Abstract
Streptococcus mutans, a primary dental pathogen, has a remarkable capacity to scavenge nutrients from the oral biofilm for its survival. Cystine is an amino acid dimer formed by the oxidation of two cysteine residues that is required for optimal growth, whereas S. mutans modulates l-cystine uptake via two recently identified transporters designated TcyABC and TcyDEFGH, which have not been fully characterized. Using a non-polar tcyABC-deficient mutant (SmTcyABC), here we report that L-cystine uptake is drastically diminished in the mutant, whereas its ability to grow is severely impaired under l-cystine starvation conditions, relative to wild type. A substrate competition assay showed that l-cystine uptake by the TcyABC transporter was strongly inhibited by dl-cystathionine and l-djenkolic acid and moderately inhibited by S-methyl-l-cysteine and l-cysteine. Using gene expression analysis, we observed that the tcyABC operon was up-regulated under cystine starvation. TcyABC has been shown to be positively regulated by the LysR-type transcriptional regulator CysR. We identified another LysR-type transcriptional regulator that negatively regulates TcyABC with homology to the B. subtilis YtlI regulator, which we termed TcyR. Our study enhances the understanding of l-cystine uptake in S. mutans which allows survival and persistence of this pathogen in the oral biofilm.
Keywords: Cystine, cysteine, transport, TcyABC, Streptococcus mutans
INTRODUCTION
As one of the primary etiological agents in dental caries, the pathogenicity of S. mutans is dependent on its ability to cope with drastic fluctuations in nutrient availability in the oral biofilm. Since these can range from nutrient abundant and starvation conditions, the remarkable adaptive capacity of S. mutans is due, in part, to its ability to detect and import nutrients vital for growth and survival. Not surprisingly, 15% of the ORFs in the UA159 genome are associated with nutrient transport, whereas more than 60 ABC type transporters exhibit specificity for different substrates including carbohydrates, amino acids and inorganic ions [1]. Cysteine, a hydrophilic amino acid is an important structural and functional component of many cellular proteins and enzymes and has been shown to be essential for growth of S. mutans under all in vitro conditions tested [2]. The dimerization of cysteine, whereby two cysteine molecules are linked by a disulfide bond upon oxidation results in formation of cystine. Both cystine and cysteine can be also be used as sources of sulfur, an indispensable element required for activity of many enzymes and involved in ion and redox metabolic pathways [3]. Cysteine biosynthesis and cystine uptake are thus important metabolic processes essential for bacterial growth and survival.
Cystine uptake has been relatively well studied in Bacillus subtilis and three different systems involved in the import of L-cystine have been identified [4]. These include two ATP-binding cassette (ABC) transporters, TcyABC and TcyJKLMN, and a symporter TcyP [4]. The TcyJKLMN, and TcyP uptake systems are high-affinity transporters while TcyABC is a low-affinity L-cystine transporter [4]. The TcyJKLMN transporter, encoded within a large operon called the ytmI operon, was found to be the most sensitive to l-cystine starvation compared with other transporters in that its expression was repressed more than 200-fold in the presence of sulfate or l-cystine [5]. In addition, the expression of the ytmI operon was induced during disulfide stress by the thiol oxidant diamide [6]. TcyP and TcyABC l-cystine transporters have also been identified in Staphylococcus aureus and were shown to be negatively regulated by the CymRSA regulator, a global regulator that controls cysteine metabolism in response to its availability [7].
Cysteine metabolism has not been extensively studied in S. mutans. However, Sperandio et al recently characterized two LysR type transcriptional regulators, CysR and HomR which activate transcription of genes involved in cysteine metabolism and transport [8]. These authors also identified two L-cystine importers, TcyABC and TcyDEFGH, whose expression was activated by CysR and HomR respectively [8]. We sought to characterize the tcyABC tri-cistronic operon encoding the TcyABC transporter in S. mutans. Mutagenesis of tcyABC severely impaired the ability of S. mutans to transport l-cystine and survive under cystine starvation conditions. We also identified a novel Lys type regulator of TcyABC which we termed TcyR. Unlike most Lys type regulators, TcyR was found to repress transcription of the tcyABC operon.
MATERIALS AND METHODS
Bacterial strains and growth conditions
S. mutans strain UA159 was used to construct mutants. Unless otherwise specified, strains were routinely cultured in Todd-Hewitt yeast extract (THYE) medium (BD Biosciences) at 37°C in air with 5% CO2 without agitation. Mutant strains were propagated in THYE agar plates supplemented with erythromycin at 10 µg per ml. Optical density (OD) was measured using an Ultrospec 3000 UV/Visible Spectrophotometer (Fisher Scientific).
Construction of mutants
S. mutans UA159 was used as the wild-type strain. The S. mutans ΔtcyA (SmTcyA), ΔtcyB (SmTcyB), ΔtcyC (SmTcyC), ΔtcyABC (SmTcyABC), and ΔtcyR (SmTcyR) mutants were constructed in UA159 by a PCR ligation-based deletion strategy as described previously [9]. Briefly, an erythromycin resistance cassette was used to disrupt the tcyC, tcyABC and tcyR coding regions in the S. mutans UA159 wild-type chromosome using primers using the primer pairs listed below. To confirm successful integration of the erythromycin gene into these coding regions, chromosomal DNA was isolated from erythromycin resistant transformants and subjected to validation using PCR and nucleotide sequence analysis. Primers used for mutagenesis (5’ to 3’) are shown below. P1-tcyA: GCTGATTTCAACTAAGGGACG, P2-tcyA: GTAAGGTAAAAGCGACCAAGG, P3-tcyA: TCAGCAGTATTTAGCGGGTG, P4-tcyA: GGTAAACCTGAGCAGTTGTCATC, P1-tcyB: CAACAGACTCAGATACAGCTCC, P2-tcyB: CCGTTAGGTAAACTGGCAAC, P3-tcyB: AAGCTGTGGAAGGAGGTGTG, P4-tcyB ACGATAAAGAATCCAACCCG, P1-tcyC: CCGATCTTGGTTCAACTGATG, P2-tcyC : CCGACAAGGGCTACAACTTC, P3-tcyC: ATTCTTGAGCAGGGAACGCC, P4-tcyC: CGGAAAAAAGCACCATCAC, P1-tcyR: TGGACTGGGCAATCTCATCACC, P2-tcyR: TGGTAACTGCTGGTTGTGTAATGTG, P3-tcyR: GAATCTCCTTTTTCTATCGCAG, P4-tcyR: TCTGTCAGGCTTCCACTATTG, Erm-F: GGCGCGCCCCGGGCCCAAAATTTGTTTGAT, Erm-R: GGCCGGCCAGTCGGCAGCGACTCATAGAAT. Note: An AscI restriction site was added at the 5’-end of the P2 primers, while an FseI restriction site was added at the 5’-end of the P3 primers. Primers were designed and analyzed with MacVector 7.2 software.
