Abstract
We provide a catalog of 3D cryo soft X-ray tomography (cryo-SXT) images obtained from ~6–12 µm thick mouse adenocarcinoma cells. Included are multiple representative images of nuclei, nucleoli, nuclear membrane, nuclear membrane channels, mitochondria, lysosomes, endoplasmic reticulum, filaments and plasma membrane, plus three structures not previously described by cryo-SXT, namely Golgi, microvilli and nuclear-membrane blebs. Sections from the 3D cryo-SXT tomograms for all the preceding structures closely resemble those seen by thin-section transmission electron microscopy (TEM). Some structures such as nuclear-membrane channels and nuclear-membrane blebs are more easily detected by cryo-SXT than TEM most likely due to their better contrast and cellular preservation in cryo-SXT combined with the ability to rapidly locate these structures within a full 3D image. We identify and discuss two current limitations in cryo-SXT: variability in image quality and difficulties in detecting weaker contrast structures such as chromatin and various nuclear bodies. Progress on these points is likely to come from the solution of several technical problems in image acquisition, plus the implementation of advanced cryo soft X-ray microscopy approaches such as phase contrast or optical sectioning.
Keywords: soft X-ray tomography, mammalian cells, ultrastructure
Introduction
Cryo soft X-ray tomography (cryo-SXT) has a number of advantages for the examination of biological specimens (Weiss et al., 2000;McDermott et al., 2009;Larabell and Nugent, 2010). Chief among these is the ability to obtain a 3D image of a moderately thick (5–15 µm) intact cell with no chemical fixation, dehydration, chemical staining or physical sectioning. Instead, cryo-SXT acquires images of cryo-preserved specimens in an absorption contrast mode. It bypasses chemical staining by operating at a wavelength of light (2.3 – 4.4 nm) called the water window at which organic material is strongly absorbing compared to water. Physical sectioning is obviated because in cryo-SXT a tilt series of images is collected of the frozen, hydrated specimen and then a tomographic reconstruction is performed to generate the final 3D image. This reconstruction can then be used to obtain virtual sections of the cell. The imaging depth in cryo-SXT is up to 15 µm, and is determined by the penetration depth of X-rays through organic material at the water-window wavelength.
Current resolution limits of cryo-SXT are approaching 10 nm (“half-pitch” resolution) for test objects (Chao et al., 2005;Rehbein et al., 2009), and down to 36 – 70 nm (equivalent to 18–35 nm “half-pitch” resolution) for biological specimens ranging in thickness from 2–12 µm thick (Carrascosa et al., 2009;Schneider et al., 2010). With these resolutions, cryo-SXT does not yet compete with conventional transmission electron microscopy (TEM) or cryo-electron microscopy, where resolutions on the order of 5 nm or less are possible (Lawson et al., 2011). However, cryo-SXT provides a complementary approach that enables examination of thicker intact cells. In this way, cryo-SXT shares some features with focused ion beam / scanning electron microscopy (FIB/SEM) (Bushby et al., 2011), which can be used to examine even thicker specimens than possible by cryo-SXT. At the moment, FIB/SEM is largely restricted to chemically fixed and stained specimens, while cryo-SXT permits examination of unstained, frozen hydrated specimens.
Recent improvements in partially coherent cryo-SXT have made it possible to visualize mammalian cells in 3D (Schneider et al., 2010). Various ultrastructural features have been identified in these initial cryo-SXT images of mammalian cells, but a detailed catalog of what can be seen is currently lacking. In addition, there is no information on the robustness of the technique and the range of variability to be expected in the images of different sub-cellular structures.
The purpose of this study is to produce an atlas of the structures seen so far in our cryo-SXT studies of a mouse adenocarcinoma cell line. This atlas includes many of the structures previously reported (Schneider et al., 2010), but now shown in more detail and with multiple examples to provide a better idea of the range of variability of these structures as detected by cryo-SXT. We also report several structures not previously described by cryo-SXT. Our goal is to provide a set of concrete examples of what is currently possible by cryo-SXT of mammalian cells, so that other researchers have a realistic understanding of the current capabilities of this technology.
Materials and methods
Cryo- SXT
We grew mouse adenocarcinoma cells on custom-made gold grids that we coated with a thin layer (0.1 µm) of cellulose nitrate. We left the cells in their normal growth medium and allowed them to settle onto the grids and adhere to the cellulose nitrate. Before cells were frozen, we added a small volume (2 µl) of a solution of gold-coated silica beads with a diameter of 270 nm that we obtained from C. Graf at the Free University in Berlin. We incubated the beads with the cells for ~1 hr to allow the beads to attach to the grids and cells. We avoided longer incubation times because this led to ingestion of some of beads into the cells.
We froze cells by first blotting the grids with Whatman #1 filter paper to leave a thin layer of fluid, and then plunged the blotted grid into liquid ethane kept at ~ −170°C. Then we examined the vitrified cells with the X-ray microscope at the Helmholtz Zentrum Berlin. We used either a 25 nm or 40 nm zone plate objective. With one of these objectives in place, we used the microscope’s tilt stage to obtain a series of images typically ranging from −60° – +60° at increments of 1°, although smaller ranges of −50° – +50° or −45° – +45° and smaller increments of 0.5° were used for a few samples to compare data collection conditions.
In principle, given the X-ray microscope’s large depth of focus, the tilt series contains a complete projection image of the specimen at every tilt angle. To obtain a virtual 3D image, we performed a tomographic reconstruction using the software package Bsoft (Heymann and Belnap, 2007). We first pre-processed the projection series by flat-fielding it. We calculated an average flat-field image from ten flat-field images acquired under the same conditions as the tilt series. We then divided each image in the tilt series by the average flat field. We also normalized all images to an average intensity of zero and a standard deviation of one in order to correct for the longer exposure times required at higher tilt angles. We eliminated camera edge artifacts by Gaussian smoothing at the image boundaries. Then, we aligned images from the corrected tilt series using the gold-coated silica beads as fiducial markers. Finally, we performed the tomographic reconstruction using a reciprocal space algorithm (Heymann et al., 2008).
For 3D rendering of the cryo-SXT data, we used ImageJ to select points defining the surfaces, and then we rendered those surfaces using the Rhino3D software package (Robert McNeel and Associates, Seattle, WA, USA).
TEM
For TEM, cells were fixed in 2% glutaraldehyde, and stained with 1% osmium tetroxide, and then 0.5% uranyl acetate, followed by dehydration and embedding in epoxy resin. Thin sections of ~90 nm were cut and stained with 0.5% uranyl acetate and lead citrate.
Results
We illustrate below the different types of structures that we have detected by cryo-SXT by providing several examples of each structure typically obtained from different cells. We also compare these structures to the corresponding structure as detected by TEM. We used the same adenocarcinoma cell line for both cryo-SXT and TEM, and examined distinct cells by each modality. The specimens for cryo-SXT were unstained and in a frozen-hydrated state, so contrast was derived from the specimen’s intrinsic X-ray absorption and scattering properties. In contrast, the TEM specimens were stained with heavy metals. Thus the comparison between these two modalities may not necessarily show identical representations of the corresponding structures.
Nucleus: chromatin
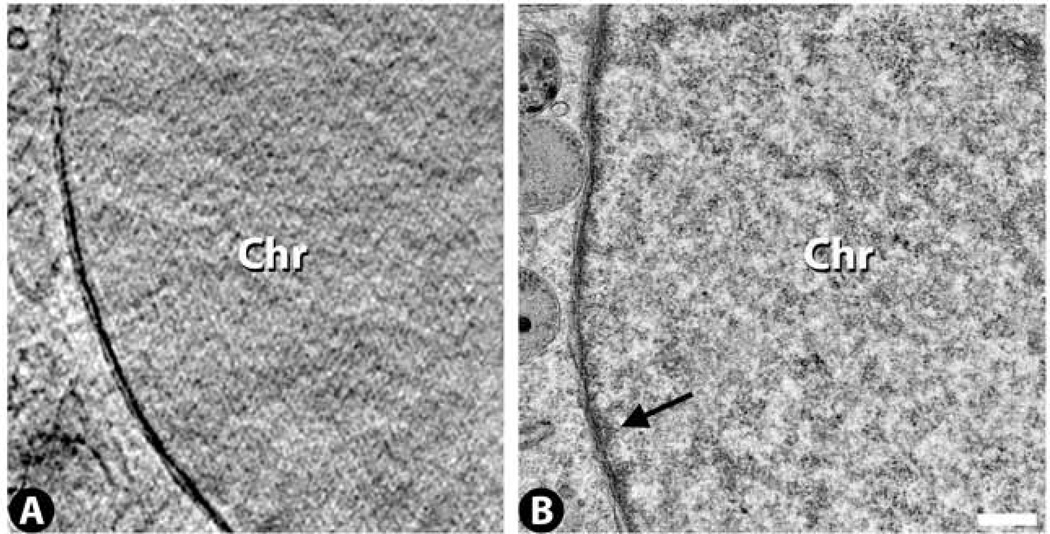
Granular structures are seen throughout the nucleus, and these are roughly similar but of lower contrast compared to what is seen in TEM by conventional uranyl acetate, lead-citrate stained nuclei of the same cell type (Fig. 1A,B). It is not clear by either of these modalities exactly how this granular structure relates to the expected chromatin fibers that should populate the nucleus. The TEM images reveal a slightly more densely stained peripheral layer of heterochromatin, which is relatively sparse in this cell type (arrow in Fig. 1B). This very thin heterochromatic layer is not detectable by cryo-SXT. Also not present in either the cryo-SXT or TEM images are various nuclear bodies such as speckles, Cajal or PML bodies. These structures are typically detected by TEM when special staining protocols are applied (Bernhard, 1969;Spector, 1993) or when special spectral imaging techniques are used (Bazett-Jones et al., 2008).
