Abstract
Enhanced mGluR5 function is causally associated with the pathophysiology of Fragile X Syndrome (FXS), a leading inherited cause of intellectual disability and autism. Here we provide evidence that altered mGluR5-Homer scaffolds contribute to mGluR5 dysfunction and phenotypes in the FXS mouse model, Fmr1 KO. In Fmr1 KO mice mGluR5 is less associated with long Homer isoforms, but more associated with the short Homer1a. Genetic deletion of Homer1a restores mGluR5- long Homer scaffolds and corrects multiple phenotypes in Fmr1 KO mice including altered mGluR5 signaling, neocortical circuit dysfunction, and behavior. Acute, peptide-mediated disruption of mGluR5-Homer scaffolds in wildtype mice mimics many Fmr1 KO phenotypes. In contrast, Homer1a deletion does not rescue altered mGluR-dependent long-term synaptic depression or translational control of FMRP target mRNAs. Our findings reveal novel functions for mGluR5-Homer interactions in the brain and delineate distinct mechanisms of mGluR5 dysfunction in a mouse model of cognitive dysfunction and autism.
Keywords: Homer1a, group I mGluRs, long-term depression, translation, ERK, PI3 Kinase, mTOR, EF2 Kinase, UP states, neocortex
Introduction
Fragile X Syndrome (FXS) is the most common inherited form of intellectual disability and a leading genetic cause of autism 1, 2. FXS is caused by transcriptional silencing of the FMR1 gene that encodes the Fragile X Mental Retardation protein (FMRP) – an RNA binding protein, that regulates translation of its interacting mRNAs 2. Most patients exhibit multiple neurological deficits, including reduced IQ, seizures, sensory hypersensitivity, social anxiety, hyperactivity and other characteristics of autism 3, 4. In the mouse model of FXS, Fmr1 knockout (KO), group 1 metabotropic receptor (mGluR1 and mGluR5) and protein synthesis- dependent plasticity is enhanced and dysregulated 5. These findings motivated the “mGluR theory of FXS” which posits that altered mGluR-dependent plasticity contributes to the pathophysiology of the disease 6-8. In support of the mGluR theory, many phenotypes in animal models of FXS are reversed by pharmacological or genetic reduction of mGluR5 or downstream signaling pathways 7, 8. Importantly, a recent report indicates that mGluR5 antagonism can be an effective therapeutic strategy in FXS patients 9.
Group 1 mGluR activation stimulates de novo protein synthesis in neurons and evidence suggests that FMRP suppresses translation of specific mRNA targets downstream of mGluR activation 2. In FXS, the loss of an FMRP-mediated “brake” is proposed to lead to excess mGluR5-driven translation of many FMRP target mRNAs which in turn, leads to an excess of mGluR- dependent plasticity 6, 8. Although mGluR5 antagonism rescues many phenotypes associated with FXS, it is unknown if this is due to excess mGluR5-driven translation.
Other evidence suggests there may be altered mGluR5 function that is upstream of translation in Fmr1 KO brains. Although total mGluR5 levels are normal in Fmr1 KO forebrain, there is less mGluR5 in the postsynaptic density (PSD) fraction and an altered balance of mGluR5 association with short and long isoforms of the postsynaptic scaffolding protein Homer 10. The N-terminal EVH1 (Ena-VASP homology) domain of Homer proteins, binds the intracellular C-terminal tail of group 1 mGluRs (mGluR5 and mGluR1a) and affects their trafficking, localization and function 11 Long, constitutively expressed forms of Homer (Homer1b, 1c, 2 and 3) multimerize through their C-terminal coiled-coil domain and localize mGluRs to the PSD through interactions with SHANK, as well as scaffold mGluRs to signaling pathways through Homer interactions with the PI3 Kinase enhancer (PIKE), Elongation Factor 2 kinase (EF2K) and the IP3 receptor 11, 12. Homer1a (H1a), a short, activity-inducible, form of Homer lacks the coiled-coiled domain and cannot multimerize with other Homers. Consequently, H1a disrupts mGluR5-long Homer complexes, alters mGluR signaling and causes constitutive, agonist-independent activity of mGluR1/5 13.
In Fmr1 KO mice mGluR5 is less associated with the long Homer isoforms and more associated with H1a 10. We hypothesized that the altered balance in mGluR5 interactions with Homer isoforms contributed to the mGluR5 dysfunction and pathophysiology of FXS. To test this hypothesis, we crossed Fmr1 KO mice with H1a KO mice and determined if H1a deletion restored mGluR5 function and Homer interactions as well as neurophysiological and behavioral phenotypes of Fmr1 KO mice. In addition, we determined if acute peptide-mediated disruption of mGluR5-Homer scaffolds in wildtype mice mimicked phenotypes of Fmr1 KO mice. Our results indicate that altered Homer isoform interactions are responsible for much, but not all, of the mGluR5 dysfunction and pathophysiology of FXS. Specifically, H1a deletion did not rescue the protein synthesis independence of mGluR-LTD or altered translational control of FMRP target mRNAs. The latter results support an essential role for FMRP in translational control of its mRNAs and mGluR-LTD. Our results provide new evidence for altered mGluR5-Homer scaffolds in Fmr1 KO phenotypes and implicate different mechanisms of mGluR5 dysfunction in distinct phenotypes. Modulation and restoration of mGluR5-Homer interactions may represent a novel therapeutic strategy for Fragile X and related cognitive and autistic disorders.
Results
Disruption of mGluR5-Homer regulates signaling to translation
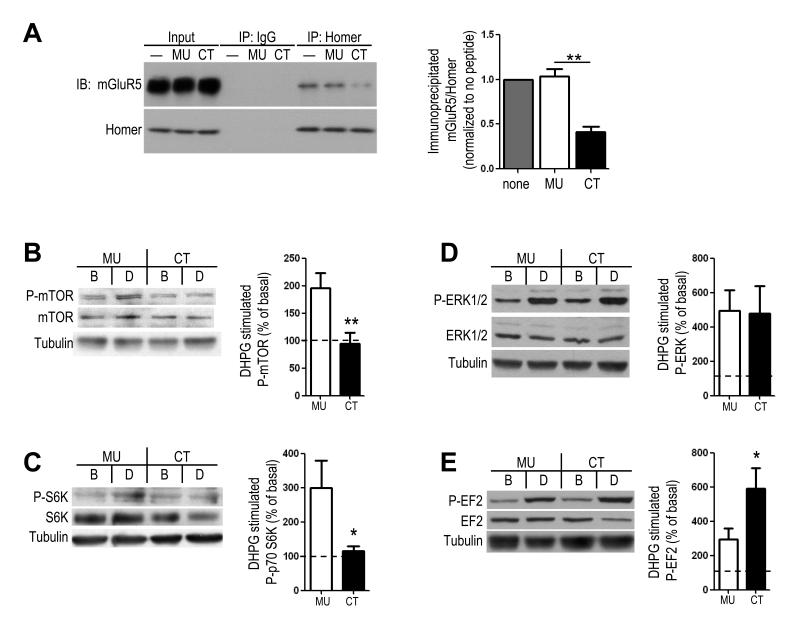
To investigate if the altered mGluR5-Homer scaffolds contribute to the mGluR5 dysfunction in Fmr1 KO mice we determined if disruption of mGluR5-Homer scaffolds with a peptide in wildtype (WT) mice mimicked mGluR5 signaling alterations in the Fmr1 KO mice. The rationale for this approach is based on data that H1a-bound mGluR5, which is increased in Fmr1 KO, is functionally equivalent to mGluR5 that cannot interact with any Homer isoform 13-15. To disrupt mGluR5-Homer, we incubated acute hippocampal slices from WT mice in a cell permeable (Tat-fused) peptide containing the Proline-rich motif (PPxxF) of the mGluR5 C-terminal tail that binds the EVH1 domain of Homer, mGluR5CT (CT; 5 μM) 14, 16, 17. mGluR5CT reduced mGluR5-Homer interactions to 41±6% of that observed in slices with no peptide treatment (n = 3 mice; p = 0.003) as determined by co-immunoprecipitation of mGluR5 and Homer (Fig. 1A). Importantly, CT peptide treatment roughly mimics the 50% decrease in mGluR5-long Homer interaction observed in Fmr1 KO hippocampal lysates (Fig. 2A). As a control, slices were incubated in a peptide with a mutated Homer binding motif, mGluR5MU (MU) 14, 16. mGluR5MU peptide had no effect on mGluR-Homer in comparison in slices with no peptide treatment (103±8% of untreated; n = 3; Fig. 1A).
Figure 1. Peptide- mediated disruption of mGluR5-Homer scaffolds in wildtype mice hippocampus bidirectionally regulates group 1 mGluR signaling to translation initiation and elongation.
A. Pretreatment of acute hippocampal slices from wildtype (WT) mice with a tat-fused peptide containing the Homer binding motif of the mGluR5 (tat-mGluR5CT; CT; YGRKKRRQRRR- ALTPPSPFR; 5 hours; 5 μM) reduces mGluR5-Homer interactions as determined using co-immunoprecipitation (co-IP) with a Pan-Homer antibody and immunoblotting for mGluR5. A control peptide containing point mutations in the Homer binding motif (tat-mGluR5-MU; YGRKKRRQRRR-ALTPLSPRR; 5 hours; 5 μM) has no effect on mGluR5-Homer co-IP in comparison to untreated slices (”-“). One-half of the input for the co-IP was run on separate blots. B-E. Disruption of mGluR5-homer interaction alters signaling to translation. Western blots of (B) phosphorylation of mTOR (S2448), (C) phosphorylation of S6K (T389), (D) phosphorylation of ERK T202/Y204 and (E) phosphorylation of EF2 (T56), in the basal (B) condition and DHPG (D) treated hippocampal slices (100 μM; 5 min) from WT mice. Slices were pretreated with the CT or MU peptide as indicated. Left: Representative Western blots of each phosphorylated and total protein as well as β-tubulin in the conditions as indicated. Right: Group data (averages ± SEM) of each protein monitored (phosphorylated/total) (normalized to basal, or untreated, slices from the same mouse). n = 4-15 slices/per condition from 3-8 mice. *p< 0.05; **p < 0.01. Full length western blots for this figure are shown in Supplementary Fig. 7.
