Abstract
Characterizing interactions between multiple brain regions is important for understanding brain function. Functional connectivity measures based on partial correlation provide an estimate of the linear conditional dependence between brain regions after removing the linear influence of other regions. Estimation of partial correlations is, however, difficult when the number of regions is large, as is now increasingly the case with a growing number of large-scale brain connectivity studies. To address this problem, we develop novel methods for estimating sparse partial correlations between multiple regions in fMRI data using elastic net penalty (SPC-EN), which combines L1- and L2-norm regularization We show that L1-norm regularization in SPC-EN provides sparse interpretable solutions while L2-norm regularization improves the sensitivity of the method when the number of possible connections between regions is larger than the number of time points, and when pair-wise correlations between brain regions are high. An issue with regularization-based methods is choosing the regularization parameters which in turn determine the selection of connections between brain regions. To address this problem, we deploy novel stability selection methods to infer significant connections between brain regions. We also compare the performance of SPC-EN with existing methods which use only L1-norm regularization (SPC-L1) on simulated and experimental datasets. Detailed simulations show that the performance of SPC-EN, measured in terms of sensitivity and accuracy is superior to SPC-L1, especially at higher rates of feature prevalence. Application of our methods to resting-state fMRI data obtained from 22 healthy adults show that SPC-EN reveals a modular architecture characterized by strong inter-hemispheric links, distinct ventral and dorsal stream pathways, and a major hub in the posterior medial cortex- features that were missed by conventional methods. Taken together, our findings suggest that SPC-EN provides a powerful tool for characterizing connectivity involving a large number of correlated regions that span the entire brain.
Keywords: Partial Correlation, inverse covariance, Elastic Net, Lasso, Regularization, Stability Selection
Introduction
Functional magnetic resonance imaging (fMRI) has emerged as a powerful technique for investigating human brain function and dysfunction. Identification of functional networks from fMRI data obtained during cognition or task-free “resting state” is critical for understanding and characterizing how different brain regions communicate with each other. In recent years, several methods have been developed to characterize functional brain networks and connectivity (Beckmann and Smith, 2004; Calhoun et al., 2001; Calhoun et al., 2003);(Bullmore and Sporns, 2009; Cole et al.; Deco et al.; Friston et al., 1997; Greicius et al., 2003; McIntosh, 2000; Rubinov and Sporns; Sun et al., 2004; Supekar et al., 2008; Tononi and Sporns, 2003).
Functional connectivity is often computed using the Pearson correlation between brain regions taken pair wise. The resulting correlation coefficients are usually converted to z-scores using the Fisher transformation and then thresholded to identify statistically significant network connections (Supekar et al., 2008). Such correlation based methods are useful for assessing context and stimulus dependent functional interactions between multiple brain regions. However, one issue with this approach is that it estimates marginal linear dependence or independence between a pair of regions without considering the influence of other regions and common driving influences. Some of these limitations can be overcome by using partial correlations which measure the linear relationship between multiple brain regions, taken pair wise, by removing the common linear influences of all other regions considered together. Under assumptions of normality, if the partial correlation between two regions is zero, then two regions are conditionally independent given temporal fluctuations in other brain regions considered (Hastie et al., 2009; Peng et al., 2009). Several previous brain imaging studies have used partial correlations for estimating functional connectivity (Hampson et al., 2002; Huang et al., 2010; Lee et al., 2011; Marrelec et al., 2007; Marrelec et al., 2006; Smith et al., 2011), but they work well only when the number of brain regions is small. Another limitation of current methods is that corrections for multiple comparisons need to be performed while identifying the significant network connections, which could greatly hamper their sensitivity especially, when the number of regions examined is large.
Estimating functional networks using partial correlations is further problematic when the number of possible connections involved in the analysis is large compared to the available observations (number of scans, in the case of fMRI). In such situations, conventional methods for estimating partial correlations result in over-fitting of the data. Therefore, currently used methods for computing partial correlation are not scalable for estimating functional brain networks involving a large number of brain regions. To overcome this problem, investigators generally restrict the number of regions involved in the analysis to a few preselected regions of interest (ROIs). Here we take advantage of recent advances in multivariate statistics to develop methods suitable for estimating functional connectivity in networks with a large number of brain regions.
The problem of estimating partial correlations is equivalent to estimating the inverse of the covariance matrix. It has been shown that partial correlations are proportional to the off-diagonal entries of the inverse covariance matrix (Hastie et al., 2009; Koller and Friedman, 2009; Meinshausen and Buhlmann, 2006; Peng et al., 2009). Under assumptions of normality, the off-diagonal elements of the covariance matrix indicate the linear conditional dependence or independence of nodes. For example, if a particular entry in the inverse covariance matrix is zero, then one can infer that the corresponding regions are conditionally independent, under assumptions of normality, given responses in the other brain regions. Partial correlations can be estimated by computing the covariance matrix from the data, and then inverting and appropriately scaling it. A reliable estimation of the inverse covariance matrix is therefore required for identifying functional networks based on fMRI data. However, the estimation of inverse covariance matrices is particularly challenging when the number of regions involved in a functional network is greater than the number of observations, namely the number of fMRI time samples acquired during an experiment. In such cases, the rank of the covariance matrix is at most the number of observations and it is necessary to use methods such as generalized inverse or pseudoinverse. However, such methods are often not accurate (Hoyle, 2010) and do not provide sparse interpretable solutions. For example, several studies have used the pseudoinverse method for estimating partial correlations at the whole brain level (Balenzuela et al., 2010; Liang et al., 2011; Liu et al., 2008; Yu et al., 2011; Zhang et al., 2011). These studies reveal a fully-connected network structure that is difficult to interpret (Supplementary Figure 1A). Importantly, network metrics which measure modular architecture of the network are likely to extract less meaningful modules, when computed on fully connected brain networks estimated using psuedoinverse methods. To overcome this problem, regularization procedures that provide sparse solutions have been proposed for estimating the inverse covariance matrices (Friedman et al., 2008, 2010; Huang et al.; Lee et al., 2011; Meinshausen and Buhlmann, 2006; Peng et al., 2009; Varoquaux et al., 2010). Based on the relationship between inverse covariance estimation and partial correlation, and between partial correlation and linear regression, regularization methods developed for linear regression have recently been extended for estimating inverse covariance matrices. For example, L1-norm regularization, also called lasso (Tibshirani, 1996), has been used previously for estimating networks or inverse covariance matrix (Friedman et al., 2008; Lee et al., 2011; Meinshausen and Buhlmann, 2006; Peng et al., 2009). One major advantage of using L1-norm regularization is that it provides sparse solutions which are very helpful in interpreting the estimated networks. Importantly, these methods do not require further statistical thresholding to infer the significant network connections. L1-norm regularization automatically finds significant network connections since the weights of insignificant connections are automatically driven to zero. Although L1-norm regularization has been used successfully, it has some limitations. One important limitation is that in the case when the number of nodes or regions is greater than the number of observations, the number of network connections L1-norm regularization can identify is at most equal to the number of observations (Zou and Hastie, 2005). Another limitation is that when the pair wise correlations are very high then L1-norm regularization tends to identify only a subset of connections. To overcome this problem, a combination of L1 and L2 norm regularization, also referred to as elastic net regularization, was proposed in the context of linear regression (Zou and Hastie, 2005). Elastic net regularization retains the desirable property of lasso by providing sparse solutions while overcoming the problems associated with large number of correlated nodes. Using L2-norm regularization in addition to L1-norm provides the necessary decorrelating step which in turn results in grouping effect that ensures selecting all the significant connections between regions (Zou and Hastie, 2005).