Figure 1.
Nuclear chromatin (Chr) images from cryo-SXT (A) and TEM (B). The arrow indicates a thin heterochromatic layer adjacent to the nuclear envelope in the TEM image. Scale bar 500 nm.
Nucleus: nucleoli
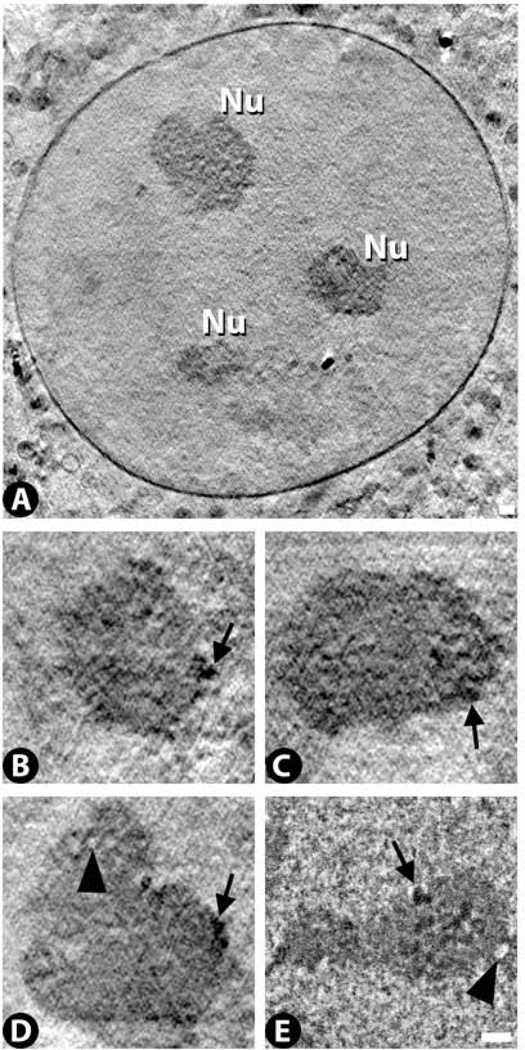
Nucleoli are readily visible by cryo-SXT, and appear as large (~2–4 µm in length) regions that are more X-ray dense than the surrounding nucleoplasm (Fig. 2A). Within nucleoli, X-ray dense and X-ray lucent regions can be identified ranging in size from ~40–300 nm in length (Fig. 2B – D). The stained TEM preparations show comparable sub-regions (Fig. 2E). The smallest X-ray dense regions may correspond to the granular component described by classical TEM morphologists, while the larger X-ray dense regions may correspond to condensed chromatin domains typically found at the periphery of nucleoli (Thiry and Lafontaine, 2005). The X-ray lucent domains may correspond to so-called nucleolar interstices which are thought to be formed by invaginations into the nucleolus of the surrounding less dense nucleoplasmic space (Thiry and Lafontaine, 2005). Indeed, these regions appear to form an interconnected network throughout much of the nucleolus in the 3D cryo-SXT images (Supplementary Material, Video 5).
Figure 2.
Nucleoli images from cryo-SXT (A – D) and TEM (E). An overview of a nucleus containing three nucleoli (Nu) is shown in (A) followed by close-up views of individual nucleoli from this same cell and one other cell in (B – D). Arrows indicate dark granular structures and arrowheads indicate less dense regions found in both the cryo-SXT and TEM images. See also Supplementary Video 5 for a through-focus image of a nucleolus. Scale bar 500 nm.
Nucleus: nuclear membrane
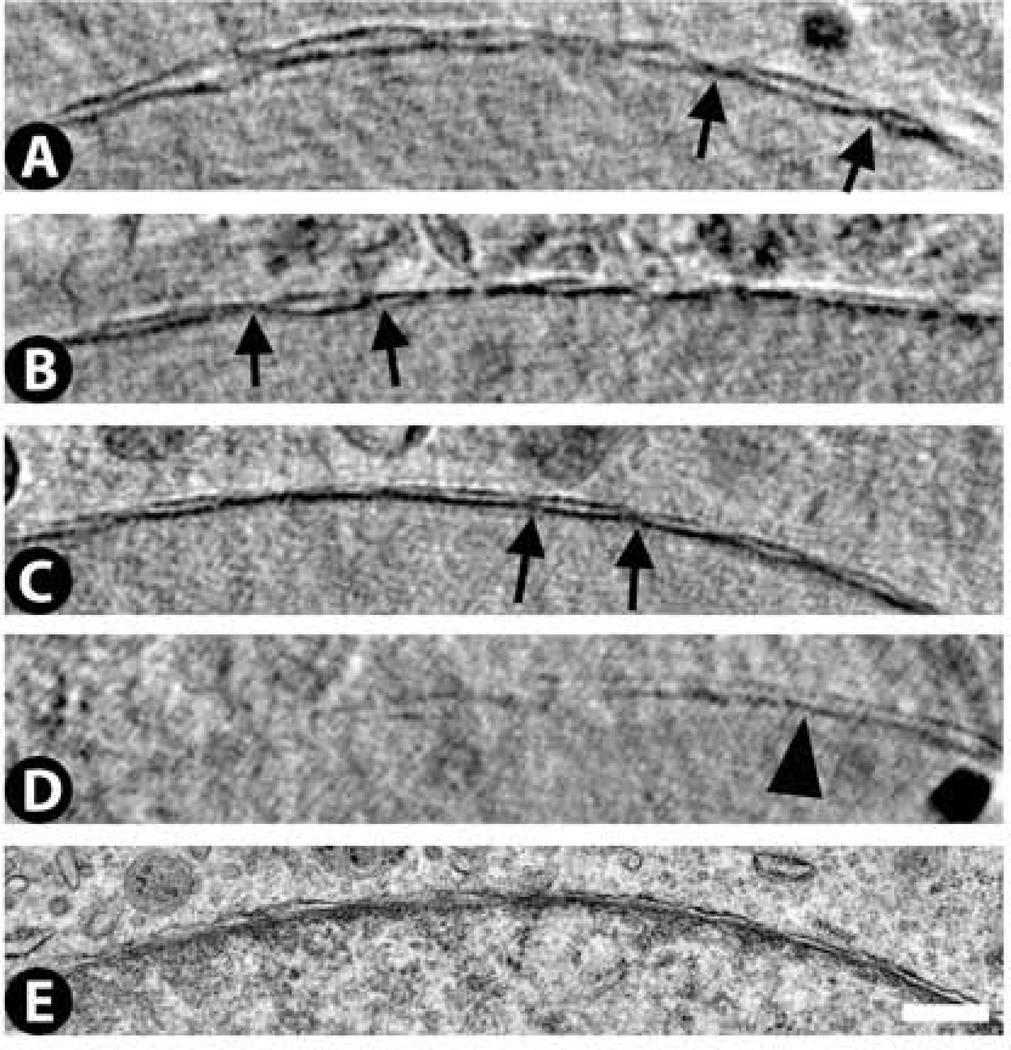
The double nuclear membrane is easily detected by cryo-SXT with the 25 nm zone plate objective. Most segments of nuclear membrane show an undulating appearance with regions of separation punctuated by regions of attachment (Fig. 3A – C). These attachment sites are probably nuclear pores, although the substructure of these pores cannot be discerned by cryo-SXT. Occasionally, some regions of the nuclear membrane can be found where attachment sites are absent and the nuclear membrane instead appears as two parallel single membranes (Fig. 3D). This may reflect a segment of membrane in which pores are absent. By TEM (Fig. 3E), a double membrane is also detected, but the outer cytoplasmic leaflet of the double membrane is ruffled in comparison to the cryo-SXT images. This is probably an artifact of dehydration, as smoother and straighter nuclear membranes, comparable to what we have seen by cryo-SXT, are also detected by cryo-electron microscopy and/or freeze substitution (Studer et al., 2008).
Figure 3.
Nuclear membrane images from cryo-SXT (A – D) and TEM (E). By cryo-SXT, the inner and outer membranes often show an undulating pattern with the points of closest contact probably indicating nuclear pores (arrows in A – C). In some cases, however, the cryo-SXT images reveal longer stretches of parallel inner and outer membranes (arrowhead in D). These features are less evident in the TEM image (E) since the outer membrane is extensively ruffled. Scale bar 500 nm.