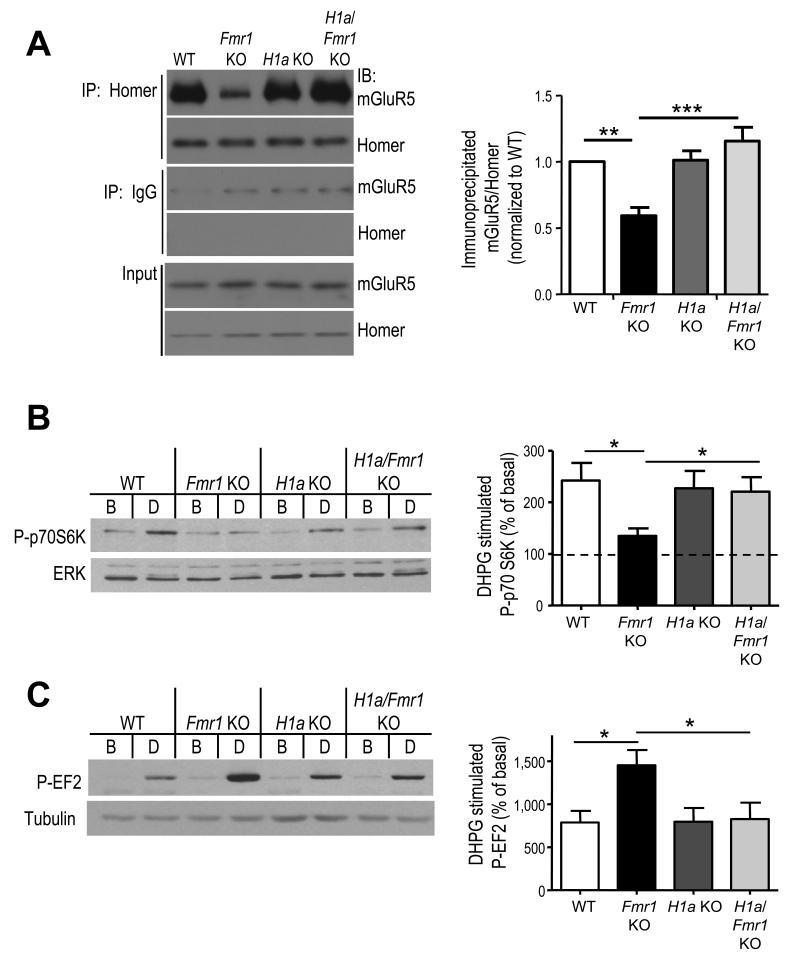
Figure 2. mGluR5-Homer scaffolds and Group 1 mGluR signaling are altered in Fmr1 KO mice and rescued by genetic deletion of Homer1a.
A. Decreased mGluR5-long Homer interactions in Fmr1 KO hippocampus are restored to WT levels in the H1a/Fmr1 double knockout littermates. The level of mGluR5 that coimmunoprecipitated with Homer was quantified and normalized to levels of immunoprecipitated Homer. Left: Representative blots from 1 set of littermates reveals a decrease in Homer/mGluR5 in Fmr1 KO that is reversed in the H1a/Fmr1 KO. One-fifth of the input for the immunoprecipitation was run on separate blots and demonstrates normal levels of mGluR5 and Homer across all genotypes. Right: Group data from independent co-IPs in 4 litters reveals a reliable decrease in mGluR5-Homer co-IP in Fmr1 KO mice and rescue by H1a deletion. B. Treatment of acute hippocampal slices from littermates of each of the four genotypes with DHPG (100 μm; 5 min) reveals a deficit in phosphorylation of p70S6K in Fmr1 KO mice that is restored to wildtype levels in H1a/Fmr1 KO. Left: Representative Western blots of P-S6K and total ERK (loading control) in the basal (B) condition and DHPG (D) treated slices. Right: Group data of P-S6K/ERK (normalized to basal levels in untreated slices from the same mouse). n = 14-21 slices/per condition from 14-21 mice. C. DHPG induced P-EF2 is enhanced in Fmr1 KO slices, an effect that is reversed in the H1a/Fmr1 KO. Left: Representative western blot of P-EF2 and β-tubulin in the basal (B), or untreated, condition and DHPG (D) treated slices from each genotype. Right: Group data (averages ± SEM) for P-EF2/tubulin (normalized to basal levels). n = 6-7 slices and mice per genotype). *p< 0.05, **p< 0.01; ***p<0.001. Full length western blots for this figure are shown in Supplementary Fig. 8.
Fmr1 KO mice display a deficit in mGluR1/5 stimulation of protein synthesis which may be a result of altered mGluR5- Homer interactions. Previously, we reported that CT-peptide mediated disruption of mGluR5-Homer interactions in rat hippocampal slices inhibited group 1 mGluR activation of the PI3K-mTOR pathway and translation initiation, but not ERK 17. Here we observed similar effects in hippocampal slices from WT mice. mGluR5CT peptide incubation blocked activation of PI3K-mTOR in response to the group 1 mGluR agonist, (RS)-3,5-Dihydroxyphenylglycine (DHPG; 100 μM; 5 min) as measured by phosphorylation of mTOR (S2448) and S6K (Thr389) (P-mTOR: MU: 196±27% of basal, n=8; CT: 94±20% of basal, n=8; p< 0.01; P-S6K; MU: 300±80% of basal, n=6; CT: 114±13% of basal, n=6; p< 0.05; Fig. 1B,C) and had no effect on ERK activation (Thr202/Tyr204; P-ERK; MU: 495±117% of basal, n=4; CT: 477±162% of basal, n=4; n.s.; Fig. 1D) 17. The CT peptide did not affect basal levels of P-mTOR, P-S6K or P-ERK (Supplementary Table 1) 18
Homer and mGluR5 each directly interact with another translational regulatory factor, Elongation Factor 2 kinase (EF2K) 12. Although phosphorylation of elongation factor 2 (EF2) by EF2K inhibits translation elongation globally, EF2K is required for translational activation of mRNAs such as such as Arc (activity-regulated cytoskeleton associated protein), and αCaMKII12. A moderate inhibition of elongation globally by EF2K is thought to release translation factors that are then available for translational activation of poorly initiated transcripts. Surprisingly, mGluR5CT-treated slices displayed a robust increase in phosphorylation of EF2 (T56) in response to DHPG (590±118% of basal; n = 15 slices, Fig. 1E) in comparison to mGluR5MU treated slices (294±63% of basal; n = 15; p< 0.05;.Supplementary Fig. 1). The CT peptide did not affect basal levels of P-EF2 (Supplementary Table 1). Taken together, our data suggest that Homer interactions facilitate mGluR activation of the PI3K-mTOR pathway to translation initiation, but dampen mGluR-induced phosphorylation of P-EF2 and thus restrain inhibition of global elongation rates. Consequently, disruption of mGluR5-Homer would be expected to block mRNA translation by blocking activation of the PI3K-mTOR pathway and translation initiation and enhancing inhibition of elongation to a level that may block elongation of all transcripts, including Arc. Consistent with this model, disrupting mGluR5-Homer interactions in hippocampal slices with CT peptide blocks DHPG-induced synthesis of Arc (MU: 122+/-5% of untreated, n=7; CT: 95+/-7% of untreated, n=7; Supplementary Fig. 1) and Elongation factor 1α (EF1α)17.
Deletion of Homer1a rescues mGluR signaling in Fmr1 KO
mGluR5-long Homer interactions are reduced in Fmr1 KO mice and we hypothesize that this contributes to altered mGluR5 function. If so, then mGluR signaling to translation in Fmr1 KO slices may mimic what is observed with CT peptide treatment of WT slices (as in Fig. 1). In support of our hypothesis, DHPG-induced activation of PI3K, mTOR and p70 ribosomal S6 kinase (S6K) is deficient in Fmr1 KO hippocampal slices and ERK activation is unaffected (Fig. 2B; 18-20). Furthermore, DHPG- induced phosphorylation of EF2 is enhanced in Fmr1 KO slices relative to WT littermates (WT: 787±135% of untreated, n=7 mice; Fmr1 KO: 1452±176% of untreated, n=6 mice; Fig. 2C). Remarkably, these alterations in mGluR signaling parallel what is observed with CT peptide treatment of WT slices (Fig. 1). Basal levels of Phospho- or total EF2 or S6K are unchanged in Fmr1 KO slices (Supplementary Table 2) 18. These results indicate that mGluR5 function is not generally enhanced or decreased in Fmr1 KO mice, but is changed in a complex way that is mimicked in WT mice by disruption of mGluR5-Homer interactions.