Only few studies to date have used regularization procedures for estimating functional networks in the context of brain imaging data (Huang et al., 2010; Lee et al., 2011; Smith et al., 2011; Varoquaux et al., 2010). Huang and colleagues and Lee and colleagues used L1-norm regularization to estimate inverse covariance matrix from positron emission tomography (PET) data but their methods have not been validated using simulations. In a recent study, Varoquaux and colleagues used group lasso regularization to estimate functional connectivity from resting state fMRI data (Varoquaux et al., 2010). Their procedure estimates inverse covariance for each subject while imposing the same functional connectivity structure for all subjects. These methods also have not been validated on simulated data. Smith and colleagues recently evaluated several network estimation methods based on simulated data (Smith et al., 2011). One of the methods evaluated was inverse covariance estimation with L1-norm regularization on networks with up to 50 nodes. Critically, no studies, except (Smith et al., 2011), have used simulations to validate these methods. Furthermore, as noted above, fMRI data has inherently high spatial correlations and adjacent regions are likely to be highly correlated. Inferring connectivity using only L1-norm regularization is therefore problematic when the regions are highly correlated and the number of possible connections is greater compared to the available observations. The main objective of our study is to address this problem by developing, testing and validating new methods for estimating stable connectivity patterns in functional networks which contain a large number of nodes.
Here, we develop new methods for estimating functional networks involving a large number of brain regions from fMRI data. Given that fMRI data has high inherent spatial correlation, we use elastic net regularization (Zou and Hastie, 2005) in contrast to lasso regularization used in the previous studies. The proposed approach overcomes the problems associated with lasso or L1-norm regularization while retaining its desirable property of providing sparse solutions. More importantly, we infer significant functional connections between brain regions using novel stability selection methods which overcomes the problems associated with choosing the optimal regularization parameters in elastic net and lasso penalties (Meinshausen and Bühlmann, 2010). In previous studies, the critical regularization parameters which influence the selection of connections between brain regions were set arbitrarily (Huang et al., 2010; Smith et al., 2011). We first test and validate the performance of our proposed methods on data simulated from two networks with varying degrees of small-world structure at different prevalence rates. We then test and validate our methods on more realistic simulations using dynamic causal models (Friston et al., 2003; Smith et al., 2011). Finally, we demonstrate the utility of our methods to uncover modules and sub-networks using resting-state fMRI data.
Methods
Notation: In the following sections, we represent matrices by upper-case and scalars and vectors by lower-case letters. Random matrices are represented by bold upper-case letters while random vectors and scalars are represented by bold lower-case letters.
Estimation of sparse partial correlation
Let fMRI observations X at p brain regions follow a multivariate normal distribution with mean μ ∈ Rp and covariance Σ ∈ Rp×p. If the m,n-th entry of the inverse covariance matrix Θ = Σ–1 is zero then, under normal assumptions, the regions m and n are said to be conditionally independent given the other regions. Also, Θ(m,n) is proportional to the regression coefficient of region n in the multiple regression of variable m on the rest of the regions (Hastie et al., 2009; Peng et al., 2009). The reverse is also true since Θ is a symmetric matrix. It can also be shown that, Θ(m,n) is proportional to the partial correlation between the two regions (Peng et al., 2009).
Given the relation between the entries of Θ and the multiple-regression, the conditional distribution of the n-th region given the other regions is
| (1) |
Where, N(μ, σ2), is a normal distribution with mean μ and variance σ2. βmn represents the regression coefficient of Xm on Xn and σnn are the residual variances. βmn and σnn are related to the entries of Θ as follows:
| (2) |
| (3) |
The partial correlation between regions m and n ρ(m, n) is then given by
| (4) |
The negative log-product - likelihood (or pseudo likelihood) for the conditional distributions up to proportionality constant can be written as:
| (5) |
where is a symmetric matrix with zeros on the diagonal and the off-diagonal elements are same as in Θ. X is N × p observation matrix with N observations and p brain regions. xn and are n-th columns of X and respectively. Let can be estimated by minimizing . However, when the number of regions (p) is much greater than the number of observations (N), which is the case in many fMRI studies, minimizing (5) directly results in over fitting of the data, which results in poor generalization. Regularization on the elements of is therefore needed to avoid over fitting (Bishop, 2006; Hastie et al., 2009).
L2-norm or ridge regularization is one such commonly used regularization method. Under L2-norm regularization, regions which are conditionally independent should have their corresponding matrix values in to be zero. However, L2-norm regularization drives these values close to zero but not exactly to zero. Additional thresholding is required to infer the underlying network. L1-norm regularization or lasso (Tibshirani, 1996), on the other hand, drives these small values to exactly zero resulting in sparse interpretable solutions. However, if the predictors in the regression are highly correlated then L1-norm regularization tends to identify only a subset of connections. Another problem using this penalty is that when the number of regions is greater than the number of subjects (p > N), at most network connections can be identified (Zou and Hastie, 2005). Therefore, L1-norm regularization cannot be used in estimating networks wherein the number of regions is greater than the number of observations. To overcome these problems Zou and Hastie proposed the elastic net (Zou and Hastie, 2005), which is a linear combination of L1 and L2-norm regularization. Similar to lasso, elastic net provides sparse solutions by accounting for correlations in the predictors. More importantly, for our purposes here, it can be used in scenarios where p > N. In this study, we use elastic net to estimate .