Northern blot analysis
S. mutans cells grown to mid-log phase (OD600 ~0.4 – 0.5) were harvested by centrifugation (4,000 × g, 15 min, 4°C) and total RNA was extracted using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Five micrograms of each RNA samples and ladder (Invitrogen) were prepared by electrophoresis on a 1% agarose-formaldehyde gel and transferred to a nylon membrane [10]. The tcyA, tcyB, and tcyC probes, generated using primers labeled with digoxigenin-dUTP with the PCR DIG Probe Synthesis Kit (Roche) as specified by the manufacturer. Transcripts were diluted with the chemiluminescent substrate CDP-star (Roche) and exposed to X-ray films (Kodak). Primers used for probe preparation are as follows (5’ to 3’): TcyA-PF: CAGGAAACAATCACTGTAGCAAC, TcyA-PR: GAATAGCAGCATAGTTAGAACCAGC, TcyB-PF: CCTCAATCAAAAGATGGGGAC, TcyB-PR: CGATAAGACGACCAACTTGTTC, TcyC-PF: TTCTGGTGCTGGGAAATCAAC, TcyC-PR: TGACCTCCTGAAAGATGGCG
Rapid amplication of 5’ cDNA ends-PCR (5’ RACE-PCR)
The 5’ RACE-PCR technique was used to define the transcriptional start site (TSS) of the tcyABC locus. Overnight cultures of S. mutans UA159 were diluted 1:50 in fresh THYE broth and incubated at 37°C until an OD600 of approximately 0.4 was reached. Total RNA was extracted using RNeasy Mini Kit. Ten micrograms of DNA-free RNA was reverse transcribed using RACE outer primer (5’-CGATAACTGATAACGTCCTG-3’) and Superscript II Reverse Transcriptase (Invitrogen) according to the supplier’s instructions. RNaseH (USB) and RNase T1 (Roche) were then added and incubated at 37°C for 30 min. The cDNA was purified using the StrataPrep PCR Purification Kit (Stratagene) following the manufacturer’s instructions. Tailing of purified cDNA using terminal deoxynucleotidyl transferase (Sigma) and dGTP/dTTP was done according to instructions. Tailed cDNAs were amplified by PCR using RACE universal primers (5’-GAATTCGAATTCCCCCCCCCCCC-3’, 5’-GAATTCGAATTCAAAAAAAAAAAA-3’) and RACE inner primer (5’-GCTGTATCTGAGTCTGTTGCTAC-3’). Amplicons were analyzed by agarose gel electrophoresis and sequenced using the RACE inner primer.
Amino acid transport assay
S. mutans cells were grown in modified Berman’s Broth (MBB) medium containing 2% trypticase, 0.1% yeast extract, 34 mM NaCl, 0.05% sodium thioglycollate, 1 mM MgSO4, 0.1 M MnSO4, buffered to pH 7.3 with MOPS buffer and supplemented with 0.1% (wt/vol) glucose [11]. Overnight cultures were diluted 1:5 in MBB medium and grown until an OD600 of approximately 0.4 was reached. Cells were harvested by centrifugation, washed twice with cold buffer A (50 mM MOPS, 50 mM KPO4, 10 mM MgSO4), resuspended in cold buffer A to a final OD600 ~ 4.0, and kept on ice until used for transport assay. The assay mixtures contained cell suspension (~109 CFU/ml), 1% glucose, and 0.1 mg/ml chloramphenicol. Reaction mixtures were preincubated for 10 min at 37°C prior to the addition of substrates. At time zero, l-[14C]cystine and cold l-cystine were added at a concentration of 4 µM (2.6 mCi/mmol) and 200 µM, respectively, and the reaction mixtures were incubated at 37°C. Samples (100 µl) were removed at regular intervals and immediately filtered through 0.22-µm pore-size membranes. The filters were washed twice with 0.5 ml of buffer A and transferred to vials containing 5 ml of a scintillation fluid for determination of radioactivity. All transport assays were carried out using three independent cultures and each time point was sampled in duplicate. The specificity of CysBPA-mediated amino acid uptake was also examined using an amino acid competition assay. Uptake of l-[14C]cystine (4 µM) was measured in the presence of 400 µM of the following unlabeled l-amino acids: arginine, cysteine, glutamine, glutamate, leucine, and methionine. As a positive control and to determine total l-[14C] cystine uptake, cells were incubated with 400 µM radio-labeled cystine with no competing substrate. As a negative control, a reaction containing no cells was incubated with l-[14C] cystine and no radioactivity was detected.