In Fmr1 KO mice, mGluR5 is more associated with H1a 10 (Supplementary Fig. 2a). Because long Homers compete with H1a for interactions with their effectors 14, 15, we hypothesized that genetic deletion of H1a in Fmr1 KO mice may restore normal mGluR5-long Homer interactions and mGluR5 function. To test this idea, Fmr1 KO mice were bred with mice with a genetic deletion of H1a to create H1a/Fmr1 double knockouts 21. H1a KO have normal levels of the long Homer isoforms 1, 2 and 3 21. In agreement with Giuffrida et al., (2005), co-immunoprecipitation of long Homer isoforms from Fmr1 KO forebrain revealed a reduced association of mGluR5 in comparison to WT littermates (Fig. 2A), whereas, co-IP of H1a revealed an increased association with mGluR5 (Supplementary Fig. 2). Total levels of mGluR5, long Homers and H1a (protein and mRNA) in Fmr1 KO hippocampi were not different from WT (Fig. 2A; Supplementary Fig. 2). Genetic deletion of H1a restored normal mGluR5-Homer interactions in Fmr1 KO mice (n = 4 mice per genotype; Fig. 2A) but did not affect levels of mGluR5, long Homers or their interactions on a WT background (Fig. 2A; Supplementary Fig. 2) 21. To determine if H1a deletion restored normal mGluR5 signaling in Fmr1 KO mice, we examined mGluR signaling to the mTOR and EF2K translational regulatory pathways. The deficit in mGluR5 signaling to mTOR in Fmr1 KO, as measured by S6K (T389) phosphorylation, (WT: 242±34% of basal, n=21; Fmr1 KO: 135±15%, n=19; Fig. 2B) was restored in H1a/Fmr1 KO (220±28% of untreated, n=18). Similarly, enhanced mGluR activation of EF2K was rescued to wildtype levels by H1a deletion (H1a/Fmr1 KO; 826±192% of basal, n=7; Fig. 2C). H1a KOs exhibited normal DHPG-induced phosphorylation of EF2 (796±159% of untreated, n=6) and S6K (227±34% of treated, n=14). There was no effect of Fmr1 or H1a genotype on basal levels of phosphorylated or total EF2 or S6K (Supplementary Table 2). Furthermore, DHPG treatment did not alter levels of total EF2 or S6K in any genotype (Supplementary Table 2) 17, 19
H1a deletion rescues enhanced translation rates in Fmr1 KO
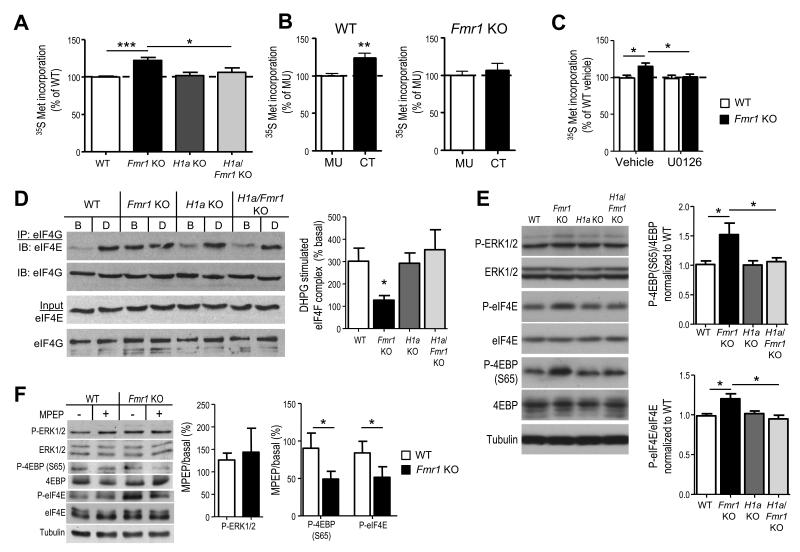
Although DHPG-induced translation is absent in Fmr1 KO mice, basal translation rates in brain are elevated 2, 8, 20, 22. Enhanced protein synthesis in hippocampal slices is reversed by pharmacological blockade of mGluR5 or ERK activation, but not by an inhibitor of PI3K or mTOR 20 (Fig. 3C; Supplementary Fig. 4). Another consequence of mGluR5-H1a interactions is constitutive, or agonist-independent mGluR5 activity 13 which may drive translation rates through ERK activation, a pathway that remains intact in Fmr1 KO mice and with Homer disruption (Fig. 1D) 18, 20. In support of this hypothesis, genetic deletion of H1a rescues enhanced translation rates as measured by 35S Met incorporation in hippocampal slice proteins (Fmr1 KO 122±4% of WT, n=14 slices from 7 mice; H1a KO 102±4% of WT, n=8 slices from 4 mice; double H1a/Fmr1 KO 106±6% of WT, n=7 slices from 4 mice; Fig. 3A). Furthermore, CT peptide-mediated disruption of mGluR5-Homer in WT slices is sufficient to mimic enhanced protein synthesis rates to that observed in Fmr1 KO slices (CT =123 ± 6% of MU treated, n=16 slices per peptide from 10 mice; p = 0.002; Fig. 3B). In contrast, mGluR5CT has no effect on protein synthesis rates in Fmr1 KO slices (CT =106 ± 8% of MU treated, n=14 (CT) or 15 (MU) slices from 8 mice; p = 0.5; Fig. 3B).
Figure 3. Altered mGluR5-Homer scaffolds in Fmr1 KO mice mediate enhanced basal translation rates and initiation complex formation.
A. Acute hippocampal slices from Fmr1 KO mice display elevated protein synthesis rate in comparison to WT littermates as measured by incorporation of 35S Met/Cys into total protein (n = 14 slices from 7 mice/genotype). Elevated protein synthesis rates in Fmr1 KO slices are reversed by H1a deletion (H1a/Fmr1 KO; n = 7 slices/ 4 mice), whereas H1a KO alone (n = 8 slices/4 mice) has no effect. B. Pretreatment of WT hippocampal slices with mGluR5CT (CT; 5 μM; 5 hours; n = 16 slices/4 mice) enhanced 35S Met/Cys incorporation in comparison to control (mGluR5MU; MU; n = 16 slices/ 4 mice) peptide. In contrast, pretreatment of Fmr1 KO hippocampal slices (n = 15 slices/ 4 mice) with CT peptide had no effect. C. Preincubation of WT or Fmr1 KO slices with an inhibitor of the upstream kinase of ERK (MAP/ERK kinase; MEK) U0126 (20 μM; 30 min) prior to 35S Met/Cys incorporation (n = 12 slices/ 6 mice per condition) equalizes protein synthesis rates. D1. Representative Western blots of eIF4E that co-immunoprecipitated with eIF4G from hippocampal slices reveal that DHPG (D) induces an increase in eIF4E association with eIF4G, forming the translation initiation (eIF4F) complex, in WT but not in Fmr1 KO littermates despite an elevated level of eIF4F complex under basal (B) conditions. H1adeletion alone has no effect on eIF4E/4G levels under basal or DHPG stimulated conditions, whereas, H1a deletion on the Fmr1 KO background reverses the enhanced eIF4F complex levels and restores DHPG induced eIF4F complex formation. D2. Quantified group data of the levels of eIF4E that coimmunoprecipitated with eIF4G (eIF4E/4G) in DHPG treated samples (normalized to the levels of eIF4E/4G in basal, or untreated slices). n = 3 mice per genotype. E1. Representative Western blots of phospho- and total ERK and translation initiation factors that are regulated by ERK, (phospho- and total eIF4E and 4EBP) from cortical homogenates in each of four genotypes. E2. Quantified group data reveals elevated P-4EBP and P-eIF4E in Fmr1 KO brains as compared to WT, which is rescued by H1a deletion. P-ERK levels were not different across any genotype (n = 4-6 mice per genotype). F1. Representative Western blots from acute hippocampal slices prepared from WT and Fmr1 KO mice treated with MPEP (10 μM) or vehicle (H2O). F2. Quantified group data of the level of phosphoproteins in MPEP treated slices expressed as a percent of basal (untreated) slices reveals a genotypic difference in P-4EBP (S65) and P-eIF4E, but not P-ERK (n = 2 slices/mouse; 4-8 mice/condition). *p< 0.05; **p< 0.01; ***p<0.001. Full length western blots for this figure are shown in Supplementary Fig. 9.
Translation initiation is the rate limiting step in translation 23. To determine if enhanced translation rates in Fmr1 KO stem from increased initiation, we measured eIF4F translation initiation complexes in hippocampal slices prepared from WT, Fmr1 KO, H1a KO and H1a/Fmr1 KO mice. The eIF4F complex is composed of the 5′ cap binding protein eIF4E, a scaffolding protein eIF4G, and the RNA helicase eIF4A 23. eIF4F complex assembly can be measured by co-immunoprecipitation of eIF4G and eIF4E and is stimulated by DHPG in WT animals 24. Therefore, we also measured eIF4F complex in DHPG-stimulated slices from each genotype. Consistent with previous reports, eIF4F complex levels are enhanced basally in Fmr1 KO slices and no longer stimulated by DHPG (Fig. 3D) 19. Like 35S Met incorporation, eIF4F complex levels in Fmr1 KO slices are restored to WT levels by H1a deletion (Fig. 3D; n = 3 mice per condition). Genetic deletion of H1a also rescues the deficit in DHPG-stimulated eIF4F complex assembly in the Fmr1 KO (Fig. 3D). These results suggest that elevated protein synthesis rates in Fmr1 KO mice are due to enhanced translation initiation that is driven by H1a-bound mGluR5. The deficit in mGluR-stimulated translation initiation in Fmr1 KO mice may be because eIF4F complex levels are saturated basally. Furthermore, mGluR-activation of mTOR is rescued in Fmr1 KO mice by H1a deletion (see Fig.2) which may also contribute to restoration of DHPG-induced eIF4F complex assembly 24.