The cost function to be minimized for elastic net regularization is:
| (6) |
Here, the parameter determines the balance between L2-norm (α = 0) and L1-norm (α = 1) regularization. The solution for α = 0 corresponds to L2-norm (ridge) regularization and for α = 1 to L1-norm (lasso) regularization. Therefore, L2-norm (ridge) and L1-norm (lasso) regularization are the special cases of elastic net. In what follows, we refer to the estimation of sparse partial correlation with elastic net penalty as SPC-EN and estimating with lasso penalty alone as SPC-L1.
We solve the above optimization problem using a cyclical coordinate descent method (Tibshirani et al., 2010). This method has previously been applied for solving lasso (Friedman et al., 2007; Krishnapuram et al., 2005; Shevade and Keerthi, 2003) and elastic net optimization problems (Friedman et al., 2010; Ryali et al.; Tibshirani et al., 2010). In this approach, the optimization problem is solved for an entire path of values of λ (for a given α), using the current estimates as “warm” starts, a strategy that has been shown to be efficient, simple and fast (Tibshirani et al., 2010). The update for each component , assuming that the other components are fixed, is given by
| (7) |
where, T(., δ) is a thresholding operator defined as
| (8) |
and
| (9) |
where, , and are the previous estimates.
Given the current estimate of , the variances are updated as (Friedman et al., 2010)
| (10) |
where, . The equations (7) and (10) are repeated until convergence.
Parameter selection using stability selection method
A major challenge in sparse estimation methods such as lasso and elastic net for high dimensional data is in determining the optimal regularization parameters such as λ in lasso penalty and α and λ in elastic net penalty (Meinshausen and Bühlmann, 2010). These parameters determine the amount of regularization that needs to be applied for selecting the non-zero variables in the model. For instance in sparse covariance estimation, these parameters determine the non-zero partial correlations and thereby conditional dependences between nodes in the network. Model selection criteria such as Akaike information criterion (AIC) and Bayesian information criterion (BIC) can be used to select regularization parameters (Bishop, 2006). These criteria provide a tradeoff between accuracy and complexity of the model and are asymptotically optimal. Previously, these criteria have been used to select the optimal regularization parameter in lasso regularization (Peng et al., 2009; Zou et al., 2007). However, for finite sample and high dimensional problems these criteria can result in erroneous results. We found that using BIC for SPC-EN always resulted in sparse solutions close to SPC-L1. To overcome this problem, we deploy new methods which combine sub-sampling procedures with variable selection methods such as lasso and elastic net. These method provides finite sample false discovery or per-comparison error rate control for selecting variables in the model (Meinshausen and Bühlmann, 2010) and estimate connections between nodes which have nonzero partial correlations or edges between them. The goal here is to estimate the set S = {(m, n), 1 ≤ (m, n) ≤ M; Θ(m, n) ≠ 0} consisting of edges between the nodes. Let the estimated set for regularization parameters (α, λ) (note that α = 1 in lasso) is denoted as . Let denote the probability that the edge (m, n) is non-zero for the regularization parameters (α, λ). Define the set of stable variables as
| (11) |
Where, πthr is a probability threshold such that 0 < πthr < 1. (Meinshausen and Bühlmann, 2010) have shown that for a given πthr, the expected number of falsely selected edges V is then bounded by
| (12) |
Where, p is the number of variables in the model and q is the average number of selected edges for a given parameter range of (α, λ). Here, we specify the required per-comparison error rate () control to be 0.05 and compute the probability threshold πthr.
To estimate the probabilities , we employ the subsampling method suggested in (Meinshausen and Bühlmann, 2010). We subsample N/2 observations randomly without replacement and estimate Sα,λ for each combination of α, λ. We repeat this 100 times as suggested in (Meinshausen and Bühlmann, 2010) to estimate the probabilities . fMRI time series observations are not independent and therefore are not exchangeable for random sub sampling. We therefore, divide the fMRI time series into consecutive non-overlapping blocks where each block consists of 10 time series samples. We can assume that the blocks are exchangeable because the autocorrelation of time series across the blocks will be weak and therefore we randomly sample the blocks without replacement instead of individual observations.
Implementation details
Fast algorithms exist for efficient implementation of sparse estimation methods (Peng et al., 2009; Tibshirani et al., 2010). In these algorithms only a set of variables, called active set, which are most likely to be in the model are updated until convergence. At the end of convergence, Karush-Kuhn-Tucker (KKT) conditions are then checked for each variable. The variables which violate KKT conditions are included in the active set and are updated until convergence. These two steps are repeated until convergence and no variable violate KKT conditions. If the solution set is very sparse such methods are found to be computationally efficient and achieve faster convergence. To further improve on this method we prescreen variables using a set of strong rules (Tibshirani et al., 2011). Only variables which survive these strong rules are optimized. We implemented this method to improve the computational efficiency of our proposed SPC-EN and SPC-L1 methods.
Simulated data from multivariate network model
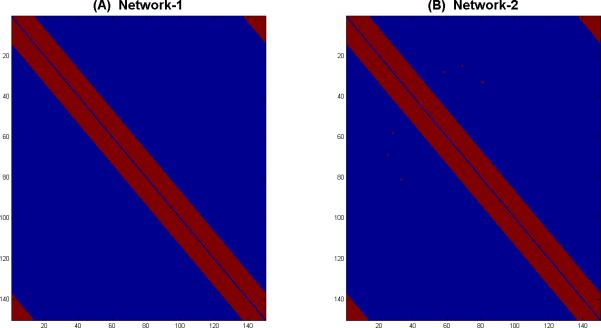
We assess the performance of SPC-EN and SPC-L1 using computer-simulated datasets. We simulate data from two networks with varying degrees of small-world structure. The networks were derived using network generation procedure described in (Watts and Strogatz, 1998). The degree of small-worldness was controlled by varying connection probability r (Watts and Strogatz, 1998). The networks used for simulation purposes were generated for r = 0.001, and r = 0.01. We chose the network to have a small-world structure, because we wanted the simulated network to be similar to real-world large-scale brain networks, which have been consistently reported to have a small-world structure (Bullmore and Sporns, 2009). We set the number of regions to p = 150 and the number of observations to N = 200. For each of two networks shown in Figure 1, we set the weight of each connection (entry of adjacency matrix A) to a random number generated from a normal distribution with mean 0.5 and standard deviation 0.1. We restrict the maximum weight to 0.6 and minimum weight to 0.4. We simulate observations at each time sample as follows:
| (13) |
Where, x(n),w(n) ∈ Rp, A is a p × p adjacency matrix of a given network and Ip is an identity matrix of dimension p. For each network, we set the percentage of connections (prevalence rate) to R where we vary R from 15% to 35% in steps of 10%. We generate 24 datasets (subjects) from each network and for every R.