Nucleus: nuclear membrane channels
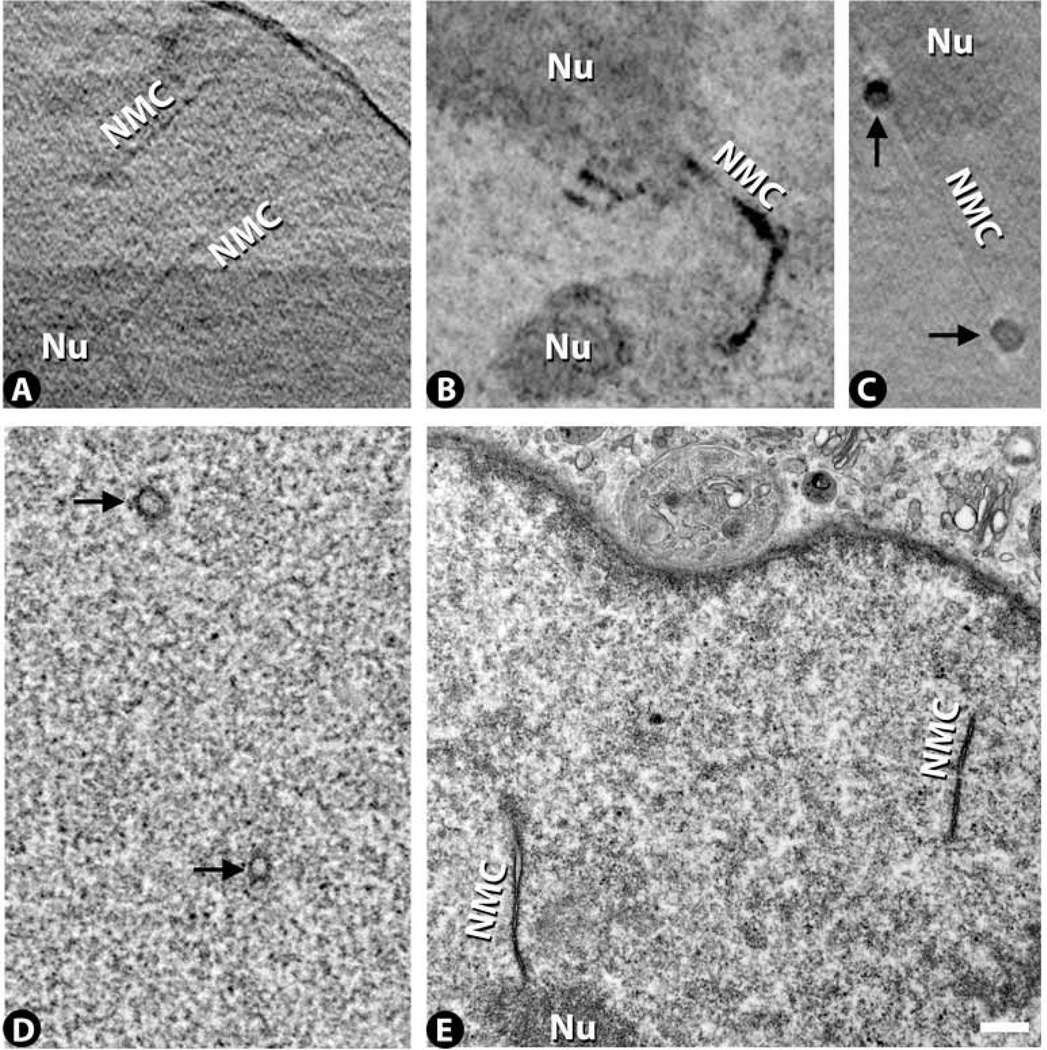
A common feature of many of the nuclei of these adenocarcinoma cells is the nuclear membrane channel. Several of these channels are present in most cells, with some cells exhibiting more than a dozen channels. The channels appear as X-ray dense regions that course through the nucleus in fairly linear paths with lengths often extending for several microns (Fig. 4A – C). The channels are often associated with nucleoli and in most cases also appear to associate with the nuclear membrane. Thus they probably reflect invaginations of the nuclear membrane bringing the cytoplasmic space deeper into the nuclear interior. The channels are quite variable in diameter. Similar channels can also be found in the TEM images (Fig. 4D,E), and have been identified before by TEM and light microscopy (Fricker et al., 1997). While light microscopy enables 3D tracking of the channels, it is at low resolution. Conversely, while electron microscopy provides high resolution images of the channel, tracking them in 3D is difficult. Thus cryo-SXT provides a good compromise that enables good 3D tracking at reasonably high resolution (see Fig. 3a in (Schneider et al., 2010) for examples of channels tracked in 3D).
Figure 4.
Nuclear membrane channel (NMC) images from cryo-SXT (A – C) and TEM (D,E). The channels exhibit varying diameters with thin channels illustrated in (A) and thick channels illustrated in (B). The channels often terminate near nucleoli (Nu). Cross sections through channels are often found (arrows - C,D). Note the thin channel seen in longitudinal section running between two thicker channels seen in cross section (C). Scale bar 500 nm.
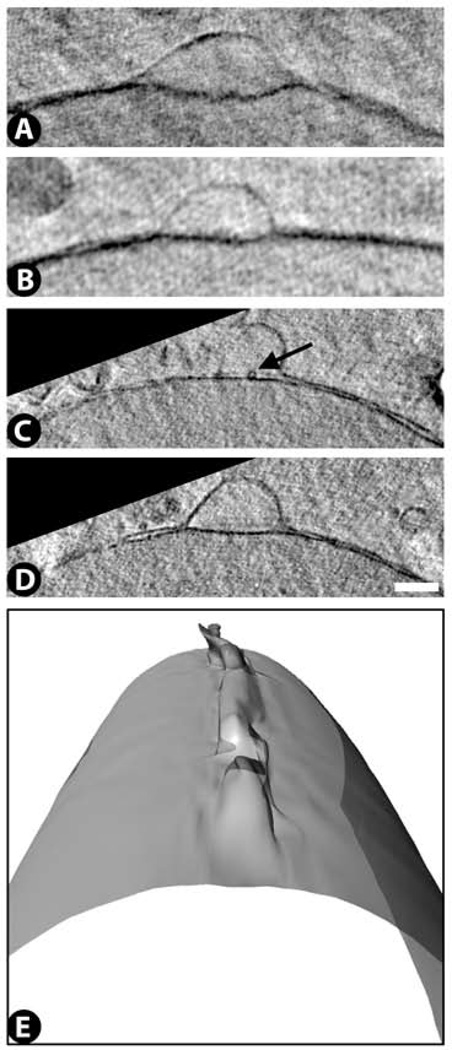
Nucleus: nuclear membrane blebs
These appear in sections as Gaussian-shaped protrusions of the outer nuclear membrane. They are ~1 µm wide and ~0.5 µm high (Fig. 5A – D). As seen in 3D, they form extended ridges up to ~2 µm in length that run along the nuclear membrane in a direction perpendicular to the cell substrate (Fig. 5E). Typically, one to two nuclear membrane blebs can be found in most nuclei. Occasionally some of these blebs show hints of small circular structures present in between the inner and outer membrane leaflets. Higher resolution images will be necessary to better characterize these sub-structures.
Figure 5.
Nuclear membrane bleb images from cryo-SXT (A – E). The blebs arise as an evagination of the outer nuclear membrane. Occasionally a small circular sub-structure can be identified in between the inner and outer membranes of the bleb (arrow in C). A rendering (E) of the outer nuclear membrane surface from panels C,D shows that these blebs can extend along the nuclear surface for up to 2 µm. See Supplementary Video 6 for a 3D movie of this surface rendering. Comparable membrane bleb structures were not detected by TEM in this study, although similar blebs have been seen in several other electron microscopy studies examining Herpes simplex virus egress (Skepper et al., 2001;Mettenleiter et al., 2009). Scale bar 500 nm.
We have not been able to identify nuclear membrane blebs in the TEM images of the same cell type, which might either reflect the difficulty of preserving membrane structures by conventional TEM procedures (Studer et al., 2008) or an artifact of the cryo-preservation procedure for cryo-SXT. Apparently similar separations of the nuclear membranes have been observed by electron microscopy in the egress of Herpes Simplex Virus – 1 capsids from the nucleus (Skepper et al., 2001;Mettenleiter et al., 2009) using either frozen-hydrated sections or high-pressure frozen / freeze-substituted sections. The electron microscopy images of these sections show a viral particle embedded between the inner and outer nuclear membranes.
Cytoplasm: mitochondria
Mitochondria are frequently observed by cryo-SXT (Fig. 6A – C,E,F). They can be recognized by their internal membranes or cristae, which in this adenocarcinoma cell are typically either linear (Fig. 6B) or curved (Fig. 6A,C). When visualized in 3D the cristae are seen to form mostly sheets, either flat or curved (Fig. 6E,F). Sometimes small connections are found between either the cristae sheets or the inner membrane of the mitochondrion (Fig. 6E,F). By cryo-SXT we cannot detect the double membrane of these cristae which by TEM (Fig. 6D) yields a separation of ~15 nm.
Figure 6.
Mitochondria images from cryo-SXT (A – C,E) and TEM (D). Cristae are evident in all images, but their double membrane structure is only visible by TEM. A rendered image (F) derived from the 3D cryo-SXT image (slice shown in E) of a mitochondrion shows that the cristae form sheets with small connections between them (arrows). See Supplementary Video 7 for a 3D movie of this surface rendering. Scale bar 500 nm.
Cytoplasm: lysosomes
Lysosomes can also be identified by cryo-SXT. They appear in these adenocarcinoma cells as circular, membrane bound organelles of ~750 nm diameter (Fig. 7A – C). They contain a number of small, X-ray dense inclusions (diameter of <50 nm), which are often grouped in clusters. Similar inclusions are present in lysosomes imaged by TEM (Fig. 7D). However, the TEM images also reveal several larger membrane bound inclusions that are either electron dense or electron lucent (Fig. 7D), which are not clearly discernable in the cryo-SXT images. This could reflect differences in imaging between cryo-SXT and TEM, but it might also reflect the broad range of lysosomal / endosomal morphologies that can be present in cells.
Figure 7.
Lysosome images from cryo-SXT (A – C) and TEM (D). Small granular inclusions (arrows) are visible by both cryo-SXT and TEM, but larger membrane-bound inclusions visible by TEM (arrowhead indicates one of several such structures in D) are not clearly visible in the cryo-SXT images. Scale bar 500 nm.