To determine how increased H1a-mGluR5 interactions lead to enhanced signaling to translation downstream of ERK, we examined phosphorylation of initiation factors known to be regulated by ERK in WT and Fmr1 KO cortical homogenates and the effects of H1a deletion. ERK phosphorylates and activates MAPK-interacting kinase (Mnk) which in turn phosphorylates the cap-binding protein eIF4E (S209) 23. ERK also phosphorylates eIF4E binding protein (4EBP) at S65, a distinct site from mTOR regulated sites (T36/45) 25. ERK- dependent phosphorylation of eIF4E and 4EBP (S65) is associated with increased translation rates in neurons and other cell types 24, 26. Consistent with a role for ERK in phosphorylation of these initiation factors in hippocampal slices, Phospho-(S209)-eIF4E and (S65)-4EBP, was strongly reduced or abolished by U0126 treatment (Supplementary Fig. 3). P-4EBP and P-eIF4E levels were enhanced in cortical homogenates from Fmr1 KO mice, an effect that was rescued by H1a deletion (Fig. 3E). As reported in hippocampal slices (Fig. 1D) 17, 20, P-ERK levels were unchanged in Fmr1 KO lysates (Fig. 3E). To determine if mGluR5 activity abnormally drives phosphorylation of ERK, eIF4E and 4EBP (S65) in Fmr1 KO mice, hippocampal slices from both WT and Fmr1 KO mice were treated with the mGluR5 inverse agonist, MPEP (10 μM). MPEP treatment did not affect P-4EBP or P-eIF4E in WT slices. However, in Fmr1 KO slices, MPEP reduced P-4EBP and P-eIF4E by ~50% (Fig. 3F). Surprisingly, MPEP had no effect on P-ERK levels in either WT or Fmr1 KO slices. These results support our hypothesis that H1a-mediated mGluR5 activity drives translation initiation through ERK phosphorylation of initiation factors. Because P-ERK levels are not affected in Fmr1 KO or with MPEP, this suggests that mGluR5 may regulate accessibility or localization of eIF4E or 4EBP with ERK, as opposed to ERK activity per se.
Altered LTD is not rescued by deletion of Homer1a
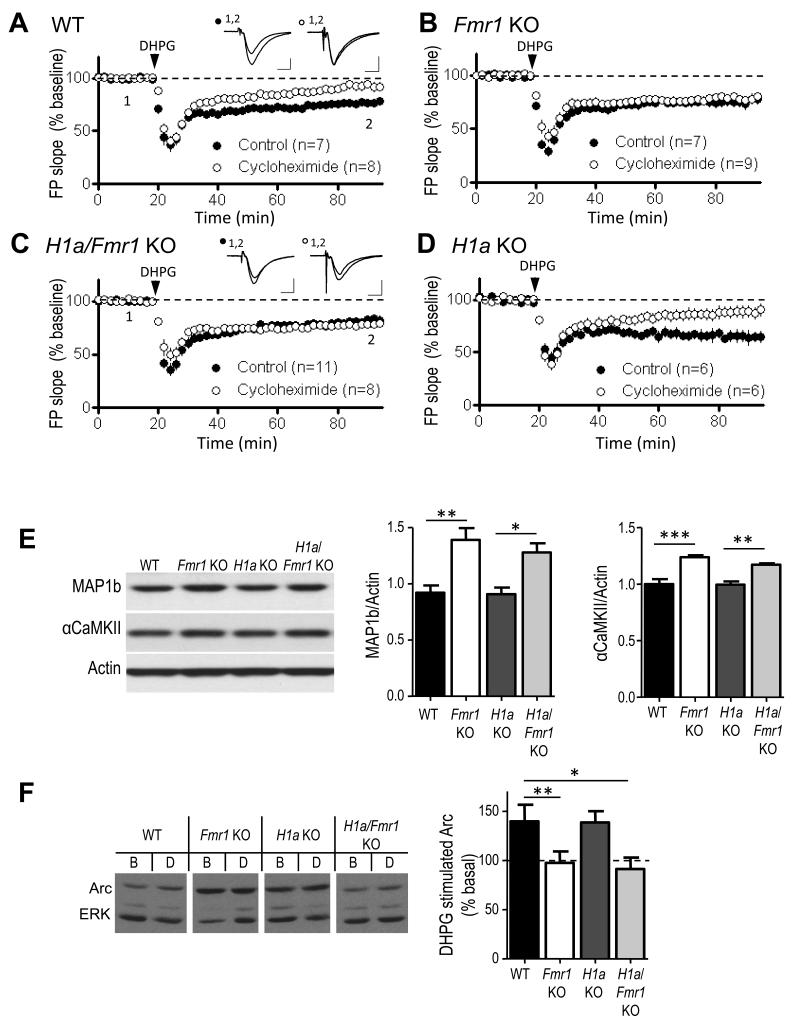
In wildtype animals, mGluR-dependent long-term synaptic depression (mGluR-LTD) within the CA1 region of the hippocampus requires dendritic protein synthesis of FMRP-interacting mRNAs such as Arc and MAP1b 5, 12, 27. Although mGluR activation induces robust LTD in Fmr1 KO mice, mGluR-induced synthesis of Arc and MAP1b is deficient and LTD is independent of new protein synthesis 5, 12. From this work it has been suggested that loss of FMRP-mediated translational suppression leads to enhanced steady state levels of “LTD proteins” which allow mGluR-LTD to persist without new protein synthesis 5. Consistent with this hypothesis, elevated levels of MAP1b and Arc have been reported in Fmr1 KO neurons 5, 28. Alternatively, H1a bound, and constitutively active mGluR5 which drives total protein synthesis rates (Fig. 3) 20 could elevate “LTD protein” levels and lead to protein synthesis-independent LTD. To distinguish between these possibilities, we determined if genetic deletion of H1a reversed the protein synthesis independence of mGluR-LTD and enhanced levels of specific FMRP target mRNAs. To test the protein synthesis dependence of mGluR-LTD, slices were preincubated in the translation inhibitor cycloheximide (60 μM). Although mGluR-LTD was reliably induced with DHPG in the double H1a/Fmr1 KO (81±3% of baseline 60-70 min after DHPG application, n=11 slices), LTD was not sensitive to cycloheximide (78±1% of baseline, n=9), similar to the Fmr1 KO (control: 74±4% of baseline, n=7; cycloheximide: 79±3% of baseline, n=9; Fig. 4B,C) 12. LTD was inhibited by cycloheximide treatment in both WT (control: 76±1% of baseline, n=7; cycloheximide 92±4% of baseline, n=8; p = 0.002) and H1a KO mice (control: 65±5% of baseline, n=6; cycloheximide: 88±5% of baseline, n=6; p = 0.01; Fig. 4A, D). These results suggest that the altered mGluR5-Homer scaffolds in Fmr1 KO mice do not mediate the protein synthesis independence of mGluR-LTD.
Figure 4. Genetic deletion of Homer1a does not reverse the protein synthesis-independence of mGluR-induced LTD or altered protein levels of FMRP target mRNAs.
A. Brief DHPG (100 μM; 5 min) induces long-term depression (LTD) of synaptic transmission in WT hippocampal slices that is reduced by the protein synthesis inhibitor cycloheximide (60μM; p<0.01). Plotted are group averages of field excitatory postsynaptic potential (fEPSPs) slope (average±SEM) normalized to pre-DHPG baseline as a function of time. Inset, average of 10 fEPSPs taken during the baseline period (1) and 55-60 min after DHPG treatment (2). Calibration = 0.5 mV, 5 ms. B, C. In Fmr1 KO and H1a/Fmr1 KO mice, DHPG-induced LTD is unaffected by cycloheximide. D. In H1a KO DHPG-induced LTD is normal and blocked by cycloheximide (p < 0.05). n = number of slices. E. Map1b and αCamKII levels are elevated in Fmr1 KO mice, and are unaffected by H1a deletion. Left: Representative western blots of Map1b, αCaMKII and actin (loading control) from hippocampal homogenates of each genotype. Right: Quantified group data of MAP1b/actin and αCaMKII/actin ratios in each genotype. (MAP1b/actin ratio; WT = 0.92±0.06; Fmr1 KO = 1.4±0.1; H1a KO = 0.8±0.06; H1a/Fmr1 KO = 1.28±0.08; αCaMKII/actin ratio; WT = 1.0±0.04; Fmr1 KO = 1.23±0.02; H1a KO = 1.0±0.03; H1a/Fmr1 KO = 1.17±0.01; n = 3-4 mice per genotype). F. DHPG-induced Arc synthesis is deficient in Fmr1 KO mice and is not rescued by H1a deletion. Left: Representative western blots of basal (B) Arc levels and DHPG (D) induced Arc from hippocampal slices of each genotype. Full length western blots for this figure are shown in Supplementary Fig.10. Group averages reveal a deficit in DHPG-induced Arc synthesis slices taken from Fmr1 KO (n = 10 mice) and H1a/Fmr1 KO (n = 11) mice. DHPG induces Arc synthesis in both WT (n = 12) and H1a KO (n = 14) littermates. *p< 0.05; **p< 0.01; ***p<0.001.
