Abstract
Background
Recently the SNP identified as rs1260326, in the glucokinase regulatory protein (GCKR) was associated with hypertriglyceridemia in adults. Since accumulation of triglycerides in hepatocytes represents the hallmark of the steatosis, we aimed to investigate whether this variant might be associated with fatty liver (hepatic fat content, HFF%). Moreover, since recently rs738409 in the PNPLA3 and rs2854116 in the APOC3 were associated with fatty liver recently, we explored how the GCKR SNP and these two variants jointly influence hepatosteatosis.
Methods and Results
We studied 455 obese children and adolescents (181 Caucasians, 139 African Americans and 135 Hispanics). All underwent an OGTT and fasting lipoprotein subclasses measurement by proton NMR. A subset of 142 children underwent a fast gradient MRI to measure the HFF%.
The rs1260326 was associated with elevated triglycerides (Caucasians p=0.00014; African Americans p=0.00417) large VLDL (Caucasians p=0.001; African Americans p=0.03) and with fatty liver (Caucasians p= 0.034; African Americans p= 0.00002; and Hispanics p= 0.016). The PNPLA3, but not the APOC3 rs2854116 SNP, was associated with fatty liver but not with triglycerides levels. There was a joint effect between the PNPLA3 and GCKR SNPs, explaining 32% of HFF% variance Caucasians (p=0.00161), 39.0% in African Americans (p=0.00000496), and 15% in Hispanics (p=0.00342).
Conclusions
The rs1260326 in GCKR is associated with hepatic fat accumulation along with large VLDL, and triglycerides levels. GCKR and PNPLA3 act together to convey susceptibility to fatty liver in obese youths.
Keywords: GCKR, PNPLA3, SNPs, obesity, youths
Introduction
Triglyceride (TG) accumulation in the liver is the earliest hallmark of Non-Alcoholic Fatty Liver Disease (NAFLD), which has emerged as the most common cause of chronic liver disease in pediatrics (1, 2). The mechanisms underlying hepatocellular accumulation of TG and its relationship to lipotoxicity are not entirely clear. Recent studies have started to unravel the genetic underpinnings that convey susceptibility to NAFLD. In particular a common missense variant (rs738409), characterized by a C-to-G substitution encoding an isoleucine-to-methionine substitution at the amino acid position 148 (I148M), in the patatin like phospholipase 3 (PNPLA3) gene has been repeatedly associated with hepatic TG accumulation, hepatocellular injury and progression of NAFLD in adults (3, 4). This observation has been replicated in children as well (5, 6). More recently two single nucleotide polymorphisms (SNPs) in the in the promoter region of the gene encoding apolipoprotein C3 (APOC3), the rs2854116 and the rs2854117, have been found to be associated with fatty liver in a group of healthy Asian Indian men (7).
Given the essential role that hepatic triglyceride accumulation has in the development of NAFLD we hypothesized that a common gene variant associated with hypertriglyceridemia might also affect hepatic triglyceride accumulation. We began our search by focusing on a SNP (rs1260326) in the glucokinase regulatory protein (GCKR) gene, previously associated with triglycerides levels in Genome Wide Association Studies (GWAS) (8, 9). The GCKR gene product, the glucokinase regulatory protein (GCKRP), regulates the glucokinase (GCK) activity competitively with respect to the substrate glucose (10, 11) inhibiting GCK activity (12). The rs1260326 is a functionally relevant SNP consisting of a C to T substitution coding for a proline to leucine substitution at the position 446 (P446L). Clues to the molecular mechanisms driving the association of this variant with high triglyceride levels come from detailed kinetic studies of the human recombinant GCKRP (13). It has been demonstrated that the GCKRP L466 variant results in a protein that has reduced regulation by physiological concentrations of fructose 6 phosphate, thus resulting indirectly in a constant increase in GCK activity. The increased GCK activity in the liver is predicted to enhance the glycolytic flux, thus promoting hepatic glucose metabolism and elevating the concentrations of malonyl CoA, a substrate for de novo lipogenesis (DNL), which may account for about 26% of fat accumulation in the liver (14). The increase in malonyl CoA will then lead to the inhibition of the carnitine-palmitoyl which in turn blocks fatty acid oxidation (13). Moreover, very recently Speliotes et al. have shown that the non-functional GCKR rs780094 variant, which is in strong linkage disequilibrium with the rs1260326 is associated with hepatic fat content in adult samples of European descent (15). Based on these premises in the present study we aimed to determine whether the rs1260326 GCKR gene variant might be associated, along with high triglycerides, also with hepatic fat accumulation and to explore the joint effect from the GCKR rs1260326 variant and SNPs previously associated with hepatic fat accumulation in PNPLA3 and APOC3. Moreover, given the potential role of the studied SNPs in the triglycerides metabolism, we aimed also to explore whether these three SNPs were associated with large VLDL levels.
Materials and Methods
Subjects
We studied 455 obese children and adolescents (181 Caucasians, 139 African Americans and 135 Hispanics; mean age 12.8 ± 2.9 years; mean z-score BMI 2.32 ± 0.51) from New Haven area (New Haven, CT) recruited through the Yale Pediatric Obesity Clinic. Caucasians tended to be older (13.4±2.94 years) than African Americans (12.9±2.83 years) and Hispanics (12.0 ±2.97 years) (p=0.002); while African Americans tended to show a higher z-score BMI (2.32±0.60) than Caucasians (2.23±0.55); and Hispanics (2.12±0.89) (p=0.056). Forty six Caucasians (31 girls), 31 African Americans (22 girls) and 37 Hispanics (23 girls) showed IGT, while 12 Caucasians (7 girls), 20 African Americans (6 girls) and 10 Hispanics (7 girls) showed type 2 diabetes (p=0.15). The prevalence of subjects showing impaired glucose tolerance (IGT) or type 2 diabetes did not differ among the groups (p=0.15).