Growth kinetics under cystine starvation
Overnight cultures were diluted 1:20 in triplicate into sterile microtitre plates containing modified minimal medium (MM) (56 mM glucose, 13.6 mM l-glutamic acid, 7 mM l-leucine, 19 mM NH4Cl, 20 mM K2HPO4, 11 mM KH2PO4, 50 mM NaHCO3, 4.9 mM MgSO4 · 7 H2O, 0.1 mM MnCl2 · 4H2O, 72 µM FeSO4 · 7 H2O, 5.5 mM sodium pyruvate, 2.6 µM riboflavin, 1.4 µM thiamine-HCl, 0.4 µM biotin, 8 µM nicotinic acid, 0.7 µM ρ-aminobenzoic acid, 1 µM calcium pantothenate, and 5 µM pyridoxal-HCl, and buffered with 0.05 M Tris-maleate (pH 7.4) to a final pH of 7.1 [12], and modified MM supplemented with 1 mM l-cystine (MMC). Growth kinetics were monitored using a Bioscreen C automated growth reader (Lab Systems) and optical density measurements obtained at 600 nm was plotted against time to obtain growth curves for each strain in specific growth medium.
Quantitative real-time PCR analysis of cysBPA expression
To determine the effects of L-cystine starvation on S. mutans tcyA, tcyB, tcyC transcription, quantitative real-time PCR was performed using the following primers listed in 5’ to 3’ direction. TcyA-F, CGTTACCCTAACCCAACGTC: TcyA-B, TCACCACCAATCTTACCCTTG, TcyB-F: TGTTCAGGTTTACCGACGTG, TcyB-B: CAAGAGAGGTTCCCTTGGTC, TcyC-F: AACCTTTTTGAGCGTCGGAC, TcyC-B: TCAGAAAGTCCAACCTTGGC, TcyR-F: ACCGAGGAGAGATTGACTTTG, TcyR-B: ACAAGCAGGAGAAGCCACTG, 16S-F: CTTACCAGGTCTTGACATCCCG, 16S-B: ACCCAACATCTCACGACACGAG. Strains were grown in modified MM supplemented with and without 1 mM l-cystine to the mid-log phase. Total RNA was harvested as described by Hanna et al [13]. A First Strand cDNA synthesis kit (MBI Fermentas) was used according to the manufacturer's specifications to generate single-stranded cDNA from 1 µg of DNA-free RNA samples. To ensure that there was no contaminating DNA, a reaction mixture without template RNA and another lacking reverse transcriptase were set-up as negative controls.
For real-time expression analysis, a relative quantification based on the relative expression of a target gene versus a reference gene was used. Comparison of the expression of each target gene between its control and test conditions was determined according to the following formula [14]: Ratio = (Etarget)ΔCt (control-test) / (Eref)ΔCt (control-test). S. mutans 16S rRNA gene was used as an internal reference as expression of this gene did not vary under the experimental assay conditions used (data not shown).
RESULTS AND DISCUSSION
Analysis of the operon encoding the TcyABC transporter
Sperandio et al recently reported a cysteine synthesis regulator, encoded by cysR, in S. mutans [8]. They also identified a potential cystine uptake system, TcyABC, encoded by NCBI locus identity tagsSMU.459, SMU.460, SMU.461, and further demonstrated that activation of tcyABC transcription was modulated by the CysR regulator [8]. Here, we sought to characterize this cystine transport system. Through a BLASTP search using the Transport Classification Database (www.tcdb.org) we found that tcyA, tcyB and tcyC encoded an amino acid ABC transporter binding protein (273 aa), an amino acid ABC transporter permease protein (267 aa) and an amino acid ABC transporter ATP-binding protein (247 aa), respectively. The tcyABC ORFs showed significant homology with the tcyJ, tcyM, and tcyN genes in B. subtilis, which are part of the ytmI operon encoding an l-cystine ABC transporter [4]. TcyA was homologous (30% identity; 72/240) to the TcyJ (YtmJ) solute binding protein, TcyB exhibited 34% identity (78/224) to the TcyM (YtmM) permease, and TcyC was homologous (53% identity; 127/238) to the B. subtilis TcyN (YtmN) ATP-binding protein.
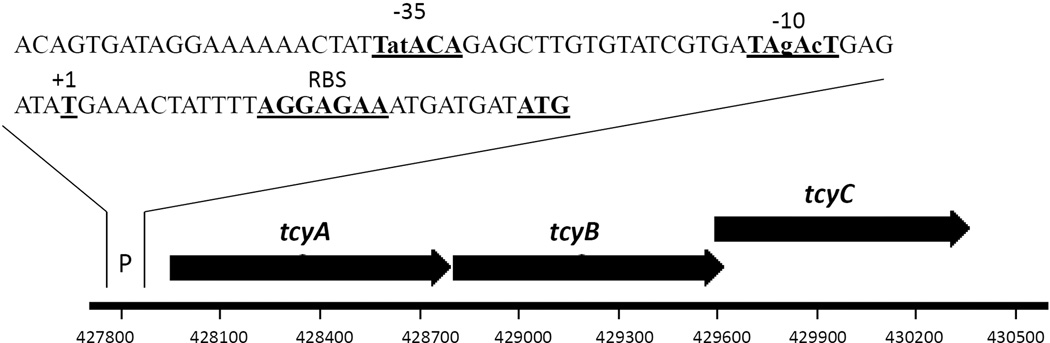
Using northern blot analysis, we detected a single mRNA transcript of ~ 2.3kb in wild-type S. mutans cells that was consistent with the co-transcription of tcyA, tcyB, and tcyC (data not shown), confirming that these genes are part of a tricistrionic operon. Using 5’RACE-PCR, we determined the transcriptional start site of the tcyABC operon to be located 8 nucleotides downstream of an inferred Pribnow box and 26 nucleotides upstream of the translational start site which is consistent with tcyABC comprising a transcriptionally discrete locus in the S. mutans UA159 genome (Figure 1).
FIGURE 1. Organization and promoter map of the tcyABC tricistronic operon in S. mutans.