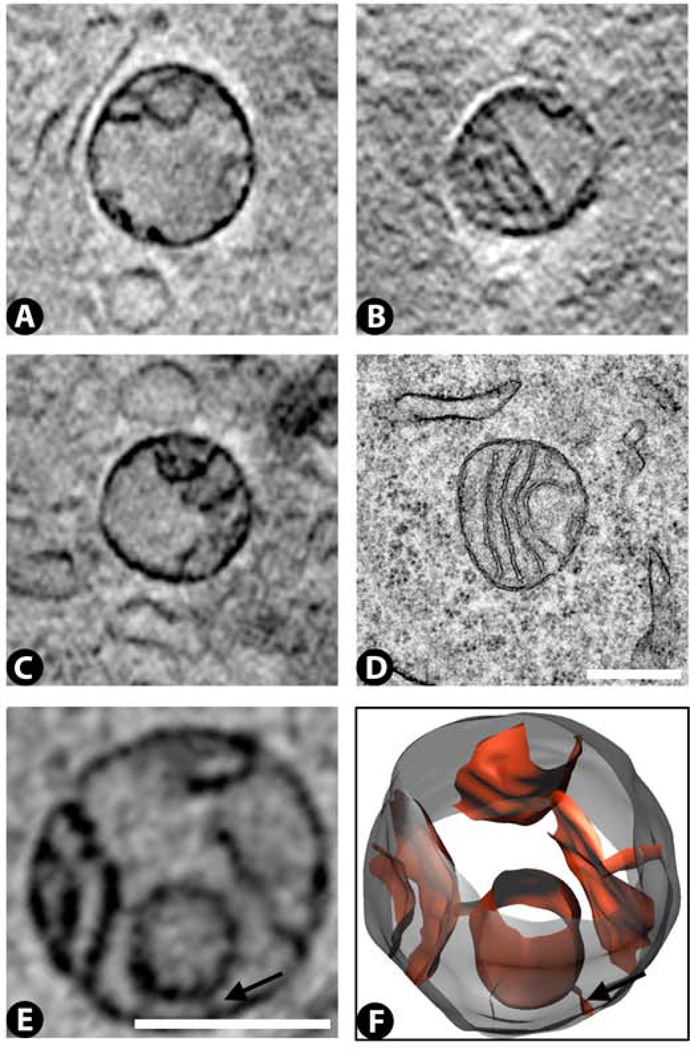
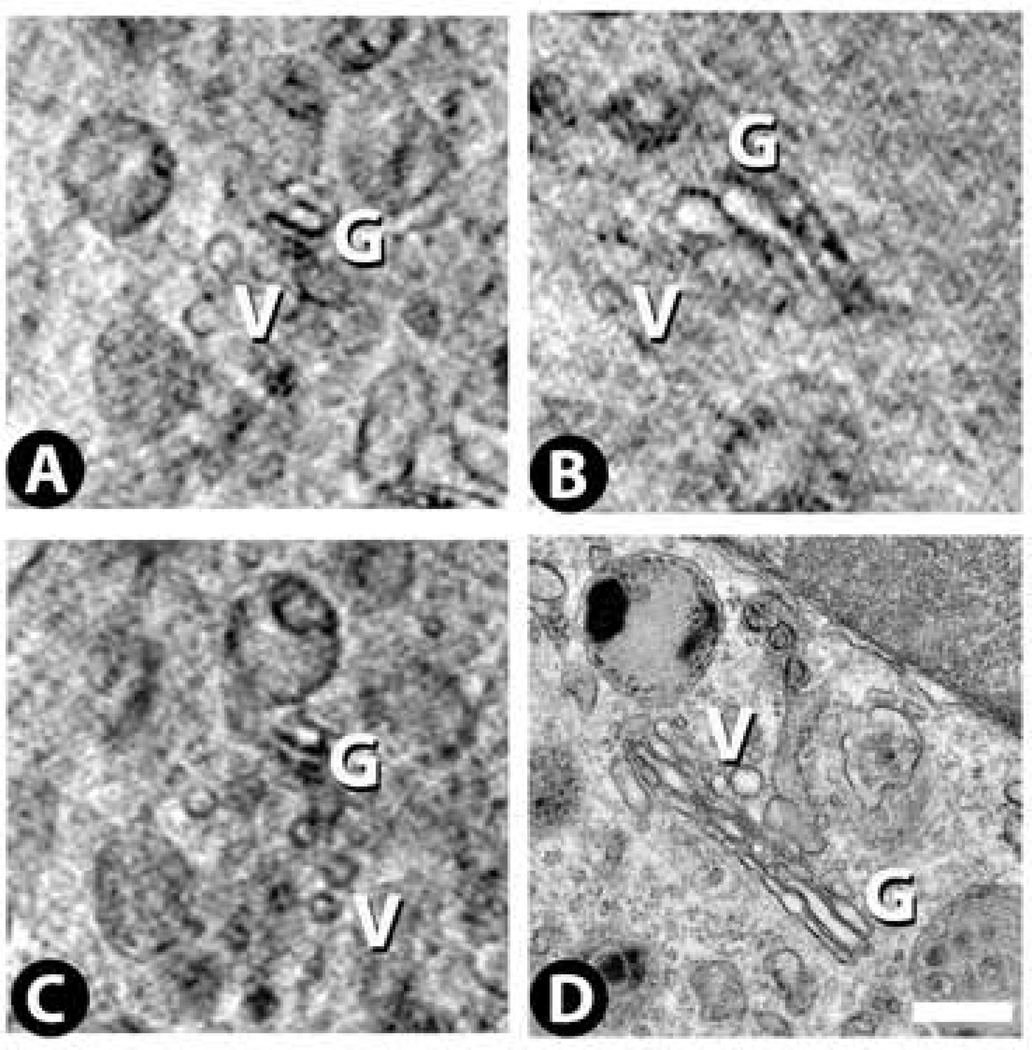
Cytoplasm: Golgi
We have been able to detect Golgi bodies in only one of the 27 cryo-SXT tomograms (Fig. 8A – C). This tomogram was obtained with the 25 nm zone plate and also showed the best preservation for all of the other cellular organelles, although these other organelles were also visible in many other tomograms obtained with either the 25 nm or 40 nm zone plates. In cryo-SXT, the Golgi appear similar to what we have seen by TEM (Fig. 8D), namely several parallel cisternae with small vesicles (~80 – 200 nm) also in the vicinity. By TEM these adenocarcinoma cells exhibit a few Golgi per cell, so they are relatively rare in this cell type.
Figure 8.
Golgi (G) images from cryo-SXT (A – C) and TEM (D). Small vesicles (V) are often found near these structures. Scale bar 500 nm.
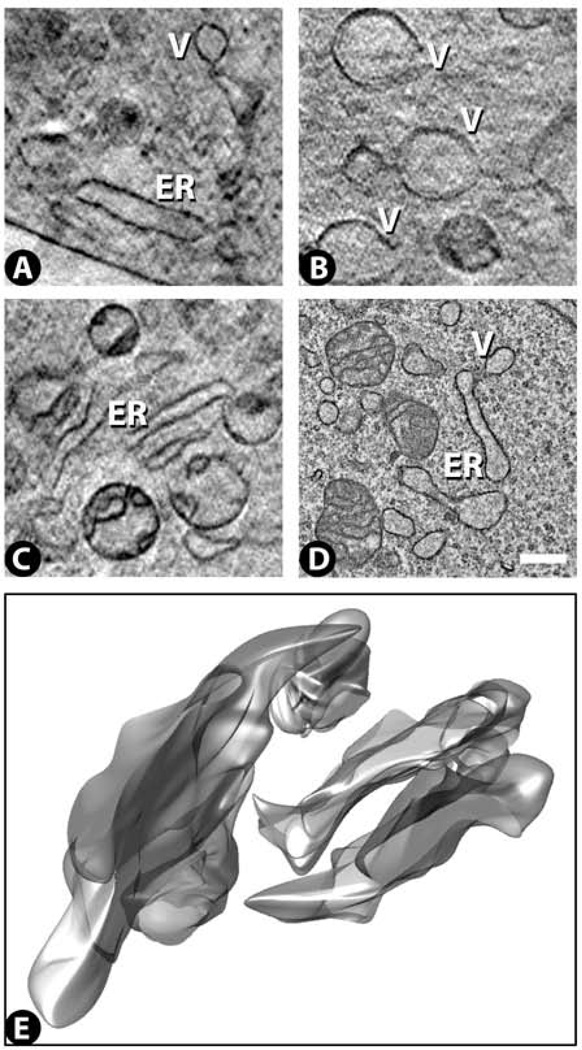
Cytoplasm: endoplasmic reticulum and large vesicles
Many of the cryo-SXT images reveal endoplasmic reticulum. These appear as membrane bound cisternae which are often elongate and present in clusters of 2–4 cisternae frequently arranged roughly in parallel (Fig. 9A,C). Typical cisternae are from ~500–1500 nm in width. Large (~900 nm), apparently empty vesicles are also detected in the cryo-SXT images (Fig. 9B), often in the neighborhood of the endoplasmic-reticulum cisternae. Similar cisternae and vesicles are also seen by TEM (Fig. 9D). 3D renderings of the cryo-SXT images show that the cisternae form roughly planar sheets that often contain bulbous protrusions (Fig. 9E).
Figure 9.
Endoplasmic reticulum (ER) images from cryo-SXT (A – C) and TEM (D). Large vesicles (V) are often found near these structures. The cisternae are seen to form extended sheets in 3D (slice in C rendered in E). See Supplementary Video 8 for a 3D movie of this surface rendering. Scale bar 500 nm.