The study was approved by the Yale University Human Investigation Committee. Parental informed consent and child assent were obtained from all participants.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes. Genotyping for GCKR rs1260326 was performed with the use of a matrix assisted laser desorption-ionization time of flight mass spectrometry on the MassARRAY platform (Sequenom). To assess the nature of genetic variation across ethnic groups at GCKR and its relationship to the carriers of the functional T-allele at rs1260326 we analyzed multi-SNP haplotypes at the GCKR gene. We genotyped 7 more SNPs extending across the 25KB GCKR gene using the Sequenom MassArray (compact) system. Details are shown in the excel supplemental file.
The PNPLA3 rs738409 variant was genotyped by automatic sequencing as previously reported (5). The SNPs in or around the APOC3 gene, rs2854116 and rs2854117 belongs to a larger linkage disequilibrium block spanning the APOA5/APOA4/APOC3/APOA1 gene region on chromosome 11 and are reported to be in linkage disequilibrium (16). Thus only the rs2854116 was genotyped and used to test the associations. Further information concerning the genotyping is provided as Supplemental Material.
Metabolic Studies
All metabolic studies were done at the Yale Center for Clinical Investigation (YCCI) at 8.00 am following a 10 – 12 h overnight fast.
A standard OGTT (1.75 g/Kg body weight, up to 75 g) was performed on all subjects. The Whole Body Insulin Sensitivity index (WBISI) was used to determine insulin sensitivity.
Imaging Studies
Imaging studies were performed in a subgroup of 142 subjects (67M/75F, 45C/45AA/52H, mean age 12.5±2.6 years; mean z-score BMI 2.22±0.70). Ninety showed a normal glucose tolerance (NGT), forty six showed impaired glucose tolerance (IGT) and six showed type 2 diabetes. This subgroup did not differ from the main cohort for age, sex, race, z-score BMI, and glucose tolerance. Details about the abdominal magnetic resonance imaging studies are provided in supplemental Materials and Methods section.
Biochemical analyses
Plasma glucose was determined using a glucose analyzer by the glucose oxidase method (Beckman Instruments, Brea, CA). Plasma insulin was measured by the Linco RIA, lipid levels using an Auto-Analyzer (model 747–200), liver enzymes, using standard automated kinetic enzymatic assays. To measure large VLDL fasting plasma samples were taken and analyzed utilizing a 400 MHz proton NMR analyzer at Liposcience (Raleigh, NC).
Statistical Analyses
The chi-square test was used to assess whether the genotypes were in Hardy Weinberg equilibrium and to test differences in genotype distribution among different ethnic groups. Prior to analyze the data all the variables were tested for normality, with non-normally distributed variables log transformed to be better approximated by normality, except for HFF% for which a square root transformation was used. Within each ethnic group the association between the genotypes and quantitative traits was evaluated by coding the genotype with an additive model of inheritance, i.e. the genotype is coded with 0, 1, or 2 corresponding to the number of minor alleles carried by each individual; age, sex, z-score BMI and glucose tolerance status were used as covariates when appropriate.
The partial correlation coefficients (r2) were used to evaluate the degree of variance explained by the genotype and to determine the combined effect of the GCKR and PNPLA3 variants on HFF %. The interaction between two genetic markers was evaluated by adding an interaction term between the two genotypes in a regression model. Unless otherwise specified, for all the data raw means and standard deviations are shown. The power analysis to evaluate whether our dataset was able to distinguish differences in our primary outcomes (triglycerides, large VLDL and HFF%) among the genotypes was performed in Quanto, assuming a gene only effect. Data concerning the power calculations is provided as supplemental material. A logistic regression was used to assess the odds of showing the metabolic syndrome as defined by Cook et al. (17) by GCKR genotype merging all the ethnic groups.
Results
Allele Frequency
The GCKR SNP rs1260326 minor allele (T) frequency was 0.446 in Caucasians, 0.129 in African Americans and 0.355 in the Hispanics (p-value for population differences in allelic frequency <.0001).
The frequency of the PNPLA3 rs738409 minor allele (G) was 0.266 in Caucasians, 0.170 in African Americans and 0.417 in the Hispanics (p-value for population differences in allelic frequency <.0001).
The frequency of the APOC3 rs2854116 C allele was 0.371 in Caucasians, 0.692 in African Americans and 0.394 in the Hispanics (p-value for population differences in allelic frequency <.0001). The allele frequencies were consistent with those shown in similar ethnic groups in the Allele Frequency Database (ALFRED, http://alfred.med.yale.edu) as well as in HAPMAP (http://hapmap.ncbi.nlm.nih.gov/). Within each ethnic group there was no evidence against the null hypothesis that the genotype distribution was in Hardy Weinberg equilibrium for all of the variants (all p>0.05).
GCKR Haplotypes and Frequencies
We also analyzed 8-SNP haplotypes extending across the 25KB GCKR gene in 10 HAPMAP populations and in our 3 local populations. (The details of these haplotype analyses can be found in the supplemental Materials and in the supplemental excel file.) We found 52 haplotypes with estimated frequencies across these populations and 13 of the 52 carry the functional T-allele. However, a relatively simple underlying pattern prevails in that the nine non-African populations have just one common haplotype that accounts for 93% to 98% of the T-alleles while in the four African populations the same haplotype accounts for 38% to 85% of the individuals carrying the T-allele. In the three studied populations 85% to 94% of the functional T-alleles present are carried on the same common haplotype background at GCKR. Although we acknowledge that the heterogeneity in nearby regulatory variants could, of course, still exist, the remarkable predominance of this one haplotype background at GCKR accounting for carriers of the functional T-allele provides some assurance of the relative genetic homogeneity of the GCKR effects observed in the three populations studied.
