOsm. Series resistance (typically 10–20 mOhms) was monitored throughout the experiment. mEPSCs were analyzed using template-based event detection and a threshold of 5 pA in Clampfit (Molecular Devices). Kolmogorov–Smirnov tests were used to determine statistical significance between cumulative probability distributions.
Statistical Analysis
Statistical analyses were carried out with Graphpad Instat (San Diego, CA) and Excel. Differences among multiple means were evaluated by one-way ANOVA and the Tukey-Kramer post hoc test. Differences between paired means were assessed with the unpaired, two-tailed Student’s t-test. The null hypothesis was rejected at the 0.05 level.
Supplementary Material
Acknowledgments
We thank Jay Baraban and Erik Anderson for thoughtful suggestions on the manuscript, and Kelsey Martin, Larry Zipursky, William Yang, and Black lab members for helpful discussion. We thank Shalini Sharma for the His-tagged PTBP1 recombinant proteins, Areum Han and Shalini Sharma for help with the EMSA experiments.. We thank Quan Lin for help with Imaris software. This work was supported in part by grants NIH RO1 GM 49662 to D.L.B., NIH R01 MH609197 to T.J.O, and NIH F32 MH84562 to E.E.G. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions: S.Z. and D.L.B. designed the studies. E.E.G and T.J.O designed and performed electrophysiological experiments. G.C. cloned and tested the shRNAs to knockdown PTBP1 and PTBP2. S.Z. performed all experiments. B.T.P. provided the Upf2fl/fl mice. S.Z., T.J.O and D.L.B wrote the paper.
References
- 1.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annual review of biochemistry. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 2.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annual review of biochemistry. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 3.Migaud M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 4.Carlisle HJ, Fink AE, Grant SG, O’Dell TJ. Opposing effects of PSD-93 and PSD-95 on long-term potentiation and spike timing-dependent plasticity. The Journal of physiology. 2008;586:5885–5900. doi: 10.1113/jphysiol.2008.163469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science (New York, N Y. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 6.Elias GM, et al. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Schluter OM, Xu W, Malenka RC. Alternative N-terminal domains of PSD-95 and SAP97 govern activity-dependent regulation of synaptic AMPA receptor function. Neuron. 2006;51:99–111. doi: 10.1016/j.neuron.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich I, Klein M, Rumpel S, Malinow R. PSD-95 is required for activity-driven synapse stabilization. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4176–4181. doi: 10.1073/pnas.0609307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beique JC, et al. Synapse-specific regulation of AMPA receptor function by PSD-95. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & development. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molecular cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annual review of biochemistry. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 14.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 16.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 18.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes & development. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyvonen MT, et al. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. RNA (New York, N Y. 2006;12:1569–1582. doi: 10.1261/rna.39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Molecular and cellular biology. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Zheng S. Identify alternative splicing events based on position-specific evolutionary conservation. PLoS One. 2008;3:e2806. doi: 10.1371/journal.pone.0002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA (New York, N Y. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spellman R, et al. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 27.Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA (New York, N Y. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Zhang W, Reed RB, Liu W, Grabowski PJ. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA (New York, N Y. 2002;8:137–149. doi: 10.1017/s1355838202015029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Molecular cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter MS, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 31.Sans N, et al. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medghalchi SM, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Human molecular genetics. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 33.Iwasato T, et al. Dorsal telencephalon-specific expression of Cre recombinase in PAC transgenic mice. Genesis. 2004;38:130–138. doi: 10.1002/gene.20009. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 35.Firestein BL, Rongo C. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Molecular biology of the cell. 2001;12:3465–3475. doi: 10.1091/mbc.12.11.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 37.Bruno IG, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Molecular cell. 42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Wang Y. Glutamate stimulates glutamate receptor interacting protein 1 degradation by ubiquitin-proteasome system to regulate surface expression of GluR2. Neuroscience. 2007;145:100–109. doi: 10.1016/j.neuroscience.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 39.Wyneken U, et al. Kainate-induced seizures alter protein composition and N-methyl-D-aspartate receptor function of rat forebrain postsynaptic densities. Neuroscience. 2001;102:65–74. doi: 10.1016/s0306-4522(00)00469-3. [DOI] [PubMed] [Google Scholar]
- 40.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Seminars in cell & developmental biology. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nature reviews. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 42.Chen L. A global comparison between nuclear and cytosolic transcriptomes reveals differential compartmentalization of alternative transcript isoforms. Nucleic acids research. 2010;38:1086–1097. doi: 10.1093/nar/gkp1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nature neuroscience. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron. 2008;58:325–332. doi: 10.1016/j.neuron.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Black DL, Zipursky SL. To cross or not to cross: alternatively spliced forms of the Robo3 receptor regulate discrete steps in axonal midline crossing. Neuron. 2008;58:297–298. doi: 10.1016/j.neuron.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calarco JA, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Zheng S, et al. NMDA-induced neuronal survival is mediated through nuclear factor I-A in mice. The Journal of clinical investigation. 2010;120:2446–2456. doi: 10.1172/JCI33144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.