Figure 1. Networks with varying degree of small-world-ness used in simulations.
(A) Small-world Network-1 with connection probability r = 0.001 and (B) Small-world Network-2 with connection probability r = 0.01 at a 35% feature prevalence rate. r is the connection probability which controls the degree of small-worldness (Watts and Strogatz, 1998).
Simulated data using DCM
To further test the performance of SPC-EN and SPC-L1, we used datasets generated using DCM (Friston et al., 2003) with network structures described by Smith and colleagues (Smith et al., 2011). We test our methods on two networks consisting of 50 and 150 nodes. The datasets for 50 node network were obtained from http://www.fmrib.ox.ac.uk/analysis/netsim. This network consists of 50 nodes and 200 time series observations. Fifty datasets were simulated from this model to represent 50 subjects. The 50-node network, (shown in Figure 2 of (Smith et al., 2011)), consists of 10 five-node rings and their interconnections. To examine the performance of our proposed methods on a larger network, we scaled up the 50-node topology to a network topology consisting of 150 nodes. This 150 node network consists of 30 five-node rings which are connected in the same fashion as in the original 50-node network. The DCM simulation script in statistical parametric mapping (SPM5) software (http://fil.ion.ucl.ac.uk/spm) with default settings was used to generate 200 fMRI time series points for each node (TR was set to 2 seconds). Each node has 33 external input onsets which were uniformly and randomly drawn from the 200 fMRI time points. The number of inputs was matched to the mean number of “up” inputs in Smith et al. (2011) with 2-second duration for the “up” state. The signal-to-noise ratio (SNR) for the output BOLD time series was set at 1 (the default value in the DCM simulation script). The weight of each connection was set in the same manner as in non-DCM datasets. In all, twenty-four different test datasets with the same network structure were generated.
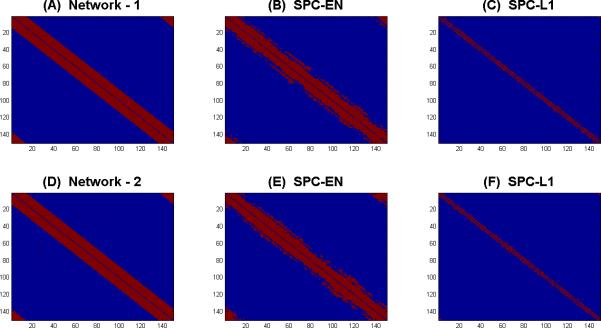
Figure 2. An illustrative example of network estimation using SPC-EN and SPC-L1.
(A) Network-1 with 35% prevalence rate. Networks estimated by (B) SPC-EN and (C) SPC-L1. Using Lasso regularization (SPC-L1) provide sparse estimation of network with low sensitivity (0.19) whereas elastic net (SPC-EN) regularization improved the sensitivity (0.81) of the estimated network. (D) Network-2 with 35% prevalence rate. Networks estimated by (E) SPC-EN and (F) SPC-L1. Using Lasso regularization (SPC-L1) provide sparse estimation of network with low sensitivity (0.18) whereas elastic net (SPC-EN) regularization improved the sensitivity (0.8) of the estimated network.
Next, we sought to examine the performance of SPC-EN and SPC-L1 on datasets generated using DCM using more realistic small-world network structures along the lines discussed in the previous section. Specifically, we attempted to generate Network-1 and Network-2 structures with 150 nodes at prevalence rates ranging from 15% to 35% in steps of 10%. However, the SPM5-based DCM simulation procedures (Smith et al., 2011) did not converge for these network structures. We therefore restricted our analysis of DCM-generated simulations to a relatively low prevalence rate of 4.9%.
Experimental data
Resting-state fMRI (rsfMRI) data were acquired from 22 adult participants. The study protocol was approved by the Stanford University Institutional Review Board. The subjects (11 males, 11 females) ranged in age from 19 to 22 y (mean age 20.4 y) with an IQ range of 97 to 137 (mean IQ: 112). The subjects were recruited locally from Stanford University and neighboring community colleges.
For the rsfMRI scan, participants were instructed to keep their eyes closed and try not to move for the duration of the 8-min scan. Functional Images were acquired on a 3T GE Signa scanner (General Electric) using a custom-built head coil. Head movement was minimized during scanning by a comfortable custom-built restraint. A total of 29 axial slices (4.0 mm thickness, 0.5 mm skip) parallel to the AC-PC line and covering the whole brain were imaged with a temporal resolution of 2 s using a T2* weighted gradient echo spiral in-out pulse sequence (Glover and Law, 2001) with the following parameters: TR = 2,000 msec, TE = 30 msec, flip angle = 80 degrees, 1 interleave. The field of view was 20 cm, and the matrix size was 64×64, providing an in-plane spatial resolution of 3.125 mm. To reduce blurring and signal loss arising from field in homogeneities, an automated high-order shimming method based on spiral acquisitions was used before acquiring functional MRI scans. A high resolution T1-weighted spoiled grass gradient recalled (SPGR) inversion recovery 3D MRI sequence was acquired to facilitate anatomical localization of functional data. The following parameters were used: TI = 300 msec, TR = 8.4 msec; TE = 1.8 msec; flip angle = 15 degrees; 22 cm field of view; 132 slices in coronal plane; 256×192 matrix; 2 NEX, acquired resolution = 1.5×0.9×1.1 mm. Structural and functional images were acquired in the same scan session.
Data were preprocessed using SPM5. The first eight image acquisitions of the task-free functional time series were discarded to allow for stabilization of the MR signal. Each of the remaining 232 volumes underwent the following preprocessing steps: realignment, normalization to the MNI template, and smoothing carried out using a 4-mm full-width half maximum Gaussian kernel to decrease spatial noise. Excessive motion, defined as greater than 3.5 mm of translation or 3.5 degrees of rotation in any plane, was not present in any of the task-free scans.