Because the analyses we report here primarily involve functional variation at the GCKR gene we have not carried out analyses estimating and correcting for background population stratification effects that might be present and that are of special concern when trying to understand the validity of associations based on non-functional SNPs.
Anthropometrics and Metabolic phenotypes by genotype
GCKR rs1260326 (table 1)
Table 1.
Clinical Characteristics of the subjects stratified by ethnicity and GCKR rs1260326 genotype
| Caucasians | African Americans | Hispanics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC (56) | CT (90) | TT (35) | p | CC (104) | CT/TT (34/1) | p | CC (58) | CT (58) | TT (19) | p | |
| Age (years) | 13.1±.85 | 13.3±2.90 | 14.4±2.93 | 0.3 | 13.1±2.9 | 12.9±2.64 | 0.8 | 12.8±2.8 | 11.8±3.1 | 12.3±2.8 | 0.2 |
| Sex (M/F) | 37/63 | 43/57 | 46/54 | 0.7 | 38/62 | 34/66 | 0.7 | 41/58 | 52/38 | 16/84 | 0.03 |
| NGT/IGT/T2D | 67/27/6 | 68/24/8 | 69/26/5 | 0.9 | 62/21/17 | 69/26/5 | 0.2 | 60/31/9 | 69/26/5 | 68/21/11 | 0.8 |
| Anthropometrics | |||||||||||
| BMI (Kg/m2) | 33.2±7.3 | 33.6±6.4 | 33.9±5.6 | 0.8 | 35.9±8.3 | 35.7±8.2 | 0.9 | 32.8±8.8 | 31.6±6.9 | 31.0±5.9 | 0.6 |
| BMI Z-score | 2.17±0.68 | 2.27±0.50 | 2.23±0.42 | 0.4 | 2.34±0.63 | 2.36±0.47 | 0.9 | 2.08±1.05 | 2.17±0.70 | 2.04±096 | 0.8 |
| Body Fat (%) | 430±10.4 | 42.3±8.3 | 42.6±6.9 | 0.9 | 45.9±10.4 | 45.7±9.0 | 0.9 | 39.6±10.9 | 41.4±9.8 | 45.1±5.3 | 0.1 |
| Lipid Profile | |||||||||||
| CHOLESTEROL *(mg/dl) | 155.9±38.5 | 164.4±46.9 | 167.7±40.7 | 0.4 | 153.5±25.5 | 159.6±27.2 | 0.3 | 160.4±37.8 | 150.5±439 | 148.4±34.8 | 0.4 |
| HDL*(mg/dl) | 43.2±10.1 | 41.6±10.5 | 39.6±11.0 | 0.3 | 45.0±10.9 | 41.8±7.3 | 0.1 | 42.1±11.3 | 42.1±9.9 | 43.2±11.5 | 0.9 |
| LDL*(mg/dl) | 93.4±31.4 | 97.7±32.6 | 92.6±38.4 | 0.6 | 93.3±21.7 | 95.4±21.6 | 0.7 | 98.9±34.2 | 87.1±39.4 | 81.8±26.2 | 0.1 |
| Glucose and Insulin levels | |||||||||||
| FASTING GLUCOSE*(mg/dl) | 93.0±10.9 | 94.6±10.1 | 93.1±8.5 | 0.5 | 96.0±11.8 | 93.5±8.4 | 0.3 | 97.0±9.6 | 94.4±9.9 | 91.8±6.9 | 0.1 |
| GLUCOSE 120*(mg/dl) | 125.7±31.1 | 130.2±33.7 | 134.0±30.4 | 0.4 | 131.3±35.8 | 127.5±26.4 | 0.6 | 131.4±27.8 | 122.2±20.4 | 135.1±37.9 | 0.1 |
| FASTING INSULIN* (µU/mL) | 39.3±28.6 | 31.9±16.3 | 30.4±14.1 | 0.1 | 39.0±22.4 | 35.9±18.7 | 0.5 | 37.2±21.5 | 33.2±22.5 | 40.9±22.4 | 0.4 |
| WBISI* | 1.62±0.93 | 1.84±1.19 | 1.71±.80 | 0.6 | 1.68±1.38 | 1.66±0.84 | 0.9 | 1.63±1.21 | 1.99±1.35 | 1.46±1.08 | 0.2 |
| Liver Enzymes | |||||||||||
| ALT*(UI/L) | 24.3±13.8 | 25.3±17.1 | 25.4±19.2 | 0.6 | 16.1±7.7 | 15.8±8.1 | 0.9 | 18.9±8.3 | 22.9±17.6 | 34.8±23.5 | 0.03 |
| AST*(UI/L) | 23.1±6.6 | 23.3±8.4 | 23.4±9.0 | 0.7 | 21.8±5.6 | 18.9±3.6 | 0.09 | 22.6±4.9 | 24.0±9.3 | 26.2±10.6 | 0.06 |
log transformed and adjusted for age, gender and z-score BMI
Within each ethnic group there were no differences among the genotypes for age, glucose tolerance status, BMI, z-score BMI and percent body fat (table 1). In the Hispanic sample, subjects homozygous for the minor allele showed a higher percentage of girls than boys (p=0.032), but this was no longer significant after adjustments for multiple comparisons.
We did not observe any difference in fasting glucose, fasting insulin, 2 hours glucose, and insulin sensitivity (WBSI) among the genotypes in any of the ethnic groups (table 1).