To determine if H1a deletion in Fmr1 KO mice rescued elevated steady state levels of LTD-promoting proteins or other FMRP target mRNAs we performed western blots of MAP1b, Arc and αCamKII in hippocampal homogenates of WT, Fmr1 KO, H1a KO and H1a/Fmr1 KO mice (Fig. 4E). MAP1b and αCamKII, were elevated in both Fmr1 KO mice and H1a/Fmr1 KO mice in comparison to WT and H1a KO mice (Fig. 4E) indicating that H1a deletion does not restore normal levels of Map1b and αCaMKII in Fmr1 KO mice. Although we did not detect elevated steady state protein levels of Arc in hippocampal homogenates of Fmr1 KO mice, we observed a deficit in DHPG-induced synthesis of Arc in hippocampal slices from Fmr1 KO mice in comparison to WT (WT: 140±17% of basal or untreated, n=12; Fmr1 KO: 98±12% of untreated, n=12; Fig. 4F). However, H1a deletion did not restore mGluR-induced synthesis of Arc in Fmr1 KO mice 12 (H1a/Fmr1 KO: 91±12% of untreated, n=11). DHPG induced synthesis of Arc was normal in H1a KO mice (139±11% of untreated, n=11). These results reveal that altered mGluR5-Homer scaffolds in the Fmr1 KO mice do not mediate abnormal mGluR-LTD or altered translational control of specific FMRP target mRNAs. Instead, these results support an essential role for FMRP interactions with its target mRNAs in mGluR-LTD and translational control of these mRNAs (Supplementary Fig. 6B) 2, 29.
mGluR5-Homer and hyperexcitable neocortical circuits
FXS patients and Fmr1 KO mice exhibit sensory hypersensitivity, epilepsy and/or audiogenic seizures suggestive of an underlying sensory circuit hyperexcitability 3, 8. We recently discovered synaptic and circuit alterations indicative of hyperexcitability in the somatosensory, barrel neocortex of Fmr1 KO mice. Neocortical slices of Fmr1 KO mice have decreased excitatory drive onto layer IV fast-spiking interneurons and prolonged thalamically-evoked and spontaneously-occurring persistent activity, or UP, states 30, 31. UP states represent a normal physiological rhythm generated by the recurrent neocortical synaptic connections and is observed in alert and slow-wave sleep states in vivo as well as neocortical slice preparations 32, 33. Importantly, genetic or pharmacological reduction of mGluR5 in Fmr1 KO mice rescues the prolonged UP states in acute slices and in vivo 31. To determine if altered mGluR5-Homer interactions contribute to altered neocortical circuit function and hyperexcitability in Fmr1 KO mice we measured spontaneously occurring UP states in acute slices from somatosensory, barrel cortex using extracellular multiunit recordings 32. As previously reported 30, 31, UP states are longer in slices of Fmr1 KO mice in comparison to WT littermates (WT: 797.4±31.47 ms, n=22 slices; Fmr1-KO: 1212±87.9 ms, n=13; Fig. 5A,B). In support for a role for altered Homer interactions, UP state duration was shortened to WT levels by H1a deletion (H1a/Fmr1 KO; 872.2±42.1 ms, n=44; Fig. 5A, B). Loss of H1a alone does not affect UP states, ruling out general alterations in excitability (H1a-KO: 767.7±35.8 ms, n=18).
Figure 5. Disruption of mGluR5-Homer interactions mediates prolonged neocortical UP states in Fmr1 KO mice.
A,B. Genetic deletion of H1a rescues prolonged UP states in the Fmr1 KO. A. Representative extracellular multiunit recordings of spontaneous, persistent activity or UP states from layer 4 of somatosensory, barrel neocortical slices from each genotype. B. Group averages reveal that the UP state duration is prolonged in the Fmr1 KO slices (n = 13) in comparison to WT (n = 22) UP state duration in the H1a/Fmr1 KO (n = 44 slices) is reduced from the Fmr1 KO and is not different from WT. There is no difference between WT and H1a single KO (n = 18), suggesting that the rescue is dependent on Fmr1, and not a general decrease in excitability due to loss of H1a. C. Representative UP state recordings from WT and Fmr1 KO slices treated with the appropriate peptide. D. Pretreatment of WT neocortical slices with mGluR5CT peptide (CT; 4 hours; 5 μM; n = 15 slices) to acutely disrupt mGluR5-Homer interactions increases UP state duration in comparison to slices pretreated with control (mGluR5MU; MU; 5 μM; n = 13) peptide. In contrast, treatment of Fmr1KO slices (n = 12) with CT peptide had no effect on UP state duration in comparison to MU treated slices (n = 15). **p< 0.01; ***p<0.001.
We next determined if CT peptide-mediated disruption of mGluR5-Homer was sufficient to prolong UP states in WT slices. Preincubation of WT neocortical slices in CT peptide increased the duration of UP states in comparison to control peptide (mGluR5MU) treatment (WT+MU: 909.8±103.7 ms, n=13 slices; WT+CT: 1408.8±156.4 ms, n=15; Fig. 5C,D). Therefore, acute disruption of mGluR5-Homer complexes is sufficient to mimic the circuit hyperexcitability in Fmr1 KO mice. In contrast, mGluR5CT had no effect on the duration of UP states in Fmr1 KO slices (KO+MU: 2014.9±117.9 ms, n=15; KO+CT: 1819.8±163.8 ms, n=12; Fig. 5C,D), likely because Homer complexes are already disrupted in these mice.
H1a deletion reverses behavioral phenotypes in Fmr1 KO
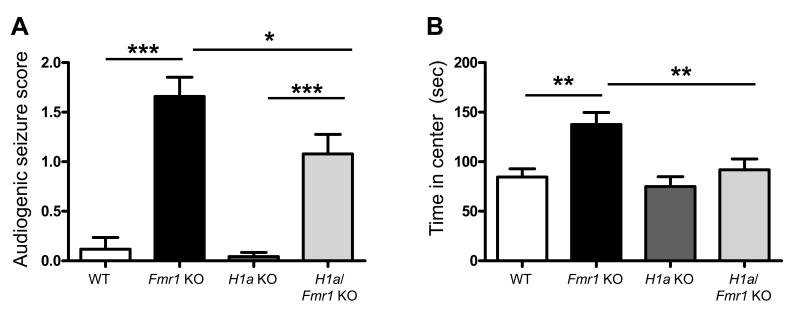
To determine if H1a deletion rescued any in vivo or behavioral phenotypes in Fmr1 KO mice, we measured the incidence of audiogenic seizures and anxiety as measured using the open field activity test across the 4 genotypes. We chose these phenotypes because they are robust in the C57BL6 strain of Fmr1 KO mice and sensitive to mGluR5 antagonists 8. Consistent with previous reports, Fmr1 KO display increased seizure incidence and severity following exposure to a loud sound relative to WT and H1a KO mice whom exhibited little or no incidence of seizure (Seizure score (0-3; 3 being most severe; see methods); WT = 0.12±0.12; n = 16 mice; H1a KO = 0.04±0.04; n = 24; Fmr1 KO=1.6 ±0.2; n = 39; p< 0.001 (Fmr1 KO vs. WT or H1a KO; Fig. 6A; Supplementary Table 3)). H1a/Fmr1 KO mice responded with a reduced incidence and severity of seizure in comparison to Fmr1 KO littermates (H1a/Fmr1 KO mice = 1.1±0.2; n = 37; p < 0.05 (H1a/Fmr1 KO vs. Fmr1 KO)). However, H1a/Fmr1 KO mice displayed an increased level of seizures in comparison to WT or H1a KO (p< 0.001; Fig. 6A). Such a partial rescue of the audiogenic seizures by H1a deletion is similar to what is observed with genetic reduction of mGluR5 in Fmr1 KO mice 7.
Figure 6. Homer1a deletion reduces audiogenic seizures and corrects open field activity in the Fmr1 KO.
A. The incidence and severity of audiogenic seizures was scored as described in methods. Fmr1 KO mice have an increased seizure score that was reduced in H1a/Fmr1 KO mice. (N = 16, 39, 24, 37 mice for WT, Fmr1 KO, H1a KO and H1a/Fmr1 KO, respectively). B. Open field activity, measured as time spent in the center of a lit open arena was increased in Fmr1 KO mice and reversed to WT levels in the H1a/Fmr1 KO mice. (N = 18, 24, 17, 17 mice for WT, Fmr1 KO, H1a KO and H1a/Fmr1 KO, respectively) *p< 0.05; **p< 0.01; ***p<0.001.
As previously reported, Fmr1 KO mice spend more time in the center of a lit open field in comparison to WT littermates which has been interpreted as reduced generalized anxiety in the mice34 (Fig. 6B; WT = 85±8 sec; n = 18 mice; Fmr1 KO = 138±12; n = 24; p< 0.01). H1a KO mice were not different than WT mice in this behavior (H1a KO = 75±10 sec; n = 17). Importantly, H1a/Fmr1 KO behave as WT mice (H1a/Fmr1 KO = 91±10 sec; n = 17), spending significantly less time than Fmr1 KO mice (p< 0.01) in the center of an open arena. There were no differences in locomotor activity between any genotype (Supplementary Figure 4). Therefore, H1a deletion completely rescued the open field activity phenotype and suggests that altered mGluR5- Homer interactions contribute to altered behavior in Fmr1 KO mice and may be relevant for altered behaviors in FXS patients.