We also identified a cysteine aminopeptidase C gene (SMU.466) downstream of the tcyABC locus. In Lactococcus lactis, the PepC aminopeptidase has broad substrate specificity and hydrolyzes napthylamide-substituted amino acids and di- and tri-peptides; its activity can be inhibited by thiol-group-blocking compounds but not by serine or metalloproteinase inhibitors [15, 16]. The presence of cysteine aminopeptidases suggests that S. mutans can access smaller peptides to free up amino acids such as cysteine.
l-cystine uptake and substrate specificity of the TcyABC system
To confirm the role of TcyABC in cystine uptake, we tested the abilities of S. mutans UA159 wild-type strain and its ΔtcyABC mutant to transport l-[14C]cystine. As shown in Figure 2, a significant (55%) decrease in l-[14C]cystine uptake was observed in the ΔtcyABC mutant SmTcyABC compared with wild-type UA159 cells (0.48 ± 0.13 nmol/mg versus 1.06 ± 0.49 nmol/mg dry cell/min, respectively) over 8 min, consistent with TcyABC being involved in l-cystine transport. In both the mutant and wild-type UA159 strains, l-cystine uptake was highest in the first 2 min, and then continued linearly at a decreased rate, tapering off in both strains after 10 min. Not surprisingly, transport of l-cystine was not completely abolished in the mutant, given that there are other transporters involved in the uptake of l-cystine in S. mutans. Studies of cystine uptake in B. subtilis have also shown the highest uptake in the first 2 min at a rate of 1.9 nmol/min/mg of protein, which then continued linearly up to 6 minutes and began to plateau, while a ΔtcyJKLMN mutant showed a decreased cystine transport rate of 1.4 nmol/min/mg of protein relative to the parent strain [4]. It is likely that that the drastic impairment of cysteine uptake in S. mutans was due to the presence of only two cysteine transport systems in this bacterium, which is in contrast to that of B. subtilis equipped with three cystine transport systems.
FIGURE 2. Time course for l-[14C]cystine uptake in S. mutans UA159 wild-type strain and its SmTcyABC mutant.
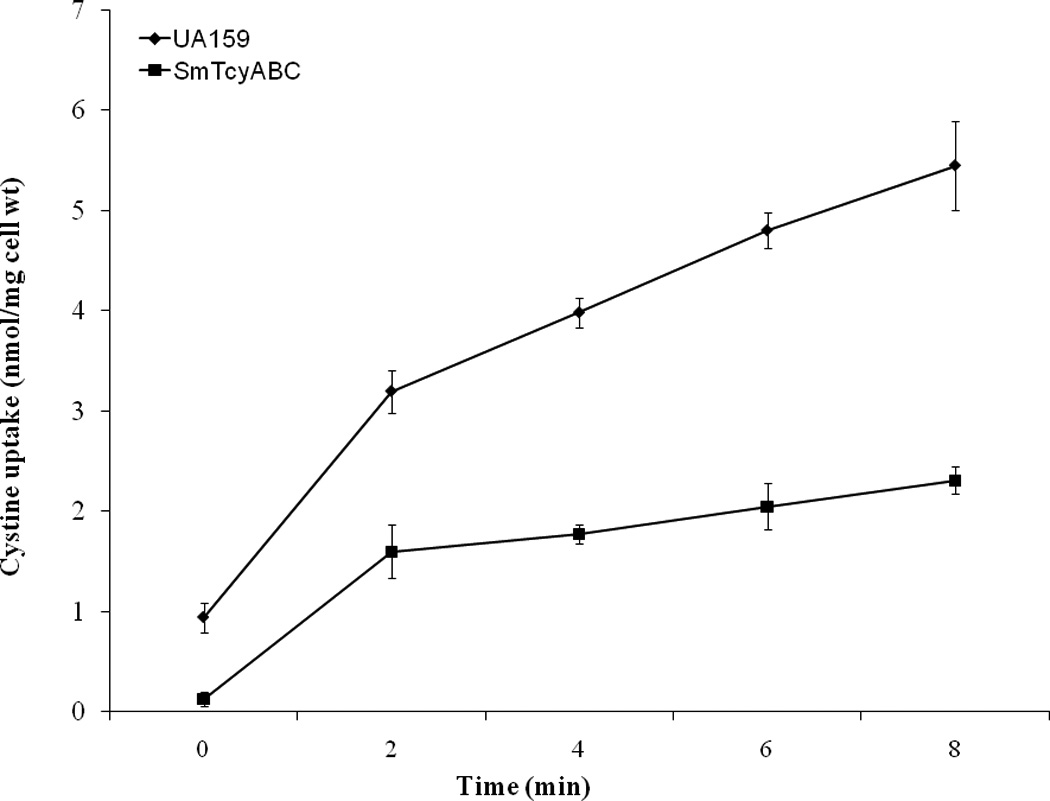
Cystine uptake was examined in the presence of 4 µM l-[14C]cystine and 200 µM of unlabeled l-cystine. Briefly, at time zero, l-[14C]cystine and cold l-cystine were added at a concentration of 4 µM (2.6 mCi/mmol) and 200 µM, respectively, and the reaction mixtures were incubated at 37°C. Samples (100 µl) were removed at regular intervals, immediately filtered and the filters washed and transferred scintillation vials for determination of radioactivity. Tansport experiments were carried out using three independent cultures and each time point was sampled in duplicate. Dry cell weights (mg) were used for normalization. Final rate of uptake was calculated as nmol of substrate per mg of cells per minute.
