Abstract
Signaling lipids control many of the most important biological pathways, typically by recruiting cognate protein binding targets to cell surfaces, thereby regulating both their function and subcellular localization. A critical family of signaling lipids is that of the phosphatidylinositol polyphosphates (PIPns), which is composed of seven isomers that vary based on phosphorylation pattern. A key protein that is activated upon PIPn binding is Akt, which then plays important roles in regulating the cell cycle, and is thus aberrant in disease. Characterization of protein–PIPn binding interactions is hindered by the complexity of the membrane environment and of the PIPn structures. Herein, we describe two rapid assays of use for characterizing protein–PIPn binding interactions. First, a microplate-based binding assay was devised to characterize the binding of effectors to immobilized synthetic PIPn headgroup–biotin conjugates corresponding to all seven isomers. The assay was implemented for simultaneous analysis of Akt-PH domain, indicating PI(3,4,5)P3 and PI(3,4)P2 as the primary ligands. In addition, density-dependant studies indicated that the amount of ligand immobilized on the surface affected the amplitude of protein binding, but not the affinity, for Akt-PH. Since the PIPn ligand motifs used in this analysis lack the membrane environment and glycerolipid backbone, yet still exhibit high-affinity protein binding, these results narrow down the structural requirements for Akt recognition. Additionally, binding detection was also achieved through microarray analysis via the robotic pin printing of ligands onto glass slides in a miniaturized format. Here, fluorescence-based detection provided sensitive detection of binding using minimal amounts of materials. Due to their high-throughput and versatile attributes, these assays provide invaluable tools for probing and perturbing protein–membrane binding interactions.
Keywords: Phospholipids, membranes, phosphoinositides, cell surface, microarray
1. Introduction
Certain lipids present within plasma and organelle membranes act as critical regulatory biomolecules that control many of the most important cellular pathways. The production of these signaling lipids is tightly controlled in both a spatial and temporal manner, as they exist at low physiological concentrations prior to upregulation caused by external stimuli. A primary mode of action involves the binding of proteins through non-covalent interactions that enforce the recruitment of effectors onto the cell membrane surface.(Cho and Stahelin, 2005; Hurley, 2006; Lemmon, 2007, 2008) These interactions not only control the function of the target protein, generally through activation upon binding, but also dictate sub-cellular localization due to the controlled presence of signaling lipids within specific membranes.(Sprong et al., 2001) Since protein–lipid binding events often act as keystone interactions that feed information into critical cellular pathways, aberrant lipid activities often correlate to debilitating diseases.
A prominent family of signaling lipids is that of the phosphatidylinositol polyphosphates (PIPns).(Best et al.; Conway and Miller, 2007) The PIPns consist of the conserved scaffold of a myo-inositol headgroup linked via a phosphodiester tether at the 1-position to the traditional glycerophospholipid backbone. The seven isomers of this family arise from variations of phosphorylation on the myo-inositol headgroup, with every permutation of phosphorylation at the 3-, 4-, and 5-positions existing in nature. Due to the key activities of the PIPns in numerous cellular pathways, defects in lipid signaling often correlate diseases, including cancer and diabetes.(Di Paolo and De Camilli, 2006; Pendaries et al., 2003; Vicinanza et al., 2008; Wymann and Schneiter, 2008) A primary example pertains to regulation of the cell cycle through the control of Akt (protein kinase B) activity.(Assinder et al., 2009; Duronio, 2008; Engelman et al., 2006; Manning and Cantley, 2007; Salmena et al., 2008; Yuan and Cantley, 2008) Here, the phosphorylation of PIPns by phosphoinositide 3-kinase (PI 3-K)(Katso et al., 2001) produces 3-phosphorylated products, PI(3,4,5)P3 and PI(3,4)P2, which recruit Akt to the membrane surface, where it is activated through multiple posttranslational phosphorylation events. Akt then feeds into a number of pathways that control cell survival. The 3-phosphatase PTEN(Maehama and Dixon, 1998) catalyzes the reverse reaction, and PI 3-K and PTEN are among the most heavily mutated oncoproteins, and tumor suppressors, respectively, in cancer.(Chow and Baker, 2006; Samuels et al., 2004) Additionally, the phosphoinositide 5-phosphatases (SHIPs) are also aberrant in disease.(Ooms et al., 2009)