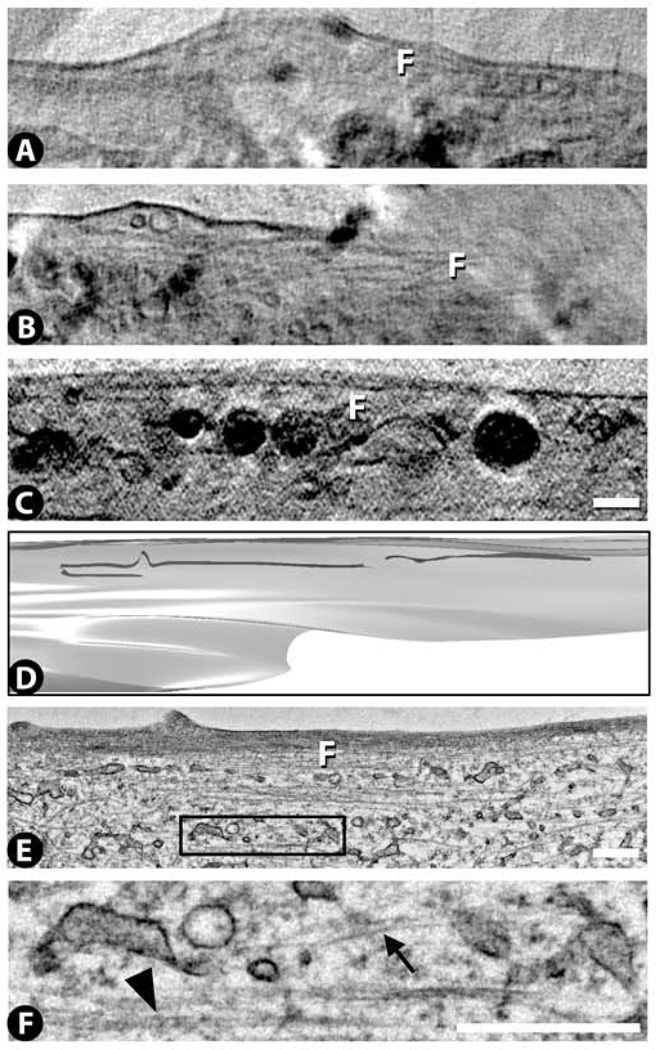
Cytoplasm: filaments
Filaments can be found in these cells by cryo-SXT, but only in thin cytoplasmic extensions that extrude from the main body of the cytoplasm (Fig. 10A – D). The filaments are roughly parallel to the long axis of the cytoplasmic extension, and are normally visible near or adjacent to the plasma membrane. Filament lengths range from ~1–5 µm, and their diameter is at the limit of resolution, as they appear to be as thick as the plasma membrane. Filaments are also found by TEM in thin cytoplasmic extensions (Fig. 10E). Here two different types of filaments can be discerned (Fig. 10F): thicker ones (~28 nm in diameter) which probably reflect microtubules and thinner ones (~6 nm in diameter) which probably reflect actin microfilaments. Thus based on the TEM images, the cryo-SXT images could conceivably be detecting both microtubules and microfilaments, but these cannot be distinguished from each other since both thicknesses are below the current resolution limit of cryo-SXT. However, we believe that the filaments that we have detected by cryo-SXT are more likely to be microtubules, since microtubules are thicker and so would produce more contrast than the thinner microfilaments.
Figure 10.
Filament (F) images from cryo-SXT (A – D) and TEM (E,F). The filaments were consistently observed in thin cytoplasmic extensions. The 3D rendered view in panel D is derived from the cryo-SXT image in panel C. See Supplementary Video 9 for a 3D movie of this surface rendering. High magnification views in TEM reveal both microfilaments (arrow) and microtubules (arrowhead) (F is a magnified view of the black rectangle in E). Scale bars 500 nm.
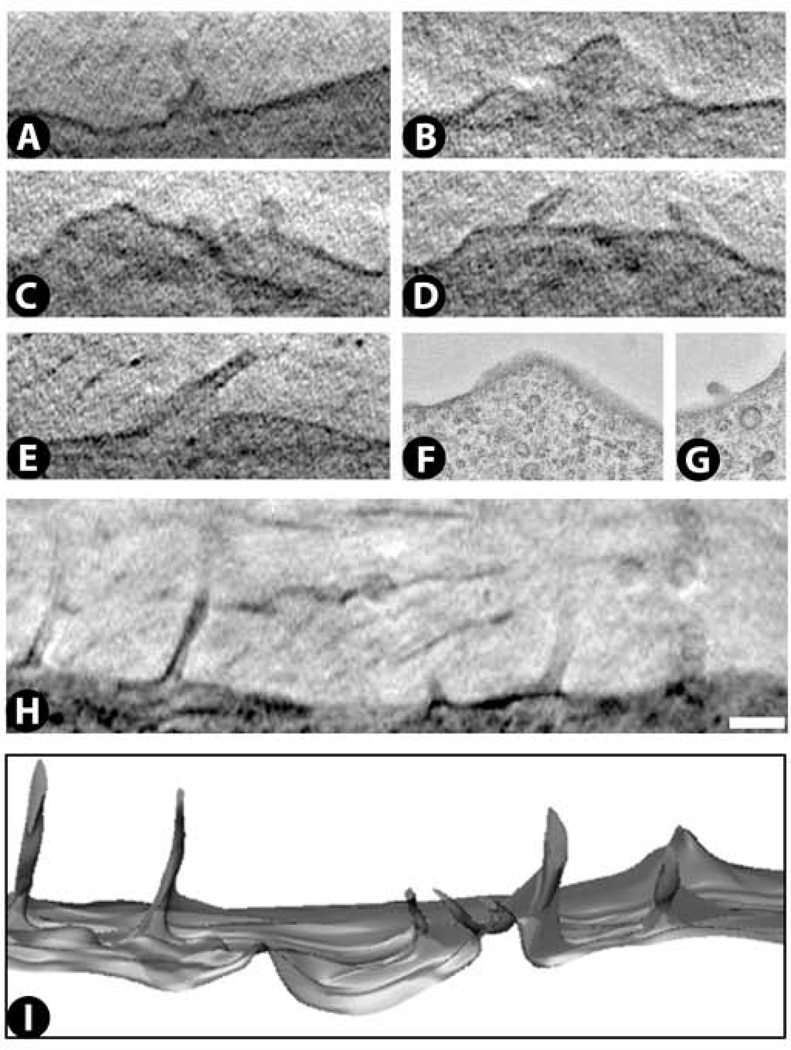
Cytoplasm: plasma membrane and microvilli
The plasma membrane is easily detected by cryo-SXT (Fig. 11A – E), although its bilayer structure is not visible due to the current resolution limit. Microvilli are also frequently detected along the cell surface (Fig. 11A – E,H,I), and these can range from rather narrow protrusions of ~100 nm in width and up to ~1 µm in length, to much broader protrusions of ~1.5 µm in width and ~ 1 µm in breadth. Similar structures can be found in the same cells by TEM (Fig. 11F,G).
Figure 11.
Microvilli and plasma membrane images from cryo-SXT (A – E) and TEM (F,G). Both thicker and thinner protrusions of the plasma membrane can be found. The number and extent of microvilli found in some cells is illustrated by the 0.45 µm thick projection image in H, and the 1.4 µm thick surface rendering of that data set in I. See Supplementary Video 10 for a 3D movie of this surface rendering. Scale bar 500 nm.
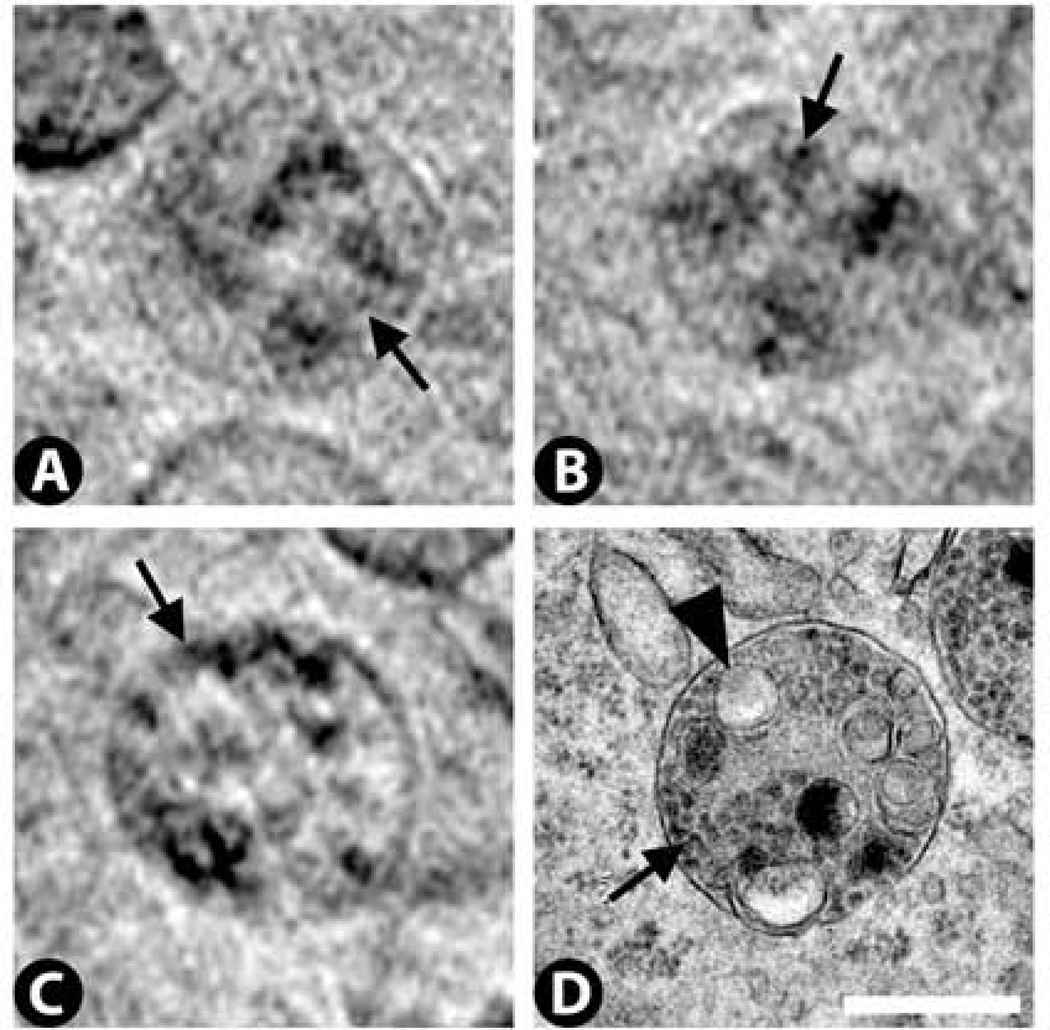
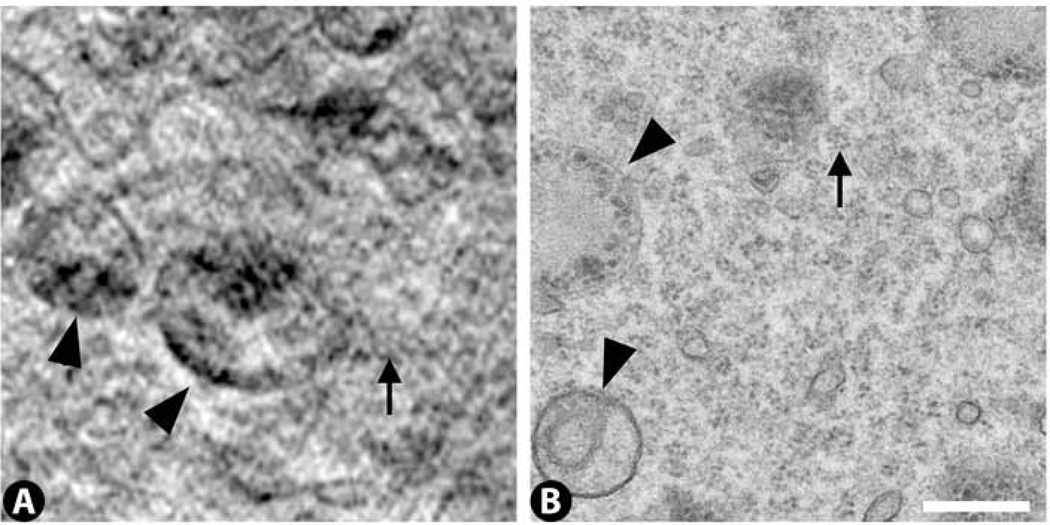
Cytoplasm: sub-structure
High magnification views of the cytoplasm in the best cryo-SXT tomograms reveal a cytoplasmic sub-structure that appears to be composed of small vesicles with an average diameter of ~50 nm (Fig. 12A). By TEM, comparable regions of cytoplasm exhibit clusters of ribosomes often arranged in circular patterns (Fig. 12B). Ribosomes are 25 – 30 nm in diameter, and so fall below the current resolution limit of 36 – 70 nm for our cryo-SXT images (Schneider et al., 2010). We would expect the ribosomes to be X-ray dense, but because they fall below the resolution limit we would not be able to distinguish adjacent ribosomes from each other. Thus, it is possible that the small vesicular pattern detected by cryo-SXT reflects the circular arrangement of ribosomes seen by TEM, but higher resolution cryo-SXT images will be required to confirm this.