The preprocessed task-free functional MRI datasets were parcellated into 90 cortical and subcortical regions using anatomical templates (Tzourio-Mazoyer et al., 2002). A time series was computed for each of the 90 regions by averaging all voxels within each region at each time point in the time series, resulting in 232 time points for each of the 90 anatomical regions of interest. These regional time series were filtered using a band pass filter (0.0083 Hz < f < 0.15 Hz). The filtered regional fMRI time series were then used to construct a 90 node whole-brain resting-state functional connectivity network for each subject, using SPC-EN and SPC-L1. High-dimensional networks such as these are difficult to interpret. A graph-theoretical framework provides a wide array of measures to quantify and thereby succinctly describe structure of the large-scale brain network represented as a graph. To examine structure of the sparse functional connectivity network, modules in the network were determined, using the module detection algorithm described in (Newman, 2006). Module detection has been applied to determine fine-grain structure of large-scale high-dimensional networks such as internet, social networks, and biological networks. In addition to determining the modules, modularity index of brain network of each subject was also computed. Modularity index is a measure of quality of division of a network in communities/modules. Networks with high modularity are characterized by modules consisting of nodes that are densely connected with other nodes within the module but sparsely connected with nodes belonging to other modules. To further characterize network structure, degree – number of edges incident on a node/region belonging to the brain network – was computed. Regions with high degree were classified as hubs.
Performance metrics
The performance of the proposed method in identifying connections between brain regions was assessed using several performance metrics, including sensitivity, false positive rate and accuracy, where:
| (14) |
| (15) |
| (16) |
TP is the number of true positives, TN is the number of true negatives, FN is the number of false negatives and FP is the number of false positives. These performance metrics are computed for each of the simulated datasets.
Results
Performance on simulated data generated using multivariate network model
We first illustrate the relative performance of SPC-EN and SPC-L1 for simulated dataset of network -1 and network-2 with prevalence rate R = 35%. The non-zero partial correlations or edges between nodes are selected using the stability selection method with FCER = 0.05.
Figure 2 shows the actual and selected network edges between nodes using SPC-EN and SPC-L1. Figure 2A shows the actual network-1, and Figure 2B and 2C respectively show the estimated networks using SPC-EN and SPC-L1. SPC-EN provides better sensitivity (0.81) compared to SPC-L1 (0.19) in identifying network edges between nodes. SPC-L1 identifies only a subset of actual connections between nodes. Similarly, Figure 2D shows the actual network-2, and Figures 2E and 2F respectively show the estimated networks using SPC-EN and SPC-L1. In this case also, SPC-EN provides better sensitivity (0.80) compared to SPC-L1 (0.18) in identifying network edges between nodes.
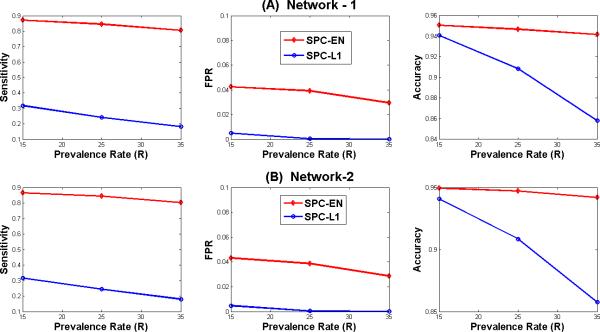
Figure 3 compares the relative performance of SPC-EN and SPC-L1 for networks-1 and -2 for three prevalence rates R in terms of sensitivity, false positive rate (FPR) and accuracy in identifying the connections between brain regions. The figure shows the average values of performance measures for 24 subjects. The sensitivity achieved by SPC-EN in identifying the connections between nodes is much better compared to SPC-L1. For example, the average sensitivity achieved by SPC-EN is about 0.85 compared to 0.24 by SPC-L1 for network 1 at prevalence rate (R) 25% as shown in the left panel of Figure 3A. Although, the false positive rates achieved by SPC-L1 are low compared to SPC-EN its sensitivity and accuracy is much poorer compared to SPC-EN as shown in Figures 3A and 3B. Additionally, the performance of SPC-EN in terms of accuracy is superior to SPC-L1 for networks 1 and 2 at three prevalence rates.
Figure 3. Sensitivity, FPR and Accuracy in identifying network connections estimated by SPC-EN and SPC-L1.
Simulated datasets based on small-world Networks 1 and 2 at prevalence rates of 15%, 25% and 35%. For both the networks, SPC-EN achieved superior performance with respect to sensitivity and accuracy at all values of connection probability r.
Performance on simulated data generated using DCM model
We applied SPC-EN on 24 datasets generated for 50-node (Smith et al., 2011) and 150-node networks. SPC-EN resulted in average sensitivity of 0.95 and average accuracy of 0.9 on the 50 node network, and average sensitivity of 0.72 and an accuracy of 0.95 on a 150 node network. SPC-L1 also showed similar performance levels. As noted in the methods section, DCM simulations did not converge for more realistic prevalence rates (> 15%) for small-world networks Network-1 and Network-2. Because of this crucial limitation in DCM-based simulations, our comparison of the relative performance of SPC-EN and SPC-L1 is based on simulations described in the previous section, where a wide range of prevalence rates could be assessed.
Performance on experimental fMRI data
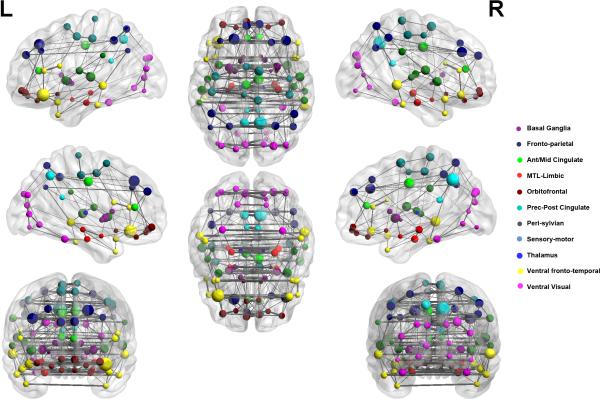
The goal of this analysis was to examine the structure of whole-brain functional connectivity and modules in fMRI data. We applied SPC-EN to resting-state fMRI data acquired from 22 healthy adults. Network structure was first determined individually in each participant and combined by group-averaging the individual network structures using a one-sample t-test. The group-averaged 90-node network plot is shown in Figure 4. The nodes of the network correspond to AAL-atlas brain regions while edges between nodes of the network correspond to strength of partial correlation between the regional fMRI time series.