Despite the significance of protein–PIPn binding interactions, numerous challenges exist that deter elucidation of the details of binding, including the complex nature of the membrane environment and of the PIPn structures, and the diverse and intricate nature of the proteins that bind to these lipids.(Cho and Stahelin, 2005; Lemmon, 2008) In the latter case, the PIPns are targeted by a continually growing list of protein binding domain families, such as the PH, PX, FYVE, ENTH, ANTH, FERM, Tubby, and PROPPIN modules. Among these domains, variations exist in the details of binding, such as the PIPn binding selectivity, the structural features required for recognition, and a range of other details such as the existence and extent of multivalency in binding. Binding modules can even exhibit broad variation in binding details within the same domain family, as is particularly evidenced with the diverse PH domain family. Further complexity results from the fact that proteins can bind multiple distinct lipids, even using a single domain. For example, the PH domain of Akt was recently found to bind to phosphatidylserine (PS) in addition to PI(3,4,5)P3.(Huang et al., 2011)
Due to the complex nature of protein–PIPn binding interactions, model systems are typically implemented to elucidate the details of binding at the molecular level. A number of effective strategies for characterizing binding have been developed, including surface plasmon resonance (SPR) analysis employing both membrane and individual lipid motifs, fluorescence quenching and polarization, monolayer penetration, vesicle pelleting, overlay assays, and calorimetry.(Cho et al., 2001; Narayan and Lemmon, 2006) In addition, a number of creative approaches for generating PIPn-presenting materials have been reported, such as solid supports that have been used for affinity purification of target proteins,(Anderson et al., 2010; Catimel et al., 2008; Catimel et al., 2009; Conway et al., 2010; Conway and Miller, 2007; Krugmann et al., 2002; Lim et al., 2002; Manifava et al., 2001; Osborne et al., 2007; Painter et al., 2001; Pasquali et al., 2007) as well as polymerized liposomes,(Ferguson et al., 2005) and PIPn-decorated dendrimers.(Richer et al., 2008; Richer et al., 2009; Webb et al., 2007) While previous PIPn-binding assays have been quite advantageous, these techniques each have associated disadvantages, which can include the inability to perform high-throughput analysis, the need for expensive and inaccessible instrumentation, and detection that can be limited to specific systems. Furthermore, since these approaches amount to model systems in which the membrane environment is simplified, it would be ideal to perform analysis in multiple contexts that mimic different aspects of the cell membrane to fully investigate binding. Finally, studies would benefit from broadly accessible and applicable assays that are amenable to high-throughput screening for efficient characterization.
Surface-based techniques for analyzing recognition have proven invaluable for probing protein–ligand binding interactions, as evidenced by studies focusing on protein–carbohydrate binding,(Liang et al., 2008; Oyelaran and Gildersleeve, 2009) which mirror certain protein–lipid binding events since many of these ligands exist as glycolipids. Carbohydrate microarray analysis has been applied to address the ligand specificities and structural requirements of recognition among proteins, to quantify the effects that multivalency has on protein binding affinity by controlling ligand density on the surface, and for detection using cell extracts to characterize variations in protein binding associated with disease.(Huang et al., 2006; Liang et al., 2007) Thus, this platform can be used for a range of studies that are not possible using traditional methods. To exploit the advantages of microarray analysis for elucidating protein–ligand binding events, we set out to develop a surface platform for the rapid analysis of protein–PIPn binding employing all seven natural isomers.
2. Experimental Procedures
2.1. Materials
Generally, reagents were purchased from Acros, Aldrich or Advanced ChemTech and used as received. Dry solvents were obtained from a Pure Solv solvent delivery system purchased from Innovative Technology, Inc. Column chromatography was performed using 230–400 mesh silica gel purchased from Sorbent Technologies and C18 (17%) reverse phase SPE columns (6 ml, 2 g) purchased from Silicycle. NMR spectra were obtained using a Varian Mercury 300 spectrometer and a Bruker Avance 400 spectrometer. Mass spectra were obtained with JEOL DART-AccuTOF spectrometer and an ABI Voyager DE Pro MALDI spectrometer with high resolution capabilities. Microplate chemiluminescence analysis was performed using a BioTek Synergy 2 multidetection microplate reader. White reacti-bind high binding capacity (HBC) streptavidin-coated 96-well microplates employed for chemiluminescence studies were purchased from Pierce Biotechnology (Rockford, IL). Mouse monoclonal anti-GST tag HRP-conjugate was purchased from Millipore (Billerica, Ma). Supersignal ELISA femto maximum sensitivity substrate was purchased from Pierce Biotechnology (Rockford, IL). The GST-tagged PH domain of Akt was expressed and purified as previously described.(Manna et al., 2007) SuperAvidin coated slides were purchased from Arrayit Corporation (Sunnyvale, CA). Slide printing was performed using a BioRad VersArray ChipWriter Pro robotic pin printer (Hercules, CA). Fluorescence slide scans were performed using a GenePix 4000B Micorarray Scanner. Rabbit anti-GST antibody and Cy3 conjugated goat anti-rabbit IgG antibody were obtained from Novus Biologicals (Littleton, CO). Synthetic procedures for modular PIPn–amine conjugates 2a–g(Gong et al., 2009a), as well as biotin-TEG-succinimidyl ester 3 and biotinylated PI(4,5)P2 analog 1c(Gong et al., 2009b) were previously described.