Figure 12.
High magnification views of the cytoplasm reveal a sub-structure by cryo-SXT (arrow in A) and clusters of ribosomes by TEM (arrow in B). The cryo-SXT sub-structure may reflect in part the ribosomal clusters, but this cannot be determined given the current resolution of the cryo-SXT images. For perspective, larger organelles (lysosomes) are indicated in both panels (arrowheads). Scale bar 500 nm.
Robustness and reproducibility in cryo-SXT
The preceding figures illustrate a selection of the best images that we have obtained so far using cryo-SXT of the adenocarcinoma cell line 3617. The images were drawn from approximately 2/3 of the 27 tomograms generated to date. However, not all of the images are of this quality, and even among the good images, there are some that are clearly better than others.
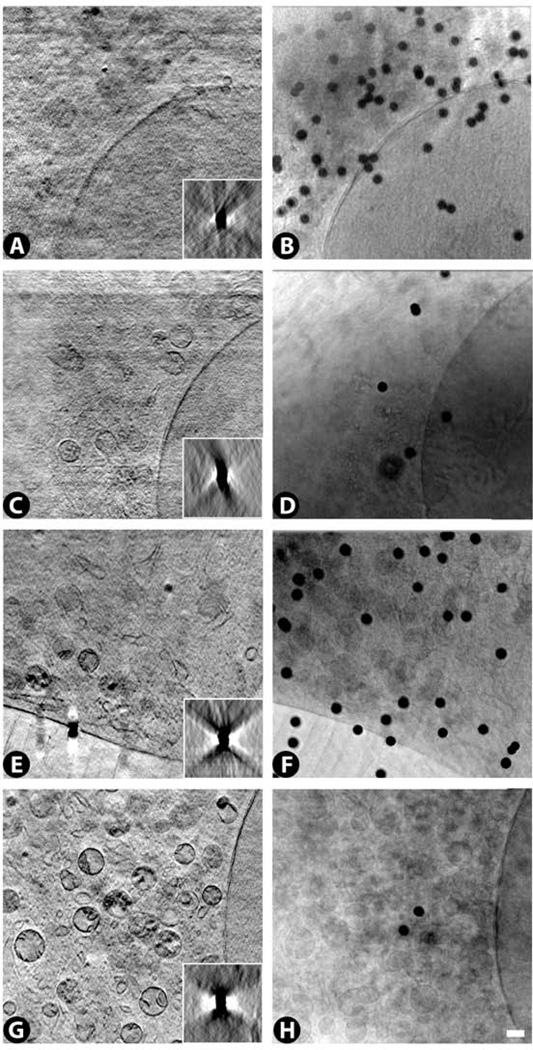
Fig. 13 illustrates the range of image variability we have seen showing virtual slices from four representative reconstructions. In the poorest reconstructed images it is difficult to discern any structures in most regions of the cytoplasm, although in a few regions, a few organelles can be detected (Fig. 13A). In somewhat better reconstructed images, membrane-bound organelles can be discerned throughout larger regions of the cytoplasm, and the internal structures of some of these organelles begin to appear (Fig. 13C). As the reconstructed images further improve, it becomes possible to see more and more clear-cut examples of organelles with better defined internal structures, including mitochondria, lysosomes and endoplasmic reticulum (Fig. 13E). In the best reconstructed images, virtually every organelle in the cytoplasm can be distinguished. These reconstructed images also reveal more cytoplasmic substructure, such as regions with smaller vesicles as well as Golgi (Fig. 13G).
Figure 13.
Virtual slices from cryo-SXT tomographic reconstructions with progressively better image quality, reflected by the increased visibility of cytoplasmic organelles (A,C,E,G). See also Supplementary Videos 1–4 for through-focus series of these four images. An equivalent trend towards more detectable cytoplasmic organelles can also be seen in the raw data (B,D,F,H), shown here at 0° tilt. Black spheres are the gold-coated beads added as fiducial markers for the reconstruction procedure. Insets show a yz slice through reconstructions of the beads. Scale bar 500 nm.
What factors could give rise to this variability? The images in Fig. 13 were obtained with the same microscope, the same zone-plate objective and the same reconstruction procedure, so the basic steps in the procedure were identical. However, there were some small changes in image acquisition that we intentionally introduced to test different conditions for data collection. Specifically, we altered the limits of the tilt range, stopping either at ±45°, ±50° or ±60°, which should provide increasingly more information for the reconstruction. However, these differences did not correlate with the image quality in Fig. 13 (Table 1). We also altered the tilt-angle step size from either 1.0° to 0.5°, but this also did not correlate with the image quality in Fig. 13. Thus, although these variables might be important at some point for optimizing the reconstructions, at the moment, they are not the key factors responsible for the image variability in Fig. 13.
Table 1.
Parameters that do not correlate with the quality of the reconstructed images in Fig. 13. The zone plate is the imaging objective. The tilt range reflects the starting and ending rotation angles of the stage during the tilt series. The step size is the angular increment between each tilt setting. The photon flux at the front of the sample is calculated from the exposure time multiplied by the beam current multiplied by the slit width (with units therefore of milliamps – meters – sec). This last parameter can be used to judge the ice-layer thickness, since the total dose to the front of the specimen will increase for thicker ice layers in order to achieve a comparably bright image on the CCD camera. Note that there is no obvious correlation of image quality with any of these parameters. This suggests that the current differences in image quality are not due to variations in the limited tilt range. It also suggests that the differences do not arise from different thicknesses of the ice layer, as good quality images were obtained from both the thinnest (lowest photon flux – third row) and thickest of the four ice layers (highest photon flux – fourth row).
In addition to these intentional changes in image acquisition, there were also some unavoidable variations in the specimen and its preparation, and in the acquisition of the tilt series, all of which could impact the reconstructions. For example, slight changes in the cryo-preservation procedure could introduce variability into the specimen. Similarly, mechanical defects in the tilt stage could introduce variability into the image acquisition. To help distinguish between these two general sources of variation, we examined the reconstructions of the gold-coated beads that were used as fiducial markers. These should not be significantly affected by cell-to-cell variations, such as the cryo-preservation, but should be sensitive to defects in image acquisition.
We found that the reconstructed bead images showed considerable variability that correlated with the quality of the cellular images. The better cellular images showed symmetrical yz views of the reconstructed beads, while the poorer cellular images showed asymmetrical yz views of the reconstructed beads (insets in Fig. 13A,C,E,G).
The asymmetrical bead images suggested problems in image acquisition, so we examined the raw data series for each dataset in Fig. 13, and found that the poorer reconstructions exhibited several defects in image acquisition (Table 2). One was lateral shifts on the order of several microns during the tilt series. This arises when the tilt stage undergoes translations while it is rotating. If the translations are large enough, some regions of the specimen move out of the field of view at certain tilt angles, thereby decreasing the desired information and increasing extraneous areas included from each image in the tilt series. A second defect present in the raw data of the poorer reconstructions was that there were fewer fiducial markers that remained in focus throughout the entire tilt series. This means that different regions of the specimen were in focus at different tilt angles. This can also arise due to translations of the stage during the tilt series, resulting for example in vertical shifts of the specimen. In addition, if the field of view is far from the stage’s tilt axis, than the region of interest in the specimen will not remain in good focus throughout the tilt series. Thus our data suggest that these various defects in image acquisition may all have contributed to the poorer reconstructions in Fig. 13.