Discussion
Two mechanisms for mGluR dysfunction in Fragile X Syndrome
Here we demonstrate a causative role for reduced Homer scaffolds in mGluR5 dysfunction in a model of human neurological disease. MGluR5 dysfunction in animal models of FXS is well established and genetic or pharmacological reduction of mGluR5 activity reduces or rescues many disease phenotypes in animal models 8 and most recently in patients 9. However, the molecular basis for mGluR5 dysfunction in FXS was essentially unknown. It has been suggested that loss of an FMRP mediated translational “brake” downstream of mGluR5 leads to enhanced mGluR5 function 6, 8, but this mechanism cannot account for the deficits in mGluR5 signaling or translation independent dysfunction of mGluR5 associated with FXS 18, 31. Our results reveal two mechanisms for mGluR5 dysfunction in Fmr1 KO mice. First, an imbalance of mGluR5 interactions from long to short Homer1a isoforms, leads to altered mGluR5 signaling, enhanced basal translation rates, neocortical hyperexcitability, audiogenic seizures and open field activity (Supplementary Fig. 6A). Because H1a bound mGluR5 is constitutively active or agonist-independent, our results strongly suggest that the therapeutic action of mGluR5 inverse agonists, such as MPEP 13, in FXS phenotypes are due, in part, to inhibition of H1a-bound, constitutively active mGluR5. Second, our results reveal that disrupted Homer scaffolds in Fmr1 KO mice cannot account for altered mGluR-LTD or abnormal translational control of FMRP target mRNAs (Fig. 4; Supplementary Fig. 6B) and implicate an essential role for FMRP binding to and translational regulation of specific mRNAs in mGluR-LTD. The discovery that altered Homer scaffolds account for much of the complex dysfunction of mGluR5 in FXS will help to develop alternative, targeted therapies for the disease and provide mechanistic links to other genetic causes of autism.
Homer scaffolds coordinate mGluR regulation of translation
Our data demonstrate novel functional roles for Homer scaffolds in coordination of mGluR-stimulated translation by facilitating activation of the PI3K-mTOR pathway and translation initiation 18, as well as limiting activation of EF2K and subsequent inhibition of elongation (Figs. 1,2). Homer scaffolds with the PI3K enhancer (PIKE), a small GTPase which binds and activates PI3K in response to mGluR activation 11. The PI3K pathway stimulates mTOR to phosphorylate eIF4E binding protein (4EBP) which in turn releases eIF4E to interact with eIF4G and form the eIF4F translation initiation complex23. Furthermore, mTOR phosphorylates p70 S6K to stimulate translation of 5′ terminal oligopyrmidine tract (5′ TOP) mRNAs that encode ribosomes and translation factors, thus increasing the translational capacity of the cell 23. mGluR activation of PI3K-mTOR-S6K pathway is blocked by mGluR5CT and the deficits in mGluR-activation of mTOR and initiation complex (eIF4F) formation in Fmr1 KO mice are restored by H1a deletion indicating a key role for Homer scaffolds in mGluR-stimulated translation initiation18.
Although somewhat counterintuitive, mGluRs stimulate phosphorylation of EF2 which in turn inhibits elongation rate 12. EF2K and moderate inhibition of elongation are necessary for mGluR-induced synthesis of Arc, as well as mGluR-LTD 12. Submaximal inhibition of global elongation may make available rate limiting factors to translate mRNAs that are poorly initiated and cannot compete effectively for these factors 35. Long Homer interactions limit EF2K activation by mGluRs and thus would be expected to temper mGluR-mediated inhibition of general elongation, and in turn, promote translation of poorly initiated mRNAs 12. Consequently, disruption of mGluR5-Homer enhances EF2K activity and would be expected to strongly inhibit elongation and block translational activation. Although we do not rescue abnormal mGluR-LTD in Fmr1 KO neurons by deletion of H1a, in WT slices mGluR5Ct peptide treatment blocks mGluR-induced synthesis of Arc (Supplementary Fig. 1C) and mGluR-LTD18. These results indicate that in WT neurons, where mGluR-LTD requires de novo protein synthesis, mGluR5-Homer interactions are necessary to properly stimulate translation and induce LTD.
In Fmr1 KO mice there is a deficit in mGluR stimulation of PI3K-mTOR and eIF4F initiation complex formation, whereas EF2K activation is dramatically enhanced. These changes would be expected to block mGluR-induced translational initiation and strongly inhibit elongation. mGluR-induced rapid synthesis of many proteins (e.g. PSD-95, EF1a, APP, Arc, CaMKII, MAP1b), is absent in Fmr1 KO mice2 that may be mediated, in part, by disrupted mGluR-Homer scaffolds and altered signaling to translation machinery. Alternatively or in addition, because FMRP interacts with these mRNAs, it is likely required for mGluR-triggered translational activation of specific target mRNAs 2, 29. The fact that H1a deletion rescues mGluR-mediated translation initiation complex formation (Fig. 3D) but not synthesis of Arc (Fig. 4), suggests a requirement for both mGluR5-Homer scaffolds and FMRP in mGluR-triggered Arc translation.
Altered mGluR5-Homer scaffolds increase translation rates
Although there is a deficit in mGluR agonist-stimulated translation in Fmr1 KO mice, steady state translation rates and levels of specific proteins are elevated 19, 20, thus reflecting the complexity of translational control. Because one function of FMRP is to suppress translation of its mRNA targets 2, an obvious possibility was that the elevated protein synthesis rates and levels directly result from loss of FMRP-mediated suppression of mRNA targets. In support of this hypothesis, H1a deletion does not reverse enhanced protein levels of Map1b and CaMKII (Fig. 4). However, elevated total protein synthesis rates and translation initiation (eIF4F) complexes were rescued by H1a deletion and mimicked in WT mice by mGluR5CT peptide treatment (Fig. 3). Thus, increased steady state translation rates in Fmr1 KO tissue are a result of altered mGluR5-Homer scaffolds that are a secondary consequence of FMRP loss. Elevated protein synthesis rates in Fmr1 KO hippocampal slices are reversed by the genetic reduction of mGluR5 (Grm5het), the mGluR5 inverse agonist MPEP, and inhibitors of ERK 8, 20 (Fig. 3C). In Fmr1 KO cortical lysates, we observed enhanced phosphorylation of translation initiation factors that are downstream of ERK (eIF4E and 4EBP) which was reversed by H1a deletion (Fig. 3E). Furthermore, blocking mGluR5 activity with MPEP strongly reduced P-eIF4E and P-4EBP in Fmr1 KO, but not WT, hippocampal slices (Fig. 3F). Together theseresults suggest that H1a-bound and constitutively active mGluR5 in Fmr1 KO neurons13 drives ERK-dependent phosphorylation of eIF4E and 4EBP which enhances eIF4F initiation complex formation and translation rates. Because we do not observe an effect of Fmr1 KO or MPEP on Phospho-ERK levels as detected by western blot, this suggests that mGluR5 drives ERK activity that is either not detectable by phosphorylation at T202/Y204 and/or regulates accessibility of eIF4E and 4EBP to ERK.
In contrast to hippocampus, recent results from neocortical synaptoneurosomes of Fmr1 KO mice demonstrate a role for PI3K activity in enhanced protein synthesis rates 22. We observed that the PI3K inhibitor wortmannin equalized translation rates between WT and Fmr1 KO hippocampal slices, but this was because wortmannin increased translation rates in WT, but not Fmr1 KO, slices (Supplementary Fig. 4). Furthermore, elevated basal phosphorylation of PI3K, mTOR and 4EBP (at the mTOR sites) was recently reported in fresh hippocampal lysates of Fmr1 KO mice, perhaps a result of increased PIKE levels 19, 20. Although we are unable to detect elevated basal activation of PI3K-mTOR pathway in our slice preparation 18 (Supplementary Table 2), persistent activation of downstream effectors of PI3K and mTOR together with constitutive mGluR5-driven ERK may elevate translation rates in Fmr1 KO. P-EF2 levels are unchanged in Fmr1 KO slices (Supplementary Table 2) suggesting that basal or constitutive mGluR5 activity is not sufficient to activate EF2K. The detailed mechanisms by which ERK, PI3K and mTOR, and EF2K contribute to elevated, basal protein synthesis rates in Fmr1 KO mice requires further study and may differ depending on the brain region, subcellular compartment or preparation.
An essential role for FMRP in proper regulation of LTD
H1a deletion and restoration of mGluR5-Homer scaffolds in Fmr1 KO mice did not rescue the protein synthesis independence of mGluR-LTD nor elevated steady state protein levels of FMRP target mRNAs, MAP1b and αCaMKII (Fig. 4). This supports the hypothesis that the protein synthesis independence of mGluR-LTD in Fmr1 KO mice is a result of loss of FMRP-mediated translational suppression of LTD promoting proteins, such as Map1b 36. Because H1a deletion rescued the elevated 35S Met incorporation, but not altered mGluR-LTD and enhanced MAP1b and CaMKII levels, this suggests that altered LTD and elevated Map1b and CamKII are not a result of elevated total protein synthesis rates. Furthermore, the fact that mGluR-triggered Arc synthesis was not rescued by H1a deletion supports an essential role for FMRP in mGluR-triggered translational activation of Arc. Recent work has implicated mGluR-triggered dephosphorylation of FMRP in translational activation of FMRP target mRNAs, such as SAPAP3 and PSD-95 2, 29.
Altered mGluR5-Homer and neocortical network dysfunction
Altered neocortical circuit function and hyperexcitability have been predicted to contribute to cognitive disorders and autism 37, 38. The epilepsy and EEG abnormalities observed in FXS patients are indicative of brain hyperexcitability 3, 8. Furthermore, FXS patients display hypersensitivity to sensory stimuli, and Fmr1 KO mice have audiogenic seizures reflecting hyperexcitability of sensory circuits 3, 8. Although UP states are a normal physiological rhythm and are not epileptiform activity, they provide an effective readout of the state of circuit function and excitability 32. Furthermore, UP states underlie the slow oscillations that occur during slow wave sleep and are implicated in memory consolidation as well as sensory processing in waking states 33. Therefore, altered neocortical UP states in the Fmr1 KO 30, 31 may be relevant to the sensory processing and cognitive abnormalities in FXS patients.