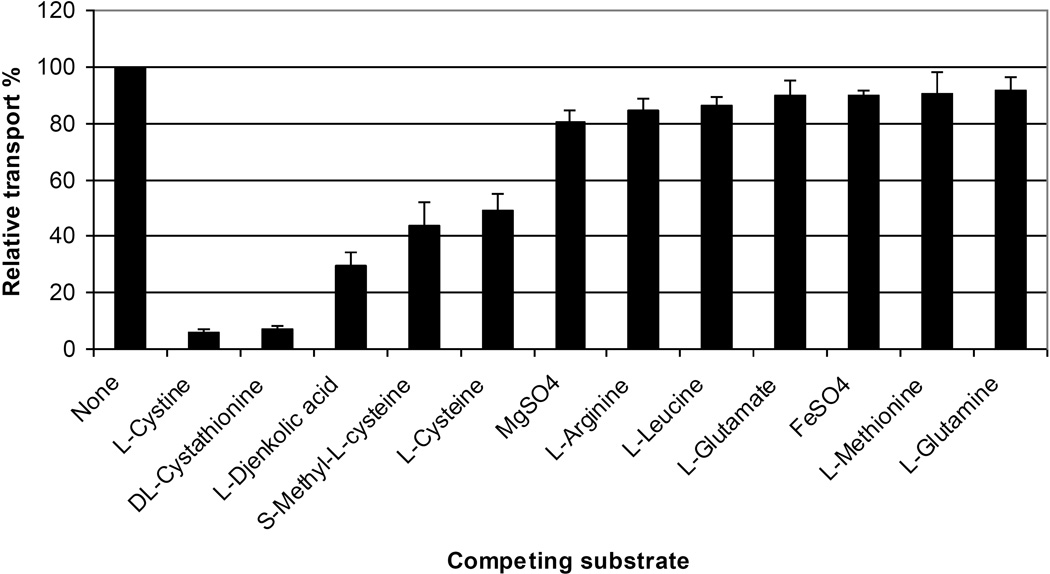
To determine the substrate specificity of the S. mutans TcyABC transport system, we measured the level of inhibition of l-[14C]cystine uptake in the presence of a 100-fold excess of different unlabeled amino acids and sulfur-containing compounds in the wild-type strain and its ΔtcyABC mutant. When unlabeled l-cystine was added in excess, a 94% decrease in l-[14C]cystine uptake was observed in the wild-type strain confirming the transporter specificity for l-cystine. In the wild-type strain, unlabeled cystathionine, djenkolic acid, S-methyl-l-cysteine, and cysteine competitively inhibited cystine uptake by > 50% (Figure 3) while arginine, glutamine, glutamate, leucine, and methionine did not effectively inhibit l-cystine uptake. In contrast, the SmCysBPA mutant showed very little l-cystine uptake and no inhibition by unlabeled l-cystine, djenkolic acid, and S-methyl-l-cysteine. Moreover, addition of an excess of these unlabeled compounds did not completely inhibit cystine uptake as determined by the relative transport of these substrates at 81.2% ± 8.3, 91.5% ± 16.2 and 87.8% ± 5.8, respectively. When other competing substrates indicated in Figure 3 were tested in the mutant, l-cystine was still transported into the cells despite the absence of the TcyABC system (data not shown), confirming the presence of other cystine transporters in S. mutans. Our results show that in addition to l-cystine transport, the TcyABC transporter participates in the uptake of l-cysteine, dl-cystathionine, l-djenkolic acid, and S-methyl-l-cysteine in S. mutans.
FIGURE 3. Relative transport % of l-[14C]cystine uptake in S. mutans UA159 in the presence of a 100-fold excess of non-radioactive compounds.
The specificity of tcyABC-mediated amino acid uptake was examined using an amino acid competition assay. Briefly, the uptake of l-[14C]cystine (4 µM) was measured in the presence of 400 µM of the following cold l-amino acids: arginine, cysteine, glutamine, glutamate, leucine, and methionine. As a positive control and to determine total l-[14C]cystine uptake, cells were incubated with radio-labeled cystine with no competing substrate. As a negative control, a reaction containing no cells was incubated with l-[14C]cystine and no radioactivity was detected. Rate of uptake was calculated using three independent experiments, each subjected to duplicate sampling procedures. Uptake was normalized using dry cell weights and final uptake was calculated as nmol of substrate per mg of cells per minute.
Regulation and expression of tcyABC under cystine starvation
The two transcriptional activators CysR and HomR are positive regulators of the TcyABC and TcyDEFGH L-cystine transport systems respectively [8]. Our search of the S. mutans UA159 genome revealed another putative LysR-type transcriptional regulator locus (SMU.2060) designated TcyR with homology (24%, 65/263) to the B. subtilis YtlI regulator. To determine the role of TcyR on the expression of the tcyABC operon, we constructed a TcyR insertion mutant (SmTcyR) and tested it under cystine starvation conditions. Gene expression was analyzed by quantitative real-time RT-PCR using cDNAs derived from S. mutans UA159 and mutant strains grown in modified MM with or without cystine. Relative to their expression in UA159, absence of TcyR resulted in an approximate 10.8-, 13.1-, and 5.2-fold induction of tcyA, tcyB, and tcyC respectively, under cystine-starvation relative to the cystine-fed state (Figure 4). These results indicate that TcyR has a negative transcriptional role on the expression of tcyABC during cystine limited conditions. LysR-type transcriptional regulators (LTTRs) are generally positive regulators in prokaryotes [17]. Interestingly, the B. subtilis YtlI regulator with which the TcyR regulator shares homology, is also a positive regulator. The negative regulator CymRSA in S. aureus [7] showed no homology to our negative regulator TcyR.
FIGURE 4. Relative transcription of genes quantified using real-time PCR.