Table 2.
Parameters that correlate with the quality of the reconstructed images in Fig. 13. Lateral shifts during the tilt series were detected by examining the raw data to identify large jumps (~2–4 µm) in the positions of the gold-coated beads used as fiducial markers during the tilt series. The # of beads in focus was also determined by following the fiducial markers through the course of the raw-data tilt series. The quality of the cryo-preservation was judged by looking for the formation of ice crystals in both the raw and reconstructed data. The thickness of the cell cytoplasm was estimated from xz views of the reconstructed specimen. Note that the reconstruction quality is correlated with each of these parameters suggesting that on average the best reconstructions will come from well-preserved, thinner specimens in which each tilt angle is well focused on the region of interest and lateral shifts of the stage are minimized.
| Reconstruction Quality |
lateral shifts during tilt series |
# beads in focus |
cryo- preservation |
thickness of cytoplasm |
|---|---|---|---|---|
| Very Bad (Fig. 13A) |
large | 0 at all tilt angles | good, but some ice on surface of ice layer | ~9 µm |
| Bad (Fig. 13C) |
large | 1 at all tilt angles | good, but some ice on surface of ice layer | ~12 µm |
| Good (Fig. 13E) |
small | 2–3 at most tilt angles | good | ~6 µm |
| Very Good (Fig. 13G) |
small | 5 at all tilt angles | good | ~6 µm |
In addition to problems with image acquisition, it is also possible that there were variations in the specimen that contributed to the variability in the reconstructions. To evaluate this possibility, we inspected single-projection views from the raw data (Fig. 13B,D,F,G) corresponding to each of the four reconstructions in Fig. 13. We found that the better reconstructions were obtained from raw data in which more cellular features could be detected. This suggests that differences between the specimens also contributed to the variability in the reconstructions.
To investigate what features in the specimen might be important, we examined several factors. We first checked the thickness of the ice layer that encapsulates the specimens. This depends on how long the specimen is blotted to remove excess fluid before it is plunge frozen. The blotting is currently done manually, so the amount of fluid removed will be variable. A thicker ice layer might be expected to reduce contrast in the image because the fraction of X-rays absorbed by the ice rather than by the specimen should increase. Although it is difficult to directly visualize the ice layer, we can estimate its thickness by measuring the total photon flux required to obtain an image of the sample, since specimens in thicker ice layers will require more photons to yield comparably bright images. This measurement shows that there is no correlation in image quality with the total photon flux reaching the front of the specimen, and therefore by inference with the thickness of the ice layer. In particular, we find that we can get good quality reconstructions with both thick and thin ice layers (Table 1).
We also checked whether there were differences in the specimens’ cryo-preservation by inspecting the raw data for signs of ice formation. We found no evidence for ice inside any of the specimens from Fig. 13, but we did see in the two poorer reconstructions some evidence for ice on the surface of the ice layer that encapsulates the specimens (Table 2). Although we do not expect this superficial ice to directly affect the image quality, it may indicate that the cryo-preservation inside of the specimen was not quite as good compared to the better cellular images in Fig. 13, where there was no evidence for ice formation on top of the ice layer.
A third feature of the specimen that could also influence the reconstruction quality is the thickness of the specimen itself. We estimated this by looking at xz views of the reconstructions. We found that the cells yielding poorer reconstructions were significantly thicker than those yielding better reconstruction (~6 µm vs. ~9–12 µm, Table 2). Specimen thickness is an important factor because the reconstruction procedures presume that the image at each tilt angle is a projection of the entire sample. This is acceptable for specimens that are less than 3 µm in thickness, which is the empirically estimated depth of field of the current X-ray microscope (Schneider et al., 2010). All the specimens examined in Fig. 13 were thicker than 3 µm, which means that the quality of the reconstruction should be affected in all cases, with more degradation possible as the specimen becomes thicker.
In sum, we have identified a number of features in the specimen and in the image acquisition that could degrade the quality of the reconstructed images and lead to variability from day to day. Future work will help identify which of these are the most important for routinely achieving the highest quality reconstructions.
Discussion
Structures seen in cryo-SXT and TEM
In general, we have found very good correspondence between structures in cryo-SXT and TEM. Notably, the major cellular organelles showed a number of similarities when cryo-SXT and TEM images were compared. Included in this list were mitochondria, lysosomes, endoplasmic reticulum, nuclear membrane, plasma membrane and filaments. This correspondence is encouraging. Cryo-SXT is still a relatively new technique, and so might be subject to various artifacts that have yet to be fully appreciated. However, cryo-SXT should eventually show less distortion and artifacts than TEM, since cryo-SXT involves only cryo-preservation while TEM typically involves chemical fixation, dehydration, staining and sectioning. Our results to date suggest that there are no major artifacts in either of these two approaches since the images we have obtained from both techniques were in general so similar.
It is important to point out that although in many cases there appears to be considerable overlap between the structural features seen by cryo-SXT and TEM, we cannot be certain at the moment if all of the sub-structures seen by these two techniques were in fact the same. It is conceivable that some of the apparent matches were accidental, since the contrast mechanisms employed by the two techniques are different. The TEM contrast is due to the affinity of particular cellular structures for the heavy metal stains of uranyl acetate, lead citrate or osmium, whereas the cryo-SXT contrast is due to the relative concentrations of organic material and water within the specimen. Thus, some sub-structures may have a high affinity for a heavy metal stain and so be readily detected by TEM, but have a relatively low ratio of organic matter to water and so be poorly detected by cryo-SXT, or vice versa. For example, some caution is warranted in comparing the small granules seen by each technique inside of lysosomes, or the dense and less dense sub-regions found by each technique inside of nucleoli. Examination of heavy metal stained specimens by cryo-SXT might help determine if some of these substructures seen in unstained specimens by cryo-SXT in fact correspond to the same structures seen in stained specimens by TEM. Cross validation of structures can also be done by comparison of cryo-SXT to images obtained by fluorescence microscopy.
Structures seen in TEM but not cryo-SXT
Despite the considerable overlap in the structural features detected by the two approaches, there were some structures detected by TEM that were not detected by cryo-SXT. In several cases, a simple explanation for their absence in the cryo-SXT images is that they were below the current resolution limit, which is 36 nm based on the Rayleigh criterion and 70 nm (the latter equivalent to 35 nm “half-pitch resolution”) based on the Fourier Ring Correlation (FRC) criterion (Schneider et al., 2010). Thus our inability to see ribosomes or distinguish microtubules from microfilaments is most likely due to their small size, since these structures are all less than 30 nm.
While ribosomes and microfilaments fall below the current resolution limit, there were other structures not detected by cryo-SXT whose dimensions should have made them large enough to detect. Included in this category were various nuclear bodies, such as Cajal bodies, PML bodies or speckles (also known as interchromatin granules), plus different membrane-bound inclusions within lysosomes. The lysosomal inclusions are readily seen by TEM, but the various nuclear bodies are not, except in some cases when specific stains are used, such as when the interchromatin granules are recognized by EDTA regressive staining due to the granules’ high RNA content (Bernhard, 1969;Spector, 1993). Notably, one EM technique that can detect these bodies is energy loss spectroscopy, where the nitrogen or phosphorus content can be used to provide better contrast to distinguish these domains (Bazett-Jones et al., 2008). Thus we may not detect these smaller structures by cryo-SXT because their contrast is not sufficiently different from the surrounding nucleus or cytoplasm to permit their identification. For example, the only nuclear body that we could readily identify by cryo-SXT was the nucleolus, which is typically 4-5x larger than other nuclear bodies and nearly 10x larger than the lysosomal inclusions. Presumably, this greater thickness of the nucleolus produces a more substantial contribution to the projection images, thereby making them visible.
Structures seen in cryo-SXT but not TEM
Just as some features could only be detected by TEM, there were a few features that could only be detected by cryo-SXT or at least were much easier to detect by cryo-SXT. Among these structures were the nuclear membrane blebs, which were readily found in many cryo-SXT images, but not in any of the TEM images. The most likely explanation is that these blebs occur infrequently, typically at most once or twice per nucleus according to our cryo-SXT images, and so would generally require serial sectioning to locate by TEM. Alternatively, it is possible that the blebs are an artifact of the cryo-preservation procedure used in cryo-SXT. Similar blebs have been detected by both cryo-electron microscopy and freeze substitution TEM in herpes-simplex-virus infected cells, where the blebs are observed to contain a virus particle inside (Skepper et al., 2001;Mettenleiter et al., 2009).
We also readily detected nuclear membrane channels by cryo-SXT. We could also identify these structures by TEM after careful examination of many thin sections, but they are much more difficult to find by this approach. While similar nuclear membrane channels had been previously detected by TEM (Fricker et al., 1997), they are not reported in most TEM studies. In contrast, they are very obvious features in many of the cryo-SXT images of nuclei, probably due to the high contrast of membranes and the ability to computationally section through the 3D image of the nucleus and recognize these structures which in most cases are not parallel to the section plane.
Finally, we found that the outer nuclear membrane was more clearly defined by cryo-SXT compared to TEM. By cryo-SXT, the outer membrane remained linear compared to the considerable ruffling seen in the TEM sections. This made it easy to see a smoothly undulating pattern of the outer membrane by cryo-SXT which defined the location of pores at fairly regular intervals along the nuclear periphery. The ruffled nature of the outer nuclear membrane in TEM is probably an artifact due to fixation and dehydration, as such ruffling is also absent in other approaches, which, like cryo-SXT, use cryo-preservation, including freeze substitution or cryo-electron microscopy (Studer et al., 2008).