Our findings indicate that the longer UP states in Fmr1 KO neocortex are mediated by enhanced, likely constitutive, activity of H1a-bound mGluR531. In support of this conclusion, prolonged UP states in the Fmr1 KO are reversed by genetic or acute, pharmacological blockade of mGluR5 31 and genetic deletion of H1a (Fig. 5A). Peptide-mediated disruption of mGluR5-Homer interactions prolongs UP states in WT, but not Fmr1 KO, slices (Fig. 5C) which suggests that regulation of Homer scaffolds, by H1a or other means, may regulate neocortical slow oscillations in the normal brain. Interestingly, H1a is induced in neocortex with sleep deprivation and contributes to the homeostatic increase in slow wave sleep that occurs in response to sleep deprivation 39. Consequently, altered UP states, Homer interactions and responses to H1a may contribute to the reported sleep problems in FXS patients 4.
How does mGluR5 activity lead to longer UP states? Prolonged UP states are not due to mGluR5-driven translation because the protein synthesis inhibitor anisomycin does not affect UP state duration in either WT or Fmr1 KO slices 31. Therefore, mGluR5 activity likely leads to prolonged UP states through posttranslational regulation of the intrinsic excitability and/or synaptic function of neocortical neurons.
Altered behavior in Fmr1 KO mice is rescued by H1a deletion
Genetic reduction of mGluR5 (heterozygosity) , or H1a deletion, completely rescue neocortical hyperexcitability (e.g. long UP states) in Fmr1 KO mice, but only partially rescue audiogenic seizures (Fig. 6A)7, 31. This suggests that hyperexcitability in other brain regions, such as the auditory brain stem, also contribute to audiogenic seizures in Fmr1 KO mice, through mGluR5 and H1a independent mechanisms 40. Importantly, mGluR5 antagonism and H1a deletion (Fig. 6B) rescue the increased open field activity in Fmr1 KO mice suggesting that abnormal Homer scaffolds contribute to behavioral symptoms associated with FXS and may represent a new therapeutic target for the disease. In contrast to initial studies7, 8, 20, recent reports have failed to rescue some Fmr1 KO mouse behaviors by reduction of mGluR5 activity41, 42, suggesting mGluR5-independent mechanisms in FXS pathology. Interestingly, in these studies inhibition of mGluR1 proved efficacious in reducing some Fmr1 KO phenotypes 41, 42. MGluR1a, is a Homer binding protein, and mGluR1a-Homer scaffolds may also be affected in Fmr1 KO mice. Alternatively, because of the diverse mRNA targets of FMRP 43 other signaling pathways are likely affected in FXS. However, because mGluR5 antagonism has proven effective in some FXS patients 9 the understanding mGluR5 function in the normal brain and its dysfunction in FXS may provide additional and more targeted treatments for the disease and provide insight into autism.
For the phenotypes we studied here, H1a deletion had no effect on the WT background, but only on the Fmr1 KO background. This is likely because H1a expression is typically low under basal conditions and is strongly induced in response to neuronal activity and experience 11. The effects of H1a induction in WT mice are expected to be mimicked by the mGluR5CT peptide, where we observe effects on mGluR5 signaling, protein synthesis rates, LTD 18 and UP state duration (Figs. 1,3,5). In contrast, CT peptide had no effect on these measures in Fmr1 KO mice 18 (Figs. 3,5) . Therefore, we would expect that experience-dependent H1a induction would affect mGluR5 function in WT, but not Fmr1 KO mice. Such insensitivity to experience-induced H1a could contribute to deficits in experience- dependent plasticity associated with FXS 7.
Mechanism of disrupted Homer scaffolds in Fmr1 KO
How does loss of FMRP lead to altered mGluR5-Homer scaffolds? Protein levels of long Homers and H1a are unchanged in total homogenates of Fmr1 KO hippocampi 10 (Supplementary Fig. 2) and FMRP is not reported to interact with mRNA for any Homer isoforms 43. Previous work reported a decrease in Tyr phosphorylation of long Homer in Fmr1 KO forebrain 10, but it is unknown if or how this affects interactions with mGluR5. Phosphorylation of Homer3 regulates interactions with other Homer effectors 44, 45. Similarly, phosphorylation of mGluR5 at the C-terminal Homer interaction domain reduces the affinity of mGluR5 for Homer 46. Therefore, posttranslational modification of mGluR5 and/or Homer in Fmr1 KO mice may underlie the decreased interactions.
Disrupted or destabilized synaptic scaffolds that affect Homer and/or mGluR5 may also contribute more generally to cognitive disorders and autism behaviors. Mutations in the Homer binding domain of SHANK3 and the Homer binding protein oligophrenin 1 are implicated in autism and intellectual disability, respectively 47, 48. Expression of a truncated SHANK3 without the Homer binding domain in mice results in degradation of SHANK3 and autistic behaviors in mice. Importantly, reduction of long Homers or induction of H1a recapitulates the degradation of synaptic SHANK3 47. An interesting possibility is that mGluR5 dysfunction, of the kind we describe here, may occur in individuals with mutations in SHANK3 or other genes that destabilize Homer scaffolds.
Methods
Animals
Congenic Fmr1 KO mice 49 were bred on the C57/BL6J background. Homer1a specific KO mice were generated as described21 and backcrossed at least 5 generations onto the C57/Bl6J mice from the UT Southwestern mouse breeding core facility. All experiments were performed on littermate controls and blind to mouse genotype. Long Evans Hooded rats were obtained from Charles River Laboratories, Wilmington, MA. The animal use protocols used in this manuscript were approved by the UT Southwestern IACUC committee.
Reagents
Drugs were prepared as stocks and stored at -20°C and used within two weeks. Tat-mGluR5ct (YGRKKRRQRRR-ALTPPSPFR) and tat-mGluR5mu (YGRKKRRQRRR-ALTPLSPRR; 5 μM) 16 were synthesized at the UT Southwestern Protein Chemistry Technology Center. The peptide was dissolved in H20 at a concentration of 5mM, aliquoted and stored at -20°C. Frozen aliquots were used within 10 days. The mixed group I mGluR agonist (RS)-3,5-dihydroxyphenylglycine (DHPG), U0126 and wortmannin were purchased from Tocris Bioscience and prepared as described 20, 22. Cycloheximide was purchased from Sigma, and freshly prepared daily by dissolving directly in ACSF.
Hippocampal slice preparation and LTD recordings
Acute hippocampal brain slices were prepared from 3-6 wk old rats or wildtype (WT), Fmr1 knockout (Fmr1 KO), Homer1a knockout (H1a-KO), or Homer1a/Fmr1 double knockout (H1a/Fmr1 KO) littermates as described previously 18. LTD recordings were performed and analyzed as described 18.
mGluR signaling in slices and western blotting
Western blotting on slices was performed as described18. Hippocampal slices were preincubated in a static incubation chamber in ACSF containing 5 μM tatmGluR5CT or MU for 4 hours prior to DHPG treatment (Fig. 1) or 30 min in MPEP (10μM; Fig. 3F). Blotting membranes were incubated with the following antibodies according to the manufacturer’s instructions: phospho-T56 EF2, total-EF2, phospho-T389 p70S6K, phospho-S2448 mTOR, total mTOR, phospho-T202/Y204 ERK, total-ERK, phospho S209-eIF4E, phospho S65-4EBP, 4EBP, eIF4G, eIF4E (all from Cell Signaling Technology), Homer (Sc-8921; Santa Cruz), mGluR5 (Millipore), β3 tubulin (Abcam), Arc (Synaptic Systems), Map1b (gift from Dr. Itzhak Fischer, Drexel University), αCaMKII (Santa Cruz, sc-5391), actin (Millipore, MAB1501). For comparison of phosphoprotein levels across conditions or genotypes immunoreactive phosphoprotein bands were normalized to total protein levels from the same slice homogenates (e.g. P-mTOR/mTOR), each of which was first normalized to loading control (either tubulin, actin or total ERK where indicated).
Coimmunoprecipitation
Hippocampus was lysed in co-immunoprecipitation buffer (50 mM Tris, pH 7.4, 120 mM NaCl, 0.5% NP40), and protein was tumbled overnight at 4°C with 1 μg of antibody (either Homer (Santa Cruz, D-3), Homer1a (Santa Cruz, M-13) or eIF4G (Cell Signaling)). Protein A/G agarose bead slurry (Thermoscientific) was added for one additional hour and the beads were then washed with co-i.p. buffer. Western blotting was performed with the Homer (Santa Cruz, E-18 sc-8921), Homer1a 21, mGluR5 (Millipore), eIF4E (Cell Signaling) or eIF4G (Cell Signaling).
Real-time RT-PCR
Hippocampi were homogenized in TRIzol reagent (Invitrogen) followed by RNA extraction according to manufacturer’s protocol. 2 μg RNA from each sample was subjected to first-strand cDNA synthesis by SuperScript III First-Strand synthesis system (Invitrogen) with two independent primers targeting on the 3′UTR of the mRNAs (Homer1a: 5′-GTG GTA AAG CTT TCC TTC AGA G-3′ and 5′-GGC ACC TCT GTG GGC CTG TGG-3′; GAPDH: 5′-GGT ATT CAA GAG AGT AGG GAG-3′ and 5′-GGG TGC AGC GAA CTT TAT TG-3′). PCR reaction was performed by GoTag Green DNA polymerase (Promega) with specific primers against Homer1a: (5′-TGA TTG CTG AAT TGA ATG TGT ACC-3′ and 5′-GAA GTC GCA GGA GAA GAT G-3′) 50 and GAPDH: (5′-AGG TCG GTG TGA ACG GAT TTG-3′ and 5′-TGT AGA CCA TGT AGT TGA GGT CA-3′).