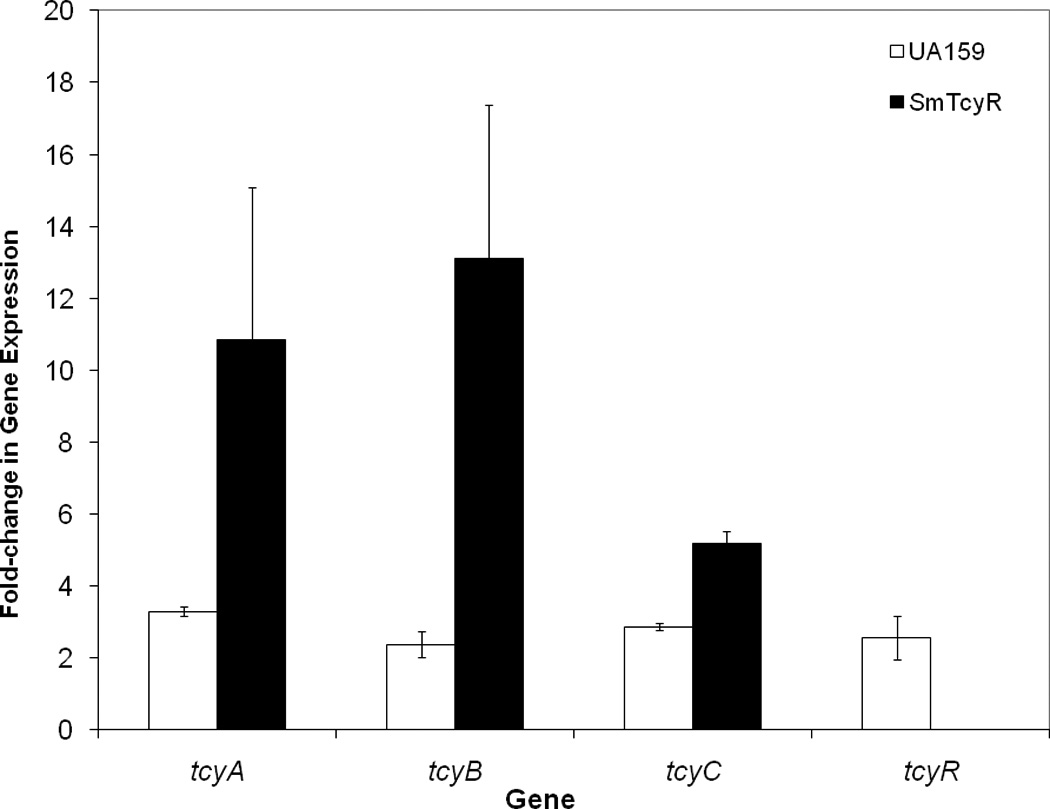
S. mutans UA159 and SmTcyR cultures were grown in Modified Minimal medium in the presence and absence of 1 mM l-cystine and used for RNA isolation and cDNA synthesis. For each strain, fold expression was calculated relative to that in cystine-added culutres whose expression was set at an user-defined value of 1.0. Results represent the mean expression from three independent experiments plus/minus std, error, each conducted in triplicate.
We also found that under cystine starvation, UA159 cells showed up-regulation of the tcyABC and tcyR genes (Figure 4). More specifically, the tcysA, tcyB, and tcyC genes were upregulated by 3.3-, 2.4-, and 2.8-fold, whereas expression of tcyR was increased by ~2.6-fold, thus suggesting induction of the transporter genes and its regulator under cystine-deprived nutritional conditions. This capability is likely advantageous under dense biofilm growth where conditions can become anaerobic and access to nutrients and free amino acids may be limited. In this environment, where cells are likely trying to scavenge cystine or cystine amino acid analogues, upregulation would be an advantage.
Growth of S. mutans under cystine starvation
To determine the effect of cystine starvation on S. mutans UA159 growth, wild type and mutant strains were grown in MM medium devoid of cysteine-HCl and growth was monitored using an automated optical density reader. Under cystine starvation, growth kinetics of both SmTycABC and TcyR mutants as well as the wild type UA 159 were drastically affected (Figure 5). In UA159, cystine starvation resulted in a lower growth yield as well as a longer doubling time (Td ~ 93.3 ± 0.7 min) compared with its growth in the presence of cystine (Td ~ 76.3 ± 1.5 min), indicating that l-cystine is required for optimal growth of S. mutans. However, growth was completely abolished in SmTycABC under cystine starvation. Supplementing the modified growth medium with 0.1 mM cystine slightly improved the drastic growth impairment of the SmTcyABC mutant (Td ~ 118.2 ± 0.8 min).
FIGURE 5. Growth of S. mutans UA159, SmTcyABC, and SmTcyR strains in modified minimal medium in the presence or absence of l-cystine.
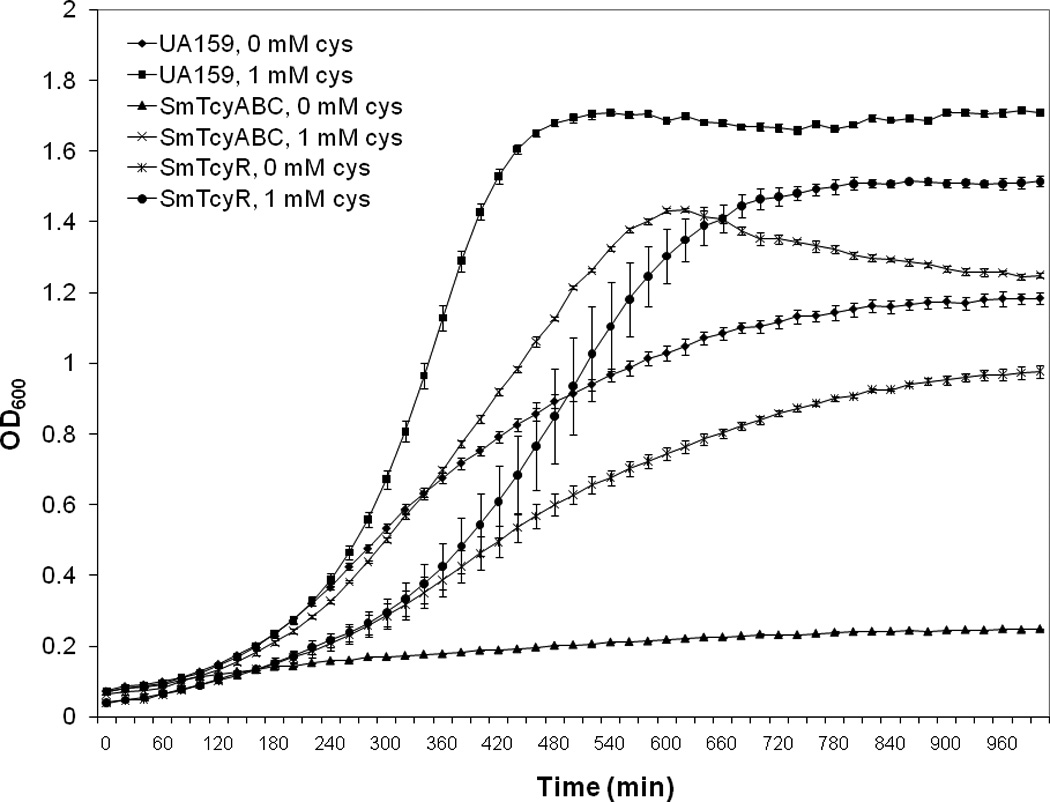
Results are representative of multiple experiments and error bars represent std. deviation.