2.2. Synthesis
2.2.1. General Procedure for the synthesis of PIP-TEG-Biotin Conjugates
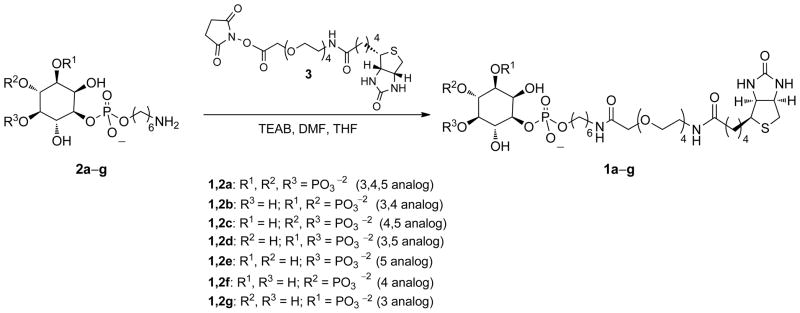
To a solution of biotin–succinimidyl ester 3 in 1 mL of N,N-dimethylformamide was added the appropriate PIPn–amine of type 2a–g in triethylammonium bicarbonate (TEAB) buffer. Next, 0.5–1.5 mL of tetrahydrofuran was added to the solution until it turned clear. The reaction mixture was then stirred at room temperature for 10 h, at which time the solvent was removed under reduced pressure. Next, acetone was added to the residue, and the white solid that formed was filtered and washed with acetone three times. The white solid was dissolved in deionized water and Chelex 100 resin (Sigma, Na+ form) was added. This solution was then stirred for 3 h and loaded onto a C18 reverse phase column (6 ml, 2 g). The corresponding fractions were lyophilized to yield 1a–g (47–99% yield).
2.2.2. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 3,4,5-tris-(disodium phosphate) (1a)
PI(3,4,5)P3–amine conjugate 2a (8 mg, 0.013 mmol) and biotin succinimidyl ester 3 (46 mg, 0.080 mmol) yielded 1a as a white solid (8 mg, 57%). 1H NMR (300 MHz, D2O): δ 4.36-4.34 (m, 1H), 4.18-4.15 (m, 1H), 3.84 (s, 2H), 3.74-3.70 (m, 4H), 3.52-3.38 (m, 18H), 3.20-3.18 (m, 2H), 3.14-3.12 (m, 2H), 3.08-3.02 (m, 3H), 2.10-2.05 (m, 2H), 1.46-1.32 (m, 8H), 1.20-1.16 (m, 6H). 31P NMR (121.5 MHz, D2O): δ 6.20 (1P), 5.11 (2P), 0.89 (1P). MALDI-MS [M+H]+: calcd for C32H63N4O25P4S 1059.2447, found 1059.2393.
2.2.3. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 3,4-bis-(disodium phosphate) (1b)
PI(3,4)P2–amine conjugate 2b (10 mg, 0.019 mmol) and biotin succinimidyl ester 3 (84 mg, 0.146 mmol) yielded 1b as a white solid (20 mg, 99%). 1H NMR (300 MHz, D2O): δ 4.38-4.36 (m, 1H), 4.20-4.17 (m, 1H), 4.13-3.99 (m, 2H), 3.83 (s, 2H), 3.76-3.60 (m, 4H), 3.48-3.38 (m, 18H), 3.17-3.13 (m, 2H), 3.02-3.00 (m, 1H), 2.77-2.73 (m, 1H), 2.56-2.52 (m, 1H), 2.04 (t, J = 6Hz, 2H), 1.44-1.30 (m, 8H), 1.18-1.10 (m, 6H). 31P NMR (121.5 MHz, D2O): δ 2.16 (1P), 1.24 (1P), −0.19 (1P). MALDI-MS [M+H]+: calcd for C32H62N4O22P3S 979.2784, found 979.2723.