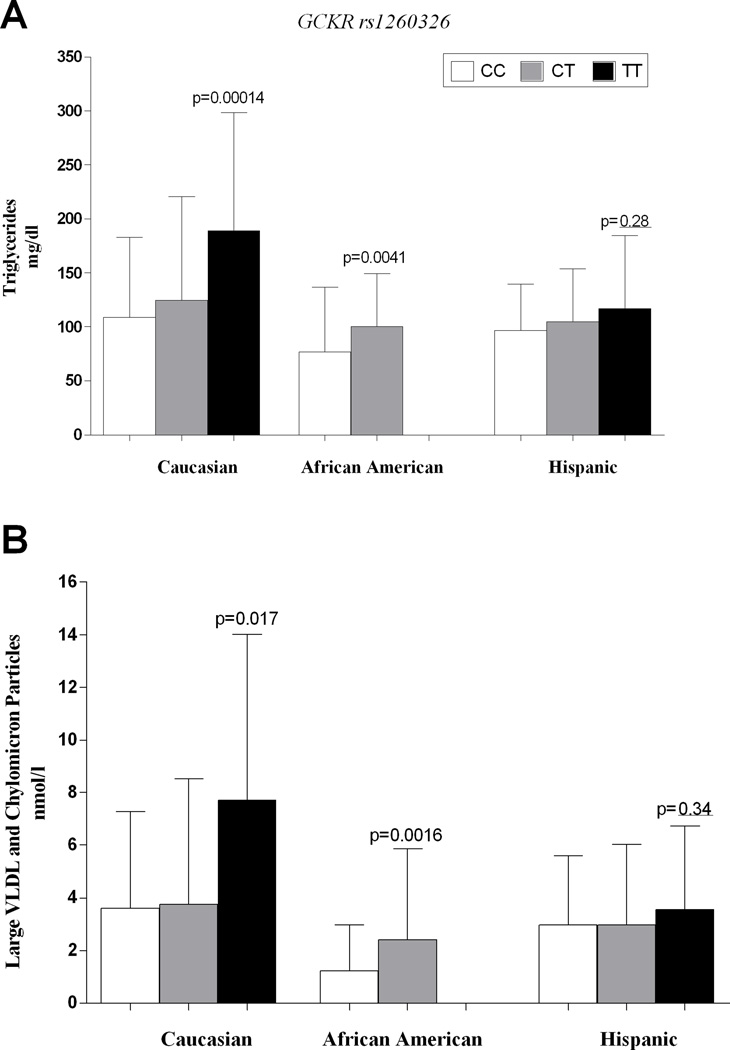
Subjects homozygous for the T allele of the GCKR rs1260326 showed higher triglyceride levels than the other genotypes independently of age, gender, z-score BMI and glucose tolerance (figure 1). The GCKR variant explained 8.12% of triglyceride variance in Caucasians (p=0.00014) and 6.4% in African Americans (p=0.00417). The differences in triglyceride levels remained statistically significant after adjusting for age, gender, z-score BMI and glucose tolerance (Caucasians adjusted p-value=0.0012; African Americans adjusted p-value=0.0048).
Figure 1.
The figure shows the triglyceride (panel A) and large VLDL and chylomicron particle levels (panel B) as well as the LDL size (panel C) according to GCKR rs1260326 genotype in all the ethnic groups.
Panel A. Triglyceride levels. The differences in triglyceride levels remained statistically significant after adjusting for age, gender, z-score BMI and glucose tolerance. Caucasian adjusted p-value=0.0012; African American adjusted p-value=0.0048.
Panel B. Large VLDL and chylomicron particle levels. The p-value after adjusting for age, gender, z-score BMI and glucose tolerance was =0.036 for Caucasians and p=0.0010 for African Americans.
In Caucasians and African American groups, subjects homozygous for the T allele showed higher levels of large VLDL (p=0.017 and 0.0016 respectively) than subjects carrying the other genotypes (figure 1); in Hispanics, although not statistically significant (p=0.28), the same trend for the total and the large VLDL levels was observed (figure 1). The p-value after adjusting for age, gender, z-score BMI and glucose tolerance was =0.036 for Caucasians and p=0.0010 for African Americans.
Moreover, subjects homozygous for the T allele had a higher prevalence of the metabolic syndrome as defined by Cook et al. (20) (CC=39.5%; CT=43.2%; TT=51.9%. Chi square= 6.38, p=0.0115) and they also had two times higher odds of showing the features of metabolic syndrome than CC homozygotes (OR 2.461; 95%CI 1.131–5.033, p=0.0181) after adjustment for age, gender, ethnicity, z-score BMI and glucose tolerance.
PNPLA3 rs738409 (Supplementary table 1)
The three genotype groups were similar in terms of age, gender, z-score BMI in all the ethnic groups as well as for the other variables listed in the Supplementary table 1. As previously reported (3) we did not observe any difference in terms of insulin resistance as measured as WBISI. Subjects homozygous for the minor allele tended to show higher ALT levels, but the difference among genotypes was not statistically significant.
APOC3 rs2854116 (Supplementary table 2)
The three genotype groups were similar in all three ethnic groups for age, gender and z-score BMI. No differences in liver enzymes, fasting glucose and insulin, 2-h glucose and WBISI were observed among the APOC3 genotypes. The African Americans carrying the C allele of the APOC3 variant showed a significant reduction in plasma triglycerides levels (p=0.010) (figure 1).