Similar to the SmTcyABC transporter mutant, the TcyR-deficient mutant (SmTcyR) had a longer doubling time (Td ~ 117.2 ± 3.8 min) under cystine-supplemented (1 mM) conditions relative to wild-type (Figure 5). In contrast to SmTcyABC, SmTcyR was able to survive under cystine-deficient conditions, although its doubling time was remarkably increased relative to wild type (Td ~ 261.0 ± 11.9 min). Also importantly, growth kinetics of SmTcyR revealed a notable increase in the lag time regardless of the presence of absence of cystine, compared with the wild-type UA159 and SmTcyABC.
We further evaluated the effect on growth by individual components of the TcyABC operon by conducting growth studies on mutants deficient in each gene. Briefly, growth kinetics were monitored for the TcyA, TcyB, and TcyC transporter mutants in modified MM without cystine (Figure 6). The most drastic effect on growth was observed for SmTcyB. Similar to TcyABC, growth of this mutant was completely abolished without cystine. Although, TcyA and TcyC were able to grow in cystine-deficient medium, their growth was tremendously impaired relative to wild type as judged by their longer doubling times; Td ~ 131.3 ± 4.8 min and 214.8 ± 21.5 min, respectively. Sperandio et al [8] also showed impaired growth in the form of pinpoint colonies when their TcyA mutant was grown in chemically defined medium with the addition of cystine as the sole sulphur source. However, they did not investigate the growth of other Tyc ABC mutants. The ability of some of our TycABC mutants to grow in the absence of cystine, albeit in an impaired fashion, suggests that the presence of other amino acids (i.e. glutamate and leucine), inorganic sulfur and/or ammonium sources were sufficient to sustain growth. S. mutans possesses amino acid biosynthetic pathways and even though most amino acids are not freely available in the environment, some strains are able to synthesize all the necessary amino acids required for survival [2, 18].
FIGURE 6. Growth of S. mutans UA159, SmTcyABC, SmTcyA, SmTcyB, and SmTcyC strains in modified minimal medium in the absence of l-cystine.
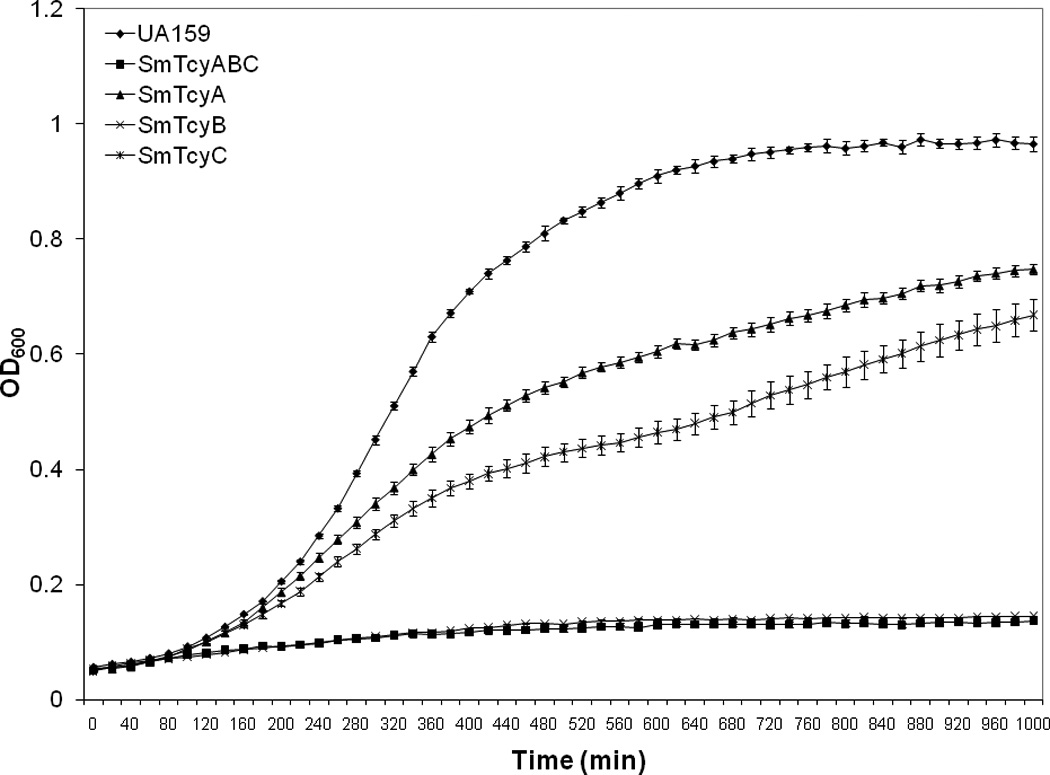
Results are representative of multiple experiments and error bars represent std. deviation.