Figure 4. Graphical representation of whole-brain functional network derived using SPC-EN.
Left and right sagittal, axial and coronal views are shown, with nodes representing cortical regions based on the AAL atlas (REF) and edges representing partial correlation between pairs of nodes. Nodes are plotted using the x, y and z coordinates of their centroids (in mm) in the MNI space and sized according to their connectivity profile, with larger nodes representing highly connected “hub” nodes. Eleven modules are shown, with nodes belonging to the same module marked in same color.
SPC-EN identified an average of 592 edges across participants. Modularity analysis revealed 11 distinct modules with the following features: (1) strong inter-hemispheric links between homologous areas across the entire brain, (2) a ventral stream module linking the visual cortex with the inferior temporal cortex and parahippocampal gyrus, (3) a dorsal stream network linking the posterior parietal cortex with the superior frontal gyrus, (4) a major hub in the precuneus node of the posterior medial cortex, and (5) extensive local coupling across regional nodes.
In contrast, SPC-L1 derived network consists of an average of 430 edges (supplementary figure S2). A two-sample t-test revealed that the number of edges in SPC-EN derived network was significantly higher compared to the SPC-L1 derived network (p < 0.01). Modularity indices, a measure of between-module connectivity versus within-module connectivity, was not significantly different between the SPC-EN (mean = 0.71 +-0.04) and SPC-L1 (mean = 0.78 +-0.03) derived networks (p = 0.24). However, the SPC-L1 derived network revealed only inter-hemispheric links and modules which were less meaningful and were strictly restricted to anatomical nearest-neighbors and their inter-hemispheric homologs. Unlike SPC-EN, SPC-L1 could not identify the ventral and dorsal streams not could it identify the precuneus as a major hub.
Discussion
We developed novel methods for estimating partial correlations between brain regions and thereby inferring functional networks in fMRI data. These partial correlations can be used to reliably identify functional brain networks in a large (>100) interacting sets of regions. SPC-EN incorporates a combination of L1 and L2 norm regularization to estimate partial correlations in the presence of highly correlated brain regions and when the number of brain regions is high compared to the number of observations or fMRI time samples. More importantly, SPC-EN computes sparse solutions which automatically identify the significant partial correlations between brain regions without additional statistical testing for significance. We develop stability selection methods which overcome the problems associated with choosing regularization parameters (α, λ in SPC-EN and λ in SPC-L1) in individual fMRI data. Below, we discuss the advantages of our method over other existing methods in detail, and demonstrate their utility in identifying functional brain networks.
Performance of SPC-EN on simulated datasets
We evaluated the performance of SPC-EN on simulated datasets generated for different small world networks and prevalence rates. L1-norm regularization employed in such methods provides sparse solutions wherein only significant connections survive while the other connections whose partial correlations are insignificant are driven exactly to zero. This is desirable since the estimated functional networks can be interpreted without further statistical testing. However, using only L1-norm has a major limitation when the number of connections between brain regions involved is greater than the number of fMRI time samples and when the pair-wise correlations between nearby regions is high, as is typically the case in experimental fMRI data. It is shown in the statistical regression literature that when the numbers of predictors (p) exceed the number of observations (N), L1-norm can select at most N predictors (Zou and Hastie, 2005). Furthermore, even in the case when N > p and the relevant predictors are highly correlated, L1-norm regularization selects only one predictor (Zou and Hastie, 2005). This is a major issue for estimating partial correlations in fMRI data, which has inherently high spatial correlation and therefore adjacent brain regions can show high levels of similarity in their temporal fluctuations. These limitations of L1-norm regularization for regression also carry to the problem of estimation of inverse covariance and partial correlation, since both problems can be posed as equivalent regression problems (Meinshausen and Buhlmann, 2006; Peng et al., 2009). These two issues can be overcome by using elastic net penalty (Zou and Hastie, 2005) which also retains the desirable property of providing sparse solutions like L1-norm regularization (lasso). Our findings on simulated datasets verify this property. As illustrated in Figures 2 and 3 SPC-EN achieved better sensitivity compared to SPC-L1.
Our simulations suggest that, among these two methods, SPC-EN achieved superior sensitivity and accuracy compared to SPC-L1 at all values of prevalence rates. Even at the lowest prevalence rate (R = 15%), SPC-EN achieved superior sensitivity compared to SPC-L1 (left panels in Figures 3A and 3B). This can be attributed to the second limitation of L1-norm regularization that because of the correlated predictors (or pair wise correlations between regions) only a subset of connections was identified. This was not the problem with SPC-EN which achieved superior sensitivity at all the values of R's. Taken together, these results demonstrate that SPC-EN is a more accurate method for inferring sparse partial correlations.
Comparison of SPC-EN with other related approaches
No previous studies have tested and validated the accuracy and stability of partial correlation methods using simulated datasets. In some of these studies where partial correlation methods were applied (Huang et al., 2010; Smith et al., 2011), optimizing the regularization parameter was not considered. The choice of these parameters is critical since they determine the significance of connections between brain regions. In this work, we used novel stability selection procedure to infer significant partial correlations between brain regions. This method overcomes the problems associated with selecting optimal weights for lasso and elastic net regularization.
Huang and colleagues (Huang et al., 2010) and Lee and colleagues (Lee et al., 2011) applied L1-norm regularization to examine functional networks estimated from PET data obtained from clinical populations. Varoquax and colleagues (Varoquaux et al., 2010) applied group lasso regularization on resting state fMRI data. This method imposes a similar structure of network connectivity on all the subjects in the group while estimating network for each subject separately. While this approach is useful for homogeneous groups where network structure can be assumed to be similar for all the subjects in the group; such assumptions may not be appropriate for investigations of clinical and pediatric populations. Beyond this, our study highlights the limitations of using L1-norm regularization when there are a large number of brain regions compared to observations. Another issue which needs to be considered is the inherently high spatial correlations in fMRI data. This is important when spatially adjacent brain regions are involved in the analysis because elastic net regularization handles this situation better than lasso regularization. None of the above studies, except one (Smith et al., 2011), has, however, tested individual methods on simulated datasets. In contrast, we tested and validated our methods on simulated data generated from two small world networks at different prevalence rates. Critically, we applied novel stability selection methods for selecting optimal regularization parameters and inferring significant connections between brain regions. Finally, Smith and colleagues compared several network estimation methods on simulated fMRI data sets (Smith et al., 2011). They found that the performance of inverse covariance estimation using L1-norm regularization and Bayesian network methods (Ramsey et al., 2009) are better compared to lag based methods such as Granger causal methods. However, the largest number of nodes used in this study was 50 and the number of observations used was about 200 with a low prevalence rate of 4.9%. Our simulations suggest that the performance of L1-norm regularization suffers at higher prevalence rates.