Association between GCKR rs1260326 and PNPLA3 rs738409 and Hepatic Fat Content (HFF%)
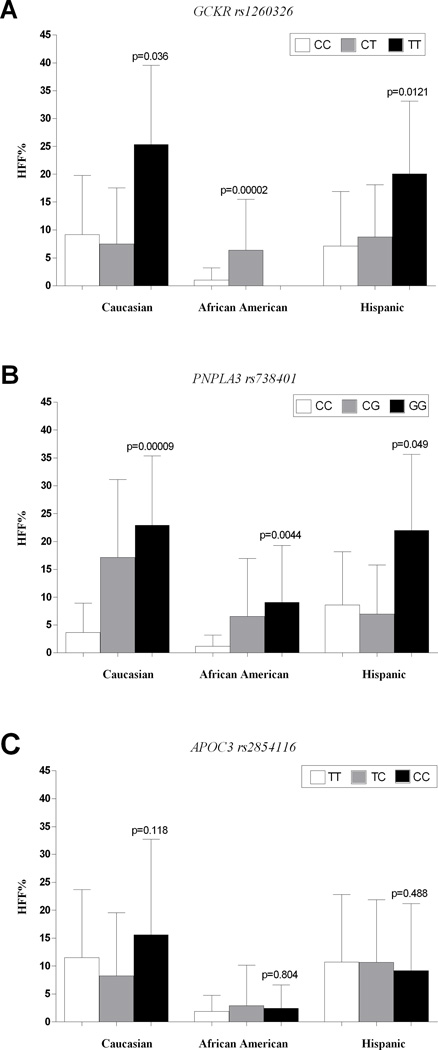
The GCKR rs1260326 was associated with higher hepatic fat content in all the ethnic groups (figure 2). The GCKR variant explained 9.2% of the HFF% variance in Caucasians (p= 0.036), 32.5% of variance (p= 0.00002) in African Americans, and 10.4% of variance in Hispanics (p=0.016). After adjusting for age, gender, z-score BMI and glucose tolerance this association between the GCKR rs1260326 variant and HFF% showed a p-value of 0.000221 and 0.0159 in African Americans and Hispanics, respectively. Although in Caucasians the association was not significant anymore under an additive model after adjusting for the covariates (p-value 0.16), when we tested the recessive inheritance model, which seemed to fit better in this group given the HFF% distribution across the genotypes, after adjusting for the covariates the p-value was 0.0279.
Figure 2.
The figure shows the HFF% according to the GCKR rs1260326 (panel A), the PNPLA3 rs738409 (panel B) and the APOC3 rs2854116 (panel C) genotypes in the three ethnic groups.
Panel A. HFF% according to GCKR rs1260326. The differences in HFF% remained statistically significant after adjusting for age, gender, z-score BMI and glucose tolerance (Caucasian adjusted p-value = 0.16 under an additive model and 0.0279 under a recessive model; African American adjusted p-value = 0.00022; Hispanic adjusted p-value= 0.0159).
Panel B. HFF% according to PNPLA3 rs738409. The differences in HFF% remained statistically significant after adjusting for age, gender, z-score BMI and glucose tolerance in all the ethnic groups (Caucasian adjusted p-value = 0.00039; African American adjusted p-value = 0.0086; Hispanic adjusted p-value = 0.09).
Panel C. HFF% according to APOC3 rs2854116. There was no difference in terms of HFF% among the APOC3 rs2854116 groups of genotype.
For the PNPLA3 variant, subjects carrying the minor (G) allele showed higher hepatic fat content in all the ethnic groups (figure 2). The PNPLA3 variant explained 29.7% of the HFF% variance in Caucasians (p= 0.00009), 6.8% in African Americans (p=0.0044), and 5.8% in Hispanics (p=0.0490). This association was statistically significant after adjustment for age, gender, z-score BMI and glucose tolerance in Caucasians (p=0.00039) and African Americans (p=0.00866), the same trend was still observed in Hispanics (p=0.09).
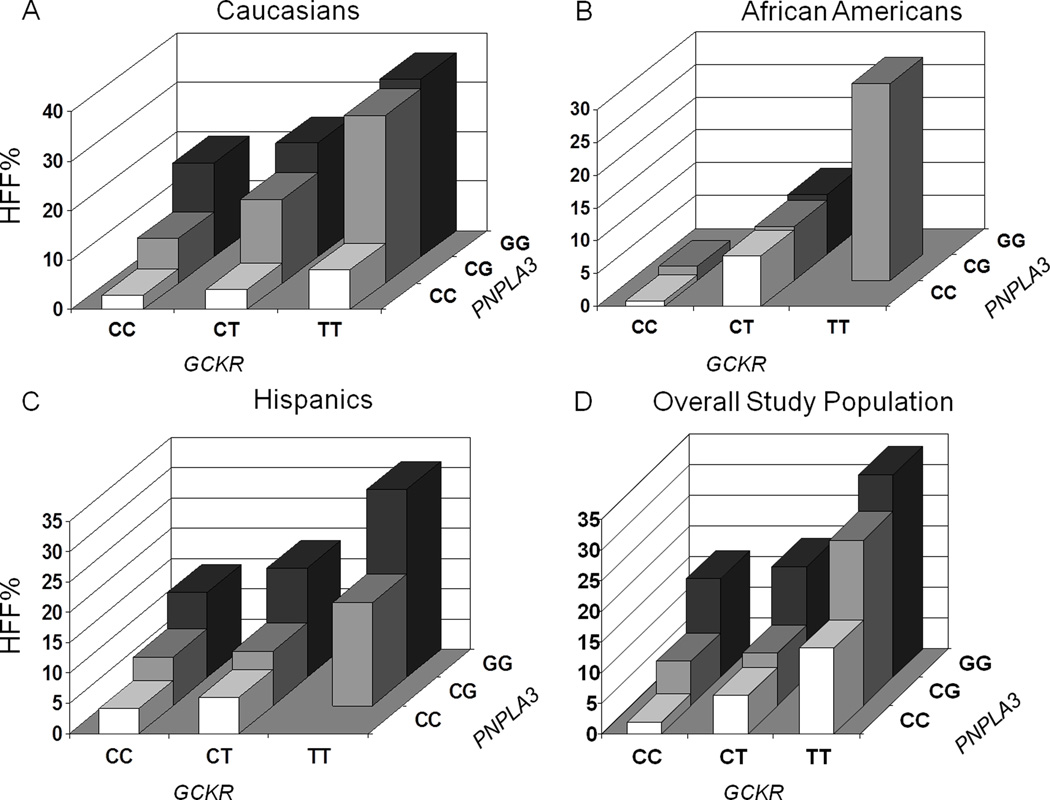
When we tested the joint effect of the two variants, we could explain 32% of the variance of HFF% in Caucasians (p=0.00161), 39.0% in African Americans (p=0.00000496), and 15% in Hispanics (p=0.00342) (figure 3).