Conclusions
The ability of S. mutans to scavenge and compete for limited nutrients in the plaque biofilm is an important aspect that confers an ecological advantage, which facilitates its survival and persistence in the oral cavity. The amino acid transport system in S. mutans UA159, encoded by the tcyABC operon that is induced under cystine-starved conditions, functions to maintain growth by transporting cystine into the cell. Furthermore, we also reveal a regulator of the tcyABC locus, encoded by tcyR, whose product appears to have a negative regulatory role on tcyABC transcription when cells are starved of L-cystine during growth. Since cystine appears to be an important nutrient for S. mutans growth, understanding the genetic pathways required for its acquisition satisfies an important step in attempts to modulate the growth and virulence of S. mutans.
ACKNOWLEDGEMENTS
We thank Dr. Joyce Azavedo for help with preparation of this manuscript. This study was supported by NIH grant R01DE013230-03 and CIHR grant MT-15431 to DGC. DGC is a recipient of a Canada Research Chair.
REFERENCES
- 1.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanesi D, Mansilla MC, Schujman GE, de Mendoza D. Bacillus subtilis cysteine synthetase is a global regulator of the expression of genes involved in sulfur assimilation. J. Bacteriol. 2005;187:7631–7638. doi: 10.1128/JB.187.22.7631-7638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burguiere P, Auger S, Hullo MF, Danchin A, Martin-Verstraete I. Three different systems participate in L-cystine uptake in Bacillus subtilis. J. Bacteriol. 2004;186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burguiere P, Fert J, Guillouard I, Auger S, Danchin A, Martin-Verstraete I. Regulation of the Bacillus subtilis ytmI operon, involved in sulfur metabolism. J. Bacteriol. 2005;187:6019–6030. doi: 10.1128/JB.187.17.6019-6030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsson J. Nutritional requirements of Streptococcus mutans. Caries Res. 1970;4:305–320. doi: 10.1159/000259653. [DOI] [PubMed] [Google Scholar]
- 6.Chapot-Chartier M-P, Nardi M, Chopin M-C, Chopin A, Gripon J-C. Cloning and sequencing of pepC a cysteine aminopeptidase gene from Lactococcus lactis subsp. cremoris AM2. Appl. Environ. Microbiol. 1993;59:330–333. doi: 10.1128/aem.59.1.330-333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppee J-Y, Auger S, Turlin E, Sekowska A, Le Caer J-P, Labas V, Vagner V, Danchin A, Martin-Verstraete I. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology. 2001;147:1631–1640. doi: 10.1099/00221287-147-6-1631. [DOI] [PubMed] [Google Scholar]
- 8.Cowman RA, Baron SS, Fitzgerald RJ. Cysteine toxicity for oral streptococci and effect of branched-chain amino acids. Infect. Immun. 1983;39:1107–1113. doi: 10.1128/iai.39.3.1107-1113.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvitkovitch DG, Gutierrez JA, Bleiweiss AS. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J. Bacteriol. 1997;179:650–655. doi: 10.1128/jb.179.3.650-655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Even S, Burguiere P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 2006;188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández M, Kleerebezem M, Kuipers OP, Siezen RJ, Kranenburg RV. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 2002;184:82–90. doi: 10.1128/JB.184.1.82-90.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara S, Kobayashi S, Nakayam H. Development of a minimal medium for Streptococcus mutans. Arch. Oral Biol. 1978;23:601–602. doi: 10.1016/0003-9969(78)90280-7. [DOI] [PubMed] [Google Scholar]
- 13.Gottschalk G. Bacterial metabolism. New York: Springer-Verlag; 1986. [Google Scholar]
- 14.Hulanicka MD, Hallquist SG, Kredich NM, Mojica-A T. Regulation of O-acetylserine sulfhydrylase B by L-cysteine in Salmonella typhimurium. J. Bacteriol. 1979;140:141–146. doi: 10.1128/jb.140.1.141-146.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kredich NM. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 1992;6:2747–2753. doi: 10.1111/j.1365-2958.1992.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 16.Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 17.Leichert LIO, Scharf C, Hecker M. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 2003;185:1967–1975. doi: 10.1128/JB.185.6.1967-1975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu P-Q, Ferro-Luzzi Ames G. In vitro disassembly and reassembly of an ABC transporter, the histidine permease. Biochemistry. 1998;95:3495–3500. doi: 10.1073/pnas.95.7.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macrina FL, Evans RP, Tobian JA, Hartley DL, Clewell DB, Jones KR. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathsam C, Eaton RE, Simpson CL, Browne GV, Valova VA, Harty DWS, Jacques NA. Two-dimensional fluorescence difference gel electrophoretic analysis of Streptococcus mutans biofilms. J. Prot. Res. 2005;4:2161–2173. doi: 10.1021/pr0502471. [DOI] [PubMed] [Google Scholar]
- 22.Saier MH., Jr. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 23.Saier MH, Jr., Goldman SR, Maile RR, Moreno MS, Weyler W, Yang N, Paulsen IT. Transport capabilities encoded within the Bacillus subtilis genome. J. Mol. Microbiol. Biotechnol. 2002;4:37–67. [PubMed] [Google Scholar]
- 24.Sato Y, Noji S, Suzuki R, Taniguchi S. Dual mechanism for stimulation of glutamate transport by potassium ions in Streptococcus mutans. J. Bacteriol. 1989;171:4963–4966. doi: 10.1128/jb.171.9.4963-4966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Ann. Rev. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 26.St. Martin EJ, Wittenberger CL. Regulation and function of ammonia-assimilating enzymes in Streptococcus mutans. Infect. Immun. 1980;28:220–224. doi: 10.1128/iai.28.1.220-224.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terleckyj B, Shockman GD. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]