Performance of SPC-EN on experimental fMRI data
We used SPC-EN to examine the structure of whole-brain functional connectivity and modules in fMRI data. Application of our method to resting-state fMRI data acquired from 22 healthy adults revealed that SPC-EN estimates large-scale brain networks with several features that reflect known features of human anatomy. These include a ventral stream network module linking the visual cortex with the inferior temporal cortex and parahippocampal gyrus, a dorsal stream network module linking the posterior parietal cortex with the superior frontal gyrus. Notably, SPC-EN estimated large-scale brain networks having high modularity suggesting that our method extracts network structures that are highly modularized consisting of tightly connected modules with sparse inter-module connectivity and therefore is better to interpret than networks derived using traditional functional connectivity methods such as Pearson correlations which produce non-modular networks consisting of large-number of spurious links. Posterior medial cortex, part of to the default mode network (Greicius et al. 2003), was found to be a major hub mediating functional interactions between the network modules. The posterior medial cortex, and the precuneus in particular, is thought to play an important role in self-referential processing imagery and episodic memory (Cavanna and Trimble, 2006). This region is also densely anatomically connected and centrally located, forming a key component of the structural core of the human brain (Hagmann et al., 2008). One study of rsfMRI reported a highly connected precuneus with short path length and low clustering suggesting a hub like role of the region in large-scale brain networks (Achard et al., 2006). These findings were, however, based on arbitrary thresholding of functional connectivity matrices and thus have the potential for high levels of error. In contrast, our finding is independent of such thresholds, as the sparse regularization procedure employed in our method drives weaker connections to exactly zero resulting in sparse interpretable connectivity matrices which do not require further thresholding. Taken together, these findings suggest that the SPC-EN method reveals anatomically-plausible resting-state brain architecture, and more importantly the findings observed are not influenced by parameter selection and yet are highly consistent with convergent findings from previous studies.
In contrast to these findings, brain networks derived using SPC-L1 lacked the features described above and instead revealed network modules which were almost entirely restricted to anatomical nearest-neighbors and their inter-hemispheric homologs (supplementary figure S2). Additionally, the SPC-L1 derived brain networks were significantly sparser compared to SPC-EN. This result mirrors findings from simulations. It is likely that a sparsity promoting method such as SPC-L1 retains strongly coupled regions and eliminates slightly less strong connections. Our results suggest that SPC-EN not only uncovers these SPC-L1 identified strong links but also preserves slightly weaker connections. This observation is consistent with theoretical evidence that the L1-regularization solution used in SPC-L1 is a lower bound to the elastic net solution used in SPC-EN.
Conclusions
We have developed a novel method for estimating functional brain networks consisting of large numbers of regions from fMRI data by estimating partial correlations between the regions. The proposed SPC-EN method uses a combination of L1 and L2-norm regularization called as elastic net which provides sparse and interpretable solutions. Using realistic simulations and experimental data, we show that using this regularization is essential for estimating networks consisting of large number of brain regions compared to fMRI time samples. An important issue, which is often ignored, with elastic net and lasso penalties, is in choosing the regularization parameters which in turn determines the selection of connections between brain regions. In this work, we have developed and used novel stability selection methods to infer significant connections between brain regions. In sum, our findings suggest that SPC-EN together with stability selection provides a more accurate and powerful tool for characterizing connectivity involving a large number of correlated brain regions.
Supplementary Material
Supplementary Figure S1: Estimation of partial correlations using (A) pseudo inverse and (B) proposed sparse partial correlation using elastic net penalty (SPC-EN). Partial correlations were estimated using, time-series of 90 nodes extracted from a single subject's resting state fMRI data. Estimation of partial correlations using pseudo inverse shows that every node is connected to every other node while the proposed method SPC-EN identifies sparse interpretable network structure. Application of network analysis on these estimated partial correlations revealed six and eleven modules respectively for partial correlations and SPC-EN. Partial correlations estimated by SPC-EN resulted in a more modular network compared to that estimated by pseudo inverse.
Supplementary Figure S2: Graphical representation of whole-brain functional network derived using SPC-L1. Left and right sagittal, axial and coronal views are shown, with nodes representing cortical regions based on the AAL atlas and edges representing partial correlation between pairs of nodes. Nodes are plotted using the x, y and z coordinates of their centroids (in mm) in the MNI space and sized according to their connectivity profile, with larger nodes representing highly connected “hub” nodes. Fifteen modules are shown, with nodes belonging to the same module marked in same color.