Figure 3.
The figure shows the joint effect of the rs738409 and rs1260326 SNPs on hepatic fat content (HFF%) in Caucasians (panel A), African Americans (panel B), Hispanics (panel C) and in the overall population (panel D).
There was no association between the APOC3 gene variants and the HFF% in any of the three ethnic groups (Caucasians p=0.118; African Americans p=0.804; Hispanics p=0.488) (figure 2).
Since the GCKR and PNPLA3 SNPs were significantly associated with HFF% we evaluated whether there was any interactive effect between the two variants in each of the three ethnic groups. No statistically significant interaction was observed between these two variants (Caucasian p=0.773; African American p=0.194; Hispanic p=0.826).
Discussion
Association between GCKR rs1260326 and PNPLA3 738109 with Fatty Liver
In the present study we found that the minor allele of the GCKR rs1260326 is associated with fatty liver and with higher serum triglycerides, large VLDL levels in obese children and adolescents. This association was evident in all three ethnic groups studied and it was independent of age, gender, z-score BMI and glucose tolerance.
Moreover, we replicated the association between the PNPLA3 rs738409 and HFF% previously described in a small group of children and adolescents (5). We were not able to replicate the association between APOC3 rs2854116 variant and fatty liver reported in male Asian Indian adults by Petersen et al (7).
These findings are in agreement with those very recently published by Speliotes et al (15). In that study, based on meta-analysis of GWAS in adult samples of European descent, the authors identified five variants including the GCKR rs780094, which is in strong linkage disequilibrium with the rs1260326 examined in this study. Interestingly, Speliotes et al. reported that the variance for HFF% explained by PNPLA3 and GCKR SNPs are 0.2% and 2.41% respectively, while the variance for HFF% observed for Caucasians in our study is 9.2% for GCKR rs1260326 and 29.7% PNPLA3 rs738409. This remarkable difference may be due to the fact that our study population is composed exclusively by obese youths. Among obese youths the effect size of each variant on the phenotype may be more pronounced than in adults because of the lack of confounding environmental factors (e.g. alcohol etc.) that may mask or attenuate the gene variant effect. Furthermore obese youths showing clinical features once believed to be exclusive of the adulthood may be a “genetically enriched” population in which the effect common variants on the phenotype may be more pronounced.
We have also observed an additive effect between the GCKR variant and the PNPLA3 rs738409 on HFF%. In fact, the additive effect of these variants explained about 32% of HFF% variance in Caucasians, 39% in African Americans and 15% in Hispanics. The lower additive effects in the Hispanics, the ethnic group with the highest prevalence of hepatic steatosis, suggests that some other genetic or environmental risk factors might account for the majority of their variance in liver fat content.
What is the mechanism by which the rs1260326 GCKR minor allele might lead to hepatic fat accumulation? A recent study has shown that the leucine in position 446 in the GCKRP protein confers a reduced capability to respond to fructose 6 phosphate, resulting indirectly in a constant increase in GCK activity in the liver, (13) which leads to higher glycolytic flux and hence increasing the glucose uptake by the liver. The increased glycolysis would raise the levels of Malonyl CoA, which in turn may favor the increase in triglyceride levels through two different mechanisms either by serving as a substrate for De Novo lipogenesis and by inhibiting carnitine-palmitoyl transferase-1, thus blocking fatty acid oxidation (13). The increase in large VLDL that we have observed is actually consistent with this pathway. The increase of large VLDLs, which represent the youngest VLDL, may be probably due to both an increased synthesis of triglycerides into the liver as consequence of an increased DNL, whose contribution to the accumulation of fat into the liver in obese adults is well established (14), and a reduced beta oxidation.
The mechanism by which the PNPLA3 variant leads to hepatic fat accumulation is still unclear. Although previous studies suggested that this variant may cause a gain of function of the protein, which would act as a lipogenic factor (18–21), More recent observations support the hypothesis that the PNPLA3 plays a role in hydrolysis of glycerolipids and thus that the rs738409 variant causes a loss of this function (22).