Metabolic labeling of hippocampal slices
Hippocampal slices were prepared as described18, 20. For these experiments (Fig. 3) the most ventral slices (2 per hippocampus) were used since basal protein synthesis rates differ between dorsal and ventral hippocampal slices 20. Slices recovered for 3.5 hours in ACSF at 32°C, and then were incubated in actinomycin D (25 μM) for 30 min. Where indicated, 20 μM U0126, or 100 nM wortmannin was added at this step. For experiments with tatmGluR5 peptides, slices were preincubated in ACSF containing 5 μM tatmGluR5CT or MU for 4 hours prior to actinomycin incubation.
Neocortical Slice preparation and UP state recordings
UP state experiments in neocortical slices were performed and analyzed as described 31.To allow time for the tatmGluR5 peptides to permeate slices, the peptide containing ACSF was perfused onto the slices in the interface chamber for 4 hr prior to recording and was supplemented with 10 μM HEPES pH7.4, 0.05% BSA, and 5 μM of the appropriate peptide. The peptide-BSA containing ACSF was not oxygenated directly, but slices were oxygenated in the interface recording chamber.
Audiogenic seizures
In order to evaluate audiogenic seizures, mice were placed in a plastic chamber (30X19X12cm) containing a 120 decibel siren (GE 50246 personal security alarm) and covered with a Styrofoam lid. A 120 dB siren was presented to mice for 5 minutes. Mice were videotaped and scored for behavioral phenotype: 0=no response, 1=wild running, 2=tonic-clonic seizures, 3=status epilepticus/death as described 7.
Behavioral measurements
Open Field Activity
Mice were placed individually into the periphery of a novel open field environment (44 cm x 44 cm, walls 30 cm high) in a dimly lit room and allowed to explore for 5 min. The animals were monitored from above by a video camera connected to a computer running video tracking software (Ethovision 3.0, Noldus) to determine the time, distance moved and number of entries into two areas: the periphery (5 cm from the walls) and the center (all areas excluding the periphery). The open field arenas were wiped and allowed to dry between mice. Time in the center was used as a measure of anxiety.
Locomotor Activity
Mice were placed individually into a new, plastic mouse cage (18 cm x 28 cm) which was located inside a dark Plexiglas box. Movement was monitored by 5 photobeams in one dimension (Photobeam Activity System, San Diego Instruments) for 2 hours, with the number of beam breaks recorded every 5 min. Data were analyzed with an ANOVA with genotype and time as the dependent variables.
Statistics
Data plotted in the figures represents the mean ± SEM. Significant differences were determined using independent or paired t-tests (for determining effects of mGluR5CT peptide). For comparisons between WT, Fmr1 KO, H1a KO and H1a/Fmr1 KO a 2-way ANOVA and Bonferroni posttests were used. Statistics on nominal data, such as seizure severity and incidence (Fig. 6A; Supplementary Table 3), a Chi squared (Fisher’s Exact test) was used. Group data is presented in the figures as mean ±SEM. *p< 0.05; **p< 0.01; ***p<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the grants from the National Institutes of Health NS045711, HD052731 (KMH), HD056370 (JRG), GM008203 (SAH), Autism Speaks (KMH), FRAXA Research Foundation and The Hartwell Foundation (JAR). We would like to thank Lorea Ormazabal and Nicole Cabalo for technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS JAR designed, performed and analyzed experiments included in Figs. 2,3,4,6 Supplementary Figs. 1,2, Tables 1, 2 and wrote a first draft of the manuscript. KAC performed and analyzed experiments in Figs. 1,3, Supplementary Figs. 2,3,4 and Table 1. SAH performed and analyzed experiments in Figs. 5,6. NPT performed and analyzed co-IP experiments for Figs. 2, 3, and Supplementary Fig. 2. WG performed and analyzed experiments for Figs. 1, 2, 3 and 4. SGB provided intellectual input on the behavioral experiments, designed and performed experiments in Fig. 6 and Supplementary Fig. 5. JJH and PFW provided intellectual input, generated and provided the H1a KO mice. JRG contributed intellectually to the overall project and in particular to the UP state experiments (Fig. 5). JRG trained and supervised SAH, designed experiments for Fig. 5, contributed funding and edited the manuscript. KMH supervised the overall project, designed experiments, contributed funding, edited figures and wrote the final version of the manuscript.
References
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry-Kravis E. Epilepsy in fragile X syndrome. Dev Med Child Neurol. 2002;44:724–728. doi: 10.1017/s0012162201002833. [DOI] [PubMed] [Google Scholar]
- 4.Hagerman R. The physical and behavioral phenotype. In: Hagerman R, Hagerman P, editors. Fragile X Syndrome: Diagnosis, Treatment, and Research. The Johns Hopkins University Press; Baltimore: 2002. pp. 3–109. [Google Scholar]
- 5.Luscher C, Huber KM. Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron. 2010;65:445–459. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Dolen G, et al. Correction of Fragile X Syndrome in Mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Jacquemont S, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 2011;3:64ra61. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- 10.Giuffrida R, et al. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–8916. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ango F, et al. Agonist-independent activation of metabotropic glutamate receptors by the intracellular protein Homer. Nature. 2001;411:962–965. doi: 10.1038/35082096. [DOI] [PubMed] [Google Scholar]
- 14.Tu JC, et al. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 15.Xiao B, et al. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 16.Mao L, et al. The scaffold protein Homer1b/c links metabotropic glutamate receptor 5 to extracellular signal-regulated protein kinase cascades in neurons. J Neurosci. 2005;25:2741–2752. doi: 10.1523/JNEUROSCI.4360-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronesi JA, Huber KM. Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse. Sci Signal. 2008;1:pe6. doi: 10.1126/stke.15pe6. [DOI] [PubMed] [Google Scholar]
- 18.Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 Leads to Excessive Protein Synthesis in the Hippocampus of a Mouse Model of Fragile X Syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu JH, et al. Homeostatic Scaling Requires Group I mGluR Activation Mediated by Homer1a. Neuron. 2010;68:1128–1142. doi: 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross C, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 24.Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbert TP, Tee AR, Proud CG. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J Biol Chem. 2002;277:11591–11596. doi: 10.1074/jbc.M110367200. [DOI] [PubMed] [Google Scholar]
- 26.Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational Control by MAPK Signaling in Long-Term Synaptic Plasticity and Memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 27.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zalfa F, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 29.Muddashetty RS, et al. Reversible Inhibition of PSD-95 mRNA Translation by miR-125a, FMRP Phosphorylation, and mGluR Signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson JR, Bartley AF, Hays SA, Huber KM. Imbalance of Neocortical Excitation and Inhibition and Altered UP States Reflect Network Hyperexcitability in the Mouse Model of Fragile X Syndrome. J Neurophysiol. 2008;100:2615–2626. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hays SA, Huber KM, Gibson JR. Altered Neocortical Rhythmic Activity States in Fmr1 KO Mice Are Due to Enhanced mGluR5 Signaling and Involve Changes in Excitatory Circuitry. J Neurosci. 2011;31:14223–14234. doi: 10.1523/JNEUROSCI.3157-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Vives MV, McCormick DA. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci. 2000;3:1027–1034. doi: 10.1038/79848. [DOI] [PubMed] [Google Scholar]
- 33.Haider B, McCormick DA. Rapid neocortical dynamics: cellular and network mechanisms. Neuron. 2009;62:171–189. doi: 10.1016/j.neuron.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZH, Smith CB. Dissociation of social and nonsocial anxiety in a mouse model of fragile X syndrome. Neurosci Lett. 2009;454:62–66. doi: 10.1016/j.neulet.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 36.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlhaas PJ, Singer W. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron. 2006;52:155–168. doi: 10.1016/j.neuron.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Mackiewicz M, Paigen B, Naidoo N, Pack AI. Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol Genomics. 2008;33:91–99. doi: 10.1152/physiolgenomics.00189.2007. [DOI] [PubMed] [Google Scholar]
- 40.Brown MR, et al. Fragile X mental retardation protein controls gating of the sodium-activated potassium channel Slack. Nat Neurosci. 2010;13:819–821. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas AM, et al. Genetic reduction of group 1 metabotropic glutamate receptors alters select behaviors in a mouse model for fragile X syndrome. Behav Brain Res. 2011;223:310–321. doi: 10.1016/j.bbr.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas AM, Bui N, Perkins JR, Yuva-Paylor LA, Paylor R. Group I metabotropic glutamate receptor antagonists alter select behaviors in a mouse model for fragile X syndrome. Psychopharmacology (Berl) 2011 doi: 10.1007/s00213-011-2375-4. [DOI] [PubMed] [Google Scholar]
- 43.Darnell JC, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizutani A, Kuroda Y, Futatsugi A, Furuichi T, Mikoshiba K. Phosphorylation of Homer3 by calcium/calmodulin-dependent kinase II regulates a coupling state of its target molecules in Purkinje cells. J Neurosci. 2008;28:5369–5382. doi: 10.1523/JNEUROSCI.4738-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang GN, et al. NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins. Science. 2008;319:476–481. doi: 10.1126/science.1151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlando LR, et al. Phosphorylation of the homer-binding domain of group I metabotropic glutamate receptors by cyclin-dependent kinase 5. J Neurochem. 2009;110:557–569. doi: 10.1111/j.1471-4159.2009.06139.x. [DOI] [PubMed] [Google Scholar]
- 47.Bangash MA, et al. Enhanced Polyubiquitination of Shank3 and NMDA Receptor in a Mouse Model of Autism. Cell. 2011 doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Billuart P, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 49.Consortium, T.D.-B.F.X. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 50.Potschka H, et al. Kindling-induced overexpression of Homer 1A and its functional implications for epileptogenesis. Eur J Neurosci. 2002;16:2157–2165. doi: 10.1046/j.1460-9568.2002.02265.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.