Acknowledgements
This research was supported by grants from the National Institutes of Health (HD047520, HD059205, HD057610, NS071221). We thank Dr. Trevor Hastie and Ryan Tibshirani for useful discussions and Dr. Stephan Smith for sharing the simulated DCM datasets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balenzuela P, Chernomoretz A, Fraiman D, Cifre I, Sitges C, Montoya P, Chialvo DR. Modular organization of brain resting state networks in chronic back pain patients. Front Neuroinform. 2010;4:116. doi: 10.3389/fninf.2010.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C, Smith S. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Bishop C. Pattern Recognition and Machine Learning. Springer; 2006. [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pekar JJ, Pearlson GD. Latency (in)sensitive ICA. Group independent component analysis of fMRI data in the temporal frequency domain. Neuroimage. 2003;20:1661–1669. doi: 10.1016/s1053-8119(03)00411-7. [DOI] [PubMed] [Google Scholar]
- Cavanna AE, Trimble MR. The precuneus: a review of its functional anatomy and behavioural correlates. Brain. 2006;129:564–583. doi: 10.1093/brain/awl004. [DOI] [PubMed] [Google Scholar]
- Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front Syst Neurosci. 4:8. doi: 10.3389/fnsys.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Hofling H, Tibshirani R. Pathwise Coordinate Optimization. Annals of Applied Statistics. 2007;1:302–332. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Applications of the lasso and grouped lasso to the estimation of sparse grahical models. Stanford University; 2010. Technical Report. [Google Scholar]
- Friston KJ, Buechel C, Fink GR, Morris J, Rolls E, Dolan RJ. Psychophysiological and modulatory interactions in neuroimaging. Neuroimage. 1997;6:218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn Reson Med. 2001;46:515–522. doi: 10.1002/mrm.1222. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Hum Brain Mapp. 2002;15:247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction. 2nd ed. Springer; New York, NY: 2009. [Google Scholar]
- Hoyle DC. Accuracy of Pseudo-Inverse Covariance Learning - A Random Matrix Theory Analysis. IEEE Trans Pattern Anal Mach Intell. 2010 doi: 10.1109/TPAMI.2010.186. [DOI] [PubMed] [Google Scholar]
- Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, Chen K, Reiman E. Learning brain connectivity of Alzheimer's disease by sparse inverse covariance estimation. Neuroimage. 2010;50:935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller D, Friedman N. Probabilistic graphical models Principles and Techniques. The MIT Press; 2009. [Google Scholar]
- Krishnapuram B, Carin L, Figueiredo MA, Hartemink AJ. Sparse multinomial logistic regression: fast algorithms and generalization bounds. IEEE Trans Pattern Anal Mach Intell. 2005;27:957–968. doi: 10.1109/TPAMI.2005.127. [DOI] [PubMed] [Google Scholar]
- Lee H, Lee DS, Kang H, Kim BN, Chung MK. Sparse brain network recovery under compressed sensing. IEEE Trans Med Imaging. 2011;30:1154–1165. doi: 10.1109/TMI.2011.2140380. [DOI] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J Neurosci. 2011;31:3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liang M, Zhou Y, He Y, Hao Y, Song M, Yu C, Liu H, Liu Z, Jiang T. Disrupted small-world networks in schizophrenia. Brain. 2008;131:945–961. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- Marrelec G, Horwitz B, Kim J, Pelegrini-Issac M, Benali H, Doyon J. Using partial correlation to enhance structural equation modeling of functional MRI data. Magn Reson Imaging. 2007;25:1181–1189. doi: 10.1016/j.mri.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Marrelec G, Krainik A, Duffau H, Pelegrini-Issac M, Lehericy S, Doyon J, Benali H. Partial correlation for functional brain interactivity investigation in functional MRI. Neuroimage. 2006;32:228–237. doi: 10.1016/j.neuroimage.2005.12.057. [DOI] [PubMed] [Google Scholar]
- McIntosh AR. Towards a network theory of cognition. Neural Netw. 2000;13:861–870. doi: 10.1016/s0893-6080(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Meinshausen N, Buhlmann P. High-dimensional graphs and variable selection with the Lasso. Annals of Statistics. 2006;34:1436–1462. [Google Scholar]
- Meinshausen N, Bühlmann P. Stability selection. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010;72:417–473. [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Wang P, Zhou N, Zhu J. Partial Correlation Estimation by Joint Sparse Regression Models. J Am Stat Assoc. 2009;104:735–746. doi: 10.1198/jasa.2009.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C. Six problems for causal inference from fMRI. Neuroimage. 2009;49:1545–1558. doi: 10.1016/j.neuroimage.2009.08.065. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Ryali S, Supekar K, Abrams DA, Menon V. Sparse logistic regression for whole-brain classification of fMRI data. Neuroimage. 51:752–764. doi: 10.1016/j.neuroimage.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevade SK, Keerthi SS. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics. 2003;19:2246–2253. doi: 10.1093/bioinformatics/btg308. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuroimage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Sun FT, Miller LM, D'Esposito M. Measuring interregional functional connectivity using coherence and partial coherence analyses of fMRI data. Neuroimage. 2004;21:647–658. doi: 10.1016/j.neuroimage.2003.09.056. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. Journal of Royal Statistical Society: Series B. 1996;58:267–288. [Google Scholar]
- Tibshirani R, Hastie T, Friedman JH. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Bien, Friedman, Hastie, Simon, Taylor, Tibshirani Strong Rules for Discarding Predictors in Lasso-type Problems (To appear in JRSSB) 2011. [DOI] [PMC free article] [PubMed]
- Tononi G, Sporns O. Measuring information integration. BMC Neurosci. 2003;4:31. doi: 10.1186/1471-2202-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Varoquaux G, Gramfort A, Poline JB, Thirion B. Advances in Neural Information Processing Systems. Vancouver, Canada: 2010. Brain covariance selection: better individual functional connectivity models using population prior. [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Yu Q, Sui J, Rachakonda S, He H, Gruner W, Pearlson G, Kiehl KA, Calhoun VD. Altered Topological Properties of Functional Network Connectivity in Schizophrenia during Resting State: A Small-World Brain Network Study. PLoS One. 2011;6:e25423. doi: 10.1371/journal.pone.0025423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang J, Wu Q, Kuang W, Huang X, He Y, Gong Q. Disrupted brain connectivity networks in drug-naive, first-episode major depressive disorder. Biol Psychiatry. 2011;70:334–342. doi: 10.1016/j.biopsych.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net Journal of Royal Statistical Society: Series B Stat. Methedology. 2005;67:301–320. [Google Scholar]
- Zou H, Hastie T, Tibshirani R. On the degrees of freedom. Annals of Statistics. 2007;35:2173–2192. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Estimation of partial correlations using (A) pseudo inverse and (B) proposed sparse partial correlation using elastic net penalty (SPC-EN). Partial correlations were estimated using, time-series of 90 nodes extracted from a single subject's resting state fMRI data. Estimation of partial correlations using pseudo inverse shows that every node is connected to every other node while the proposed method SPC-EN identifies sparse interpretable network structure. Application of network analysis on these estimated partial correlations revealed six and eleven modules respectively for partial correlations and SPC-EN. Partial correlations estimated by SPC-EN resulted in a more modular network compared to that estimated by pseudo inverse.
Supplementary Figure S2: Graphical representation of whole-brain functional network derived using SPC-L1. Left and right sagittal, axial and coronal views are shown, with nodes representing cortical regions based on the AAL atlas and edges representing partial correlation between pairs of nodes. Nodes are plotted using the x, y and z coordinates of their centroids (in mm) in the MNI space and sized according to their connectivity profile, with larger nodes representing highly connected “hub” nodes. Fifteen modules are shown, with nodes belonging to the same module marked in same color.