Association between GCKR rs1260326 and triglycerides
Along with the increase of liver fat content and triglycerides levels, subjects carrying the T allele showed larger VLDL size and a greater prevalence of the metabolic syndrome. Thus this phenotype might suggest that rs1260326 SNP increases the future cardiovascular risk in these obese children and adolescents. Recent studies have shown in two large adult populations that although the rs1260326 SNP is clearly associated with high triglycerides levels, it doesn’t seem to be associated with an increased risk to develop myocardial infarction and ischemic heart disease (23, 24). In only one study, an association between the rs1260326 minor allele and an increased intima-media thickness in a group of 455 subjects with metabolic syndrome was observed (25), while the association between the rs780094, which is in strong linkage disequilibrium with the rs1260326, was not observed in a large group of patients enrolled in the ARIC study (26). Thus, whether the rs1260326 variant, confers an increased risk to develop cardiovascular disease in the long term is still unclear. Considering that subjects homozygous for the T allele also have fatty liver, as indicated by the elevated hepatic fat content fraction, makes the whole scenario even more complex. Indeed, hepatic steatosis per se is a strong risk factor for insulin resistance in both adults and children (27) and it is associated with an adverse cardiovascular lipoprotein profile (28). Moreover, recent reports have clearly shown that nonalcoholic fatty liver disease (NAFLD), of which steatosis represents the first step, in overweight and obese children is associated with multiple cardiovascular risk factors (29) and that children with NAFLD may develop end-stage liver disease with the consequent need for liver transplantation (2). How might we explain that a SNP that causes an increase in TG levels and hepatic fat accumulation does not seem to be associated with long term adverse cardiovascular risk? The answer may lie in the effect of rs1260326 on glucose metabolism. As stated earlier, the P446L variant is predicted to cause a permanent increase of GCK activity leading to an increase in glycolysis, which in turn will cause a lowering of plasma glucose. Several studies have, in fact, shown that subjects homozygous for the GCKR rs1260326 minor allele have lower plasma glucose levels, lower serum insulin and lower HOMA-IR than the other genotypes. With these observations in mind, one could speculate that the beneficial effect of the GCKR rs1260326 on insulin resistance would balance out the increase in triglycerides and in general the adverse cardiovascular profile.
In the present study we did not observe any effects of the variants on glucose or insulin levels in the different ethnic groups. This finding is in agreement with a recent meta-analysis of over 6000 children by Baker et al. who have shown a lower effect estimates of the GCKR rs780094 (which is in strong linkage disequilibrium with the rs1260326) on the glucose levels than that seen in adults (30). Thus, the possibility exists that association of these variants with glucose might increase with age (30).
Lack of association between APOC3 rs2854116 and hepatic fat content
Consistent with a recent reports (31–33), but in contrast with the study of Petersen’s et al (6), we did not observe any association between the APOC3 variant and fatty liver. The lack of an association between the APOC3 genotype and the hepatic fat content may be due to the different ethnic background of the studied groups. Moreover, the effect of the APOC3 variant seems to be driven by the insulin resistance (6), but our population was composed by obese children and adolescents all of whom show some degree of insulin resistance. Thus, the presence of obesity induced insulin resistance may mask the effect of the APOC3 variant on hepatic fat accumulation thus explaining the difference between studies.
Limitations and Conclusions
We are aware that this study has some limitations. The small sample size represents probably the main pitfall of the study, raising the possibility of false negative results. Moreover, the lack of histological data did not allow us to explore whether the rs1290329 might be implicated in the progression of liver damage and in particular in the development of a clinically relevant state such as liver fibrosis. This latter, indeed, does not depend only on liver fat accumulation, but other factors such as insulin resistance, play a pivotal role in its development (34).
In conclusion, the rs1260326 SNP in the GCKR gene is associated with hepatic fat accumulation and influence of particle size of VLDL. These results suggest that the GCKR variant may lead to hepatic fat accumulation through an increased hepatic triglycerides production. The presence of both the GCKR and PNLPA3 genetic variants act together to confer susceptibility to fatty liver in obese youths.
Supplementary Material
Acknowledgments
Funding Sources. This work was supported by the American Hearth Association (AHA) (11CRP5620013 to N.S.) and the National Institutes of Health (NIH) (grants R01-HD-40787, R01-HD-28016, and K24-HD-01464 to S.C.).
Footnotes
Disclosures. None
Contributor Information
Nicola Santoro, Email: nicola.santoro@yale.edu.
Clarence K. Zhang, Email: clarence.k.zhang@yale.edu.
Hongyu Zhao, Email: hongyu.zhao@yale.edu.
Andrew J Pakstis, Email: andrew.pakstis@yale.edu.
Grace Kim, Email: grace.kim@yale.edu.
Romy Kursawe, Email: romy.kursawe@yale.edu.
Daniel J. Dykas, Email: daniel.dykas@yale.edu.
Allen E. Bale, Email: allen.bale@yale.edu.
Cosimo Giannini, Email: cosimo.giannini@yale.edu.
Bridget Pierpont, Email: bridget.pierpont@yale.edu.
Melissa M. Shaw, Email: melissa.m.shaw@yale.edu.
Groop Leif, Email: leif.groop@med.lu.se.
Sonia Caprio, Email: sonia.caprio@yale.edu.