2.2.4. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 3,5-bis-(disodium phosphate) (1d)
PI(3,5)P2 amine conjugate 2d (10 mg, 0.019 mmol) and biotin succinimidyl ester 3 (46 mg, 0.080 mmol) yielded 1d as a white solid (17 mg, 72%). 1H NMR (300 M Hz, D2O): δ 4.44-4.39 (m, 1H), 4.27-4.22 (m, 1H), 4.07-4.05 (m, 2H), 3.88 (s, 2H), 3.76-3.70 (m, 4H), 3.58-3.42 (m, 18H), 3.21-3.18 (m, 2H), 3.08-3.03 (m, 1H), 2.83-2.77 (m, 1H), 2.60-2.58 (m, 1H), 2.08 (t, J = 6 Hz, 2H), 1.50-1.35 (m, 8H), 1.24-1.16 (m, 6H). 31P NMR (121.5 MHz, D2O): δ 1.64 (1P), 0.30 (1P), −0.31 (1P). MALDI-MS [M+H]+: calcd for C32H62N4O22P3S 979.2784, found 979.2639.
2.2.5. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 5-disodium phosphate) (1e)
PI(5)P–amine conjugate 2e (10 mg, 0.023 mmol) and biotin succinimidyl ester 3 (46 mg, 0.080 mmol) yielded 1e as a colorless crystal (14 mg, 65%). 1H NMR (300 MHz, D2O): δ 4.55-4.45 (m, 1H), 4.35-4.25 (m, 1H), 4.19-4.10 (m, 1H), 3.98-3.92 (m, 2H), 3.85-3.75 (m, 2H), 3.70-3.45 (m, 20H), 3.32-3.22 (m, 4H), 3.16-2.12 (m, 1H), 2.89-2.85 (m, 1H), 2.18-2.14 (m, 2H), 1.70-1.40 (m, 8H), 1.40-1.20 (m, 6H). 31P NMR (121.5 MHz, D2O): δ 3.75 (1P), −0.30 (1P). MALDI-MS [M+H]+: calcd for C32H61N4O19P2S 899.3121, found 899.2996.
2.2.6. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 4-disodium phosphate) (1f)
PI(4)P–amine conjugate 2f (10 mg, 0.023 mmol) and biotin succinimidyl ester 3 (83 mg, 0.144 mmol) yielded 1f as a colorless crystal (20 mg, 93%). 1H NMR (300MHz, D2O): δ 4.48-4.36 (m, 1H), 4.21-4.17 (m, 1H), 4.02-3.99 (m, 2H) 3.86-3.82 (s, 2H), 3.76-3.68 (m, 4H), 3.56-3.36 (m, 18H), 3.18-3.14 (m, 2H), 3.04-3.00 (m, 1H), 2.79-2.73 (m, 1H), 2.58-2.51 (m, 1H), 2.08-2.00 (t, J = 6Hz, 2H), 1.44-1.32 (m, 8H), 1.19-1.07 (m, 6H).). 31P NMR (121.5 MHz, D2O): δ 3.99 (1P), −0.11 (1P). MALDI-MS [M+H]+: calcd for C32H61N4O19P2S 899.3121, found 899.3103.
2.2.7. 1-(6-(14-Biotinamido-3,6,9,12-tetraoxatetradecanamido)hexyl sodium phosphate)-1D-myo-inositol 3-disodium phosphate) (1g)
PI(3)P–amine conjugate 2a (12 mg, 0.027 mmol) and biotin–succinimidyl ester 3 (46 mg, 0.080 mmol) yielded 1g as a colorless crystal (12 mg, 47%). 1H NMR (300 MHz, D2O): δ 4.50-4.44 (m, 1H), 4.31-4.27 (m, 1H), 3.80 (s, 2H), 3.65-3.54 (m, 18H), 3.51-3.47 (m, 4H), 3.27-3.23 (m, 4H), 3.13-3.09 (m, 1H), 2.89-2.83 (m, 1H), 2.66-2.62 (m, 1H), 2.14 (t, J = 6Hz, 2H), 1.65-1.38 (m, 8H), 1.38-1.16 (m, 6H). 31P NMR (121.5 MHz, D2O): δ 4.15 (1P), −0.08 (1P). MALDI-MS [M+H]+: calcd for C32H61N4O19P2S 899.3121, found 899.3007.