Structures not clearly seen in either cryo-SXT or TEM
Chromatin was not well resolved by either cryo-SXT or TEM. In both modalities, the nuclear sub-structure appears granular. There is hope that with a 3D modality like cryo-SXT it may eventually be possible to track chromatin fibers inside the nucleus, but at the moment, fiber-like structures are not clearly visible. However, our ability to detect filaments in the cytoplasm shows that thin filaments can be identified by cryo-SXT, and that the current contrast mechanism in cryo-SXT, namely the water window, is not limited to the detection of membrane structures. However, like the nuclear bodies and lysosomal inclusions discussed above, chromatin filaments might be harder to detect in the nucleus due to its thickness (6–12 µm in these adenocarcinoma cells), yielding lower contrast in the projection images. Consistent with this possibility, we have only detected filaments by cryo-SXT in the thin cytoplasmic extensions (<1 µm thickness) of these adenocarcinoma cells.
Future improvements in cryo-SXT
Based on our data, we can suggest three major areas for improvement in cryo-SXT. The first area is contrast. We speculated that low contrast could be responsible for our inability to detect several different structures by cryo-SXT, including different nuclear bodies, chromatin and some lysosomal inclusions. These structures are relatively small compared to the thickness of the cell, and they may be difficult to distinguish from their surroundings in projection images. This suggests that other contrast mechanisms in addition to the water window, such as phase contrast, or element specific filtering are worth investigating in cryo-SXT (Schneider, 1998).
The second area for improvement in cryo-SXT is resolution. We could not detect a number of finer features such as ribosomes and the double membrane of mitochondrial cristae. Fortunately, it is not the X-ray wavelength of 2.4 nm that we used that imposes a resolution limit for detecting such structures, but rather the X-ray optics which currently employ a zone plate for the objective whose outer width sets the achievable resolution. Thus improvements in nanofabrication are expected to yield higher resolution X-ray objectives (Rehbein et al., 2011).
The third and final area for improvement is in the reliability and robustness of cryo-SXT. We found that reconstruction quality can vary considerably, even though the same cell type is being examined with the same freezing procedure. This variation did not appear to depend on the tilt angle range, the tilt angle increment or the thickness of the specimen ice layer. However, we did find correlations with four other factors: 1.) positioning of the region of interest on the stage’s tilt axis; 2.) translations of the stage during its rotation; 3.) superficial ice on the surface of the ice layer; 4.) specimen thickness.
At the moment, it is not certain which of the preceding problems contribute most significantly to the poorer reconstructions, but all of the problems can be remedied with varying degrees of effort. Better positioning of the region of interest on the stage’s tilt axis is straightforward to accomplish before the tilt series begins. Correction of stage translations is also possible using feedback software to readjust stage position during image acquisition. Good cryo-preservation is an art, but with experience becomes increasingly reproducible. The most difficult issue is specimen thickness. If this would prove to be significant, then optical sectioning approaches for X-ray microscopy might be implemented. These could include either deconvolution processing of the current X-ray images, or confocal designs for X-ray microscopy to collect thinner optical sections which would not require further processing.
In sum, we have shown that X-ray microscopy can yield excellent images of many sub-cellular structures using only the natural contrast of water and organic material. We have also identified some limitations of the technique, all of which can be addressed. Thus we can expect further improvements in the capabilities of this approach.
Supplementary Material
Video 1: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the lowest quality level. Corresponds to Fig. 13A.
Video 10: Rendered plasma membrane with microvilli shown in a 360° rotation. Corresponds to Fig. 11I.
Video 2: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the second lowest quality level. Corresponds to Fig. 13C.
Video 3: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the second highest quality level. Corresponds to Fig. 13E.
Video 4: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the highest quality level. Corresponds to Fig. 13G.
Video 5: Through focal series of a nucleolus by cryo-SXT. Note that the electron lucent regions within the nucleolus sometimes appear to form a partially connected network. These regions may correspond to the nucleolar interstices identified by some TEM studies, which suggest that these reflect invaginations of the surrounding nucleoplasm into the nucleolar space.
Video 6: Rendered outer membrane surface of a nuclear membrane bleb. The movie zooms in on this structure followed by a panning rotation. Corresponds to Fig. 5E.
Video 7: Rendered mitochondrion shown in a 360° rotation. Outer membrane is gray and cristae are red. Corresponds to Fig. 6F.
Video 8: Rendered endoplasmic reticulum shown in a 360° rotation. Corresponds to Fig. 9E.
Video 9: Rendered filaments in a cytoplasmic extension shown in a 360° rotation. Filaments are red and membrane is blue/gray. Corresponds to Fig. 10D.
Acknowledgments
We thank Michael Kruhlak and Richard Leapman for advice and comments on the manuscript. This work was funded in part by the Human Frontier Science Program Research Grant Ref. RGP0053/2005-C, the German Federal Ministry of Education and Research under contract number 05KS4BY1/7, the intramural program of the National Institutes of Health, including the National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Cancer Institute, Center for Cancer Research, including Science Applications International Corporations Frederick contract number HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bazett-Jones DP, Li R, Fussner E, Nisman R, Dehghani H. Elucidating chromatin and nuclear domain architecture with electron spectroscopic imaging. Chromosome Res. 2008;16:397–412. doi: 10.1007/s10577-008-1237-3. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- 3.Bushby AJ, P'ng KM, Young RD, Pinali C, Knupp C, et al. Imaging three-dimensional tissue architectures by focused ion beam scanning electron microscopy. Nat Protoc. 2011;6:845–858. doi: 10.1038/nprot.2011.332. [DOI] [PubMed] [Google Scholar]
- 4.Carrascosa JL, Chichon FJ, Pereiro E, Rodriguez MJ, Fernandez JJ, et al. Cryo-X-ray tomography of vaccinia virus membranes and inner compartments. J Struct Biol. 2009;168:234–239. doi: 10.1016/j.jsb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Chao W, Harteneck BD, Liddle JA, Anderson EH, Attwood DT. Soft X-ray microscopy at a spatial resolution better than 15 nm. Nature. 2005;435:1210–1213. doi: 10.1038/nature03719. [DOI] [PubMed] [Google Scholar]
- 6.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larabell CA, Nugent KA. Imaging cellular architecture with X-rays. Curr Opin Struct Biol. 2010;20:623–631. doi: 10.1016/j.sbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDermott G, Le Gros MA, Knoechel CG, Uchida M, Larabell CA. Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends Cell Biol. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: an update. Virus Res. 2009;143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Rehbein S, Guttmann P, Werner S, Schneider G. Soft X-ray microscopy at HZB: zone plate development and imaging using the 3rd order of diffraction. In: McNulty I, Eyberger C, Lai B, editors. The 10th International Conference on X-ray Microscopy, AIP Conference Proceeding; 2011. pp. 32–37. 32–37. [Google Scholar]
- 14.Rehbein S, Heim S, Guttmann P, Werner S, Schneider G. Ultrahigh-resolution soft-x-ray microscopy with zone plates in high orders of diffraction. Phys Rev Lett. 2009;103 doi: 10.1103/PhysRevLett.103.110801. 110801-1-110801-4. [DOI] [PubMed] [Google Scholar]
- 15.Schneider G. Cryo X-ray microscopy with high spatial resolution in amplitude and phase contrast. Ultramicroscopy. 1998;75:85–104. doi: 10.1016/s0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 16.Schneider G, Guttmann P, Heim S, Rehbein S, Mueller F, et al. Three-dimensional cellular ultrastructure resolved by X-ray microscopy. Nat Methods. 2010;7:985–987. doi: 10.1038/nmeth.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skepper JN, Whiteley A, Browne H, Minson A. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment --> deenvelopment --> reenvelopment pathway. J Virol. 2001;75:5697–5702. doi: 10.1128/JVI.75.12.5697-5702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 19.Studer D, Humbel BM, Chiquet M. Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol. 2008;130:877–889. doi: 10.1007/s00418-008-0500-1. [DOI] [PubMed] [Google Scholar]
- 20.Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Weiss D, Schneider G, Niemann B, Guttmann P, Rudolph D, et al. Computed tomography of cryogenic biological specimens based on X-ray microscopic images. Ultramicroscopy. 2000;84:185–197. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the lowest quality level. Corresponds to Fig. 13A.
Video 10: Rendered plasma membrane with microvilli shown in a 360° rotation. Corresponds to Fig. 11I.
Video 2: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the second lowest quality level. Corresponds to Fig. 13C.
Video 3: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the second highest quality level. Corresponds to Fig. 13E.
Video 4: Through focal series of a reconstructed cytoplasm and nucleus by cryo-SXT exemplifying the highest quality level. Corresponds to Fig. 13G.
Video 5: Through focal series of a nucleolus by cryo-SXT. Note that the electron lucent regions within the nucleolus sometimes appear to form a partially connected network. These regions may correspond to the nucleolar interstices identified by some TEM studies, which suggest that these reflect invaginations of the surrounding nucleoplasm into the nucleolar space.
Video 6: Rendered outer membrane surface of a nuclear membrane bleb. The movie zooms in on this structure followed by a panning rotation. Corresponds to Fig. 5E.
Video 7: Rendered mitochondrion shown in a 360° rotation. Outer membrane is gray and cristae are red. Corresponds to Fig. 6F.
Video 8: Rendered endoplasmic reticulum shown in a 360° rotation. Corresponds to Fig. 9E.
Video 9: Rendered filaments in a cytoplasmic extension shown in a 360° rotation. Filaments are red and membrane is blue/gray. Corresponds to Fig. 10D.