References
- 1.Mencin AA, Lavine JE. Nonalcoholic fatty liver disease in children. Curr Opin Clin Nutr Metab Care. 2011;14:151–157. doi: 10.1097/MCO.0b013e328342baec. [DOI] [PubMed] [Google Scholar]
- 2.Feldstein AE, Charatcharoenwitthaya P, Treeprasertsuk S, Benson JT, Enders FB, Angulo P. The natural history of non-alcoholic fatty liver disease in children: a follow-up study for up to 20 years. Gut. 2009;58:1538–1544. doi: 10.1136/gut.2008.171280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romeo S, Huang-Doran I, Baroni MG, Kotronen A. Unravelling the pathogenesis of fatty liver disease: patatin-like phospholipase domain-containing 3 protein. Curr Opin Lipidol. 2010;21:247–252. doi: 10.1097/mol.0b013e328338ca61. [DOI] [PubMed] [Google Scholar]
- 5.Santoro N, Kursawe R, D'Adamo E, Dykas DJ, Zhang CK, Bale AE, et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52:1281–1290. doi: 10.1002/hep.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valenti L, Alisi A, Galmozzi E, Bartuli A, Del Menico B, Alterio A, et al. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1274–1280. doi: 10.1002/hep.23823. [DOI] [PubMed] [Google Scholar]
- 7.Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 9.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matschinsky FM. Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes. 1990;39:647–652. doi: 10.2337/diab.39.6.647. [DOI] [PubMed] [Google Scholar]
- 11.Matschinsky FM. Regulation of pancreatic beta-cell glucokinase: from basics to therapeutics. Diabetes. 2002;51:S394–S404. doi: 10.2337/diabetes.51.2007.s394. [DOI] [PubMed] [Google Scholar]
- 12.Slosberg ED, Desai UJ, Fanelli B, St Denny I, Connelly S, Kaleko M, et al. Treatment of type 2 diabetes by adenoviral-mediated overexpression of the glucokinase regulatory protein. Diabetes. 2001;50:1813–1820. doi: 10.2337/diabetes.50.8.1813. [DOI] [PubMed] [Google Scholar]
- 13.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3′ untranslated region polymorphisms. Proc Natl Acad Sci U S A. 1993;90:4562–4566. doi: 10.1073/pnas.90.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook S, Auinger P, Li C, Ford ES. Metabolic syndrome rates in United States adolescents, from the National Health and Nutrition Examination Survey 1999–2002. J Pediatr. 2008;152:165–170. doi: 10.1016/j.jpeds.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Chang B, Li L, Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52:1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning JD, Cohen JC, Hobbs HH. Patatin-like phospholipase domain-containing 3 and the pathogenesis and progression of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1189–1192. doi: 10.1002/hep.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011 doi: 10.1074/jbc.M111.290114. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varbo A, Benn M, Tybjærg-Hansen A, Grande P, Nordestgaard BG. TRIB1 and GCKR polymorphisms, lipid levels, and risk of ischemic heart disease in the general population. Arterioscler Thromb Vasc Biol. 2011;31(4):51–57. doi: 10.1161/ATVBAHA.110.216333. [DOI] [PubMed] [Google Scholar]
- 24.Kozian DH, Barthel A, Cousin E, Brunnhöfer R, Anderka O, März W, et al. Glucokinase-activating GCKR polymorphisms increase plasma levels of triglycerides and free fatty acids, but do not elevate cardiovascular risk in the Ludwigshafen Risk and Cardiovascular Health Study. Horm Metab Res. 2010;42:502–506. doi: 10.1055/s-0030-1249637. [DOI] [PubMed] [Google Scholar]
- 25.Mohás M, Kisfali P, Járomi L, Maász A, Fehér E, Csöngei V, et al. GCKR gene functional variants in type 2 diabetes and metabolic syndrome: do the rare variants associate with increased carotid intima-media thickness? Cardiovasc Diabetol. 2010;9:79. doi: 10.1186/1475-2840-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen-Torvik LJ, Li M, Kao WH, Couper D, Boerwinkle E, Bielinski SJ, et al. Association of a fasting glucose genetic risk score with subclinical atherosclerosis: The Atherosclerosis Risk in Communities (ARIC) study. Diabetes. 2011;60:331–335. doi: 10.2337/db10-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Adamo E, Cali AM, Weiss R, Santoro N, Pierpont B, Northrup V, et al. Central role of fatty liver in the pathogenesis of insulin resistance in obese adolescents. Diabetes Care. 2010;33:1817–1822. doi: 10.2337/dc10-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Adamo E, Northrup V, Weiss R, Santoro N, Pierpont B, Savoye M, et al. Ethnic differences in lipoprotein subclasses in obese adolescents: importance of liver and intraabdominal fat accretion. Am J Clin Nutr. 2010;92:500–508. doi: 10.3945/ajcn.2010.29270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwimmer JB, Pardee PE, Lavine JE, Blumkin AK, Cook S. Cardiovascular risk factors and the metabolic syndrome in pediatric nonalcoholic fatty liver disease. Circulation. 2008;118:277–283. doi: 10.1161/CIRCULATIONAHA.107.739920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker A, Sharp SJ, Timpson NJ, Bouatia-Naji N, Warrington NM, Kanoni S, Beilin LJ, et al. Association of genetic Loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60:1805–1812. doi: 10.2337/db10-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozlitina J, Boerwinkle E, Cohen JC, Hobbs HH. Dissociation between APOC3 variants, hepatic triglyceride content and insulin resistance. Hepatology. 2011;53:467–474. doi: 10.1002/hep.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenti L, Nobili V, Al-Serri A, Rametta R, Leathart JB, Zappa MA, et al. The APOC3 T-455C and C-482T promoter region polymorphisms are not associated with the severity of liver damage independently of PNPLA3 I148M genotype in patients with nonalcoholic fatty liver. J Hepatol. 2011 doi: 10.1016/j.jhep.2011.03.035. in press. [DOI] [PubMed] [Google Scholar]
- 33.Sentinelli F, Romeo S, Maglio C, Incani M, Burza MA, Scano F, et al. Lack of effect of apolipoprotein C3 polymorphisms on indices of liver steatosis, lipid profile and insulin resistance in obese Southern Europeans. Lipids Health Dis. 2011;10:93. doi: 10.1186/1476-511X-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dongiovanni P, Valenti L, Rametta R, Daly AK, Nobili V, Mozzi E, et al. Genetic variants regulating insulin receptor signalling are associated with the severity of liver damage in patients with non-alcoholic fatty liver disease. Gut. 2010;59:267–273. doi: 10.1136/gut.2009.190801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.