2.3. Binding Analysis
2.3.1. Akt-PH microplate binding analysis using PIPn-biotin conjugates 1a–g
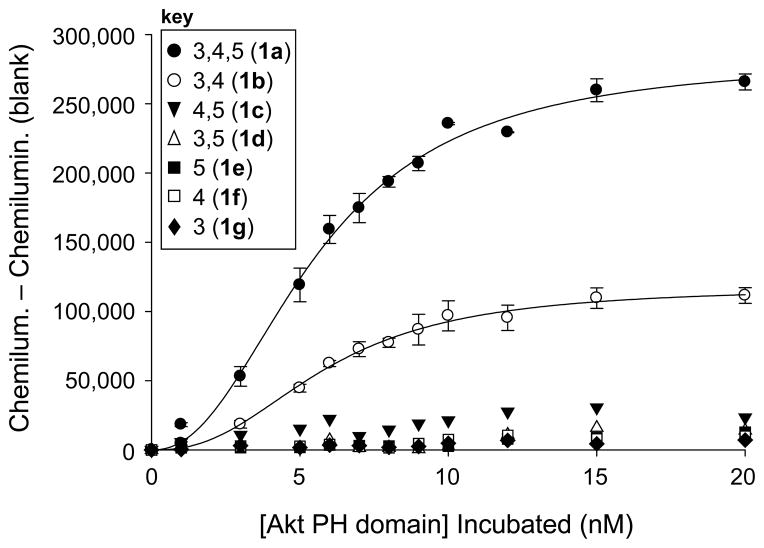
Each binding study involved side-by-side analysis of each headgroup isomer through immobilization onto separate rows of a streptavidin-coated 96-well microplate. First, the wells to be used were incubated with 200 μL of the wash buffer for the experiment, which consisted of 20 mM Tris buffer at pH 8.0 with 0.05% tween as a blocking additive to avoid nonspecific surface adsorption, for 30 minutes. This was removed, followed by the addition of 100 μL of solutions of biotin-TEG-PIPn ligands of type 1a–g in wash buffer into different rows (500 nM for studies in Figure 1; 500, 400, 300, and 200 nM for studies in Figure 2). These solutions were incubated for 1h, removed, and the wells were washed with 3 × 250 μL wash buffer. Next, solutions of Akt-PH in wash buffer with varying concentrations (0, 1, 3, 5, 6, 7, 8, 9, 10, 12, 15, 20 nM) were added. These solutions were incubated for 1h, removed, and the wells were washed with 3 × 250 μL wash buffer. Following this, 200 μL solutions of HRP-tagged anti-GST antibody (100 ng/mL) in wash buffer were added to each well. After a 1 h incubation, these solutions were removed, and the wells were washed with 3 × 250 μL wash buffer. Chemiluminescence detection of bound antibody was next performed using supersignal ELISA femto maximum sensitivity substrate. For detection, a 1:1 mixture of the substrate and peroxide solutions were mixed, and 100 μL of the resulting solution was added to each well. The microplate was then immediately placed in the microplate reader, and the chemiluminescence at 425 nm was repeatedly measured for 10 m. Several microplate readings from each experiment were then averaged, and the entire experiment was rerun at least 3 times and averaged to produce the data depicted in Figures 1 and 2. In order to curve fit the data, for each data point, the blank well containing no added protein, which generally yielded minimal signal, was subtracted from wells containing added protein. The data were analyzed by non-linear least-square analysis using the program SigmaPlot to obtain two different curve fits, a sigmoidal fit as well as a single site saturation curve. While the date fit better to a sigmoidal fit, as is depicted in Figures 1–2, both fits provided comparable results in terms of binding affinity. Sigmoidal properties typically indicate the presence of more than one equilibria or cooperativity,(Connors, 1987) and have previously been reported for similar protein–PIPn binding interactions.(Ferguson et al., 2005)
Figure 1.
Binding curves obtained from Akt-PH microplate binding results using analogs corresponding to all seven PIPn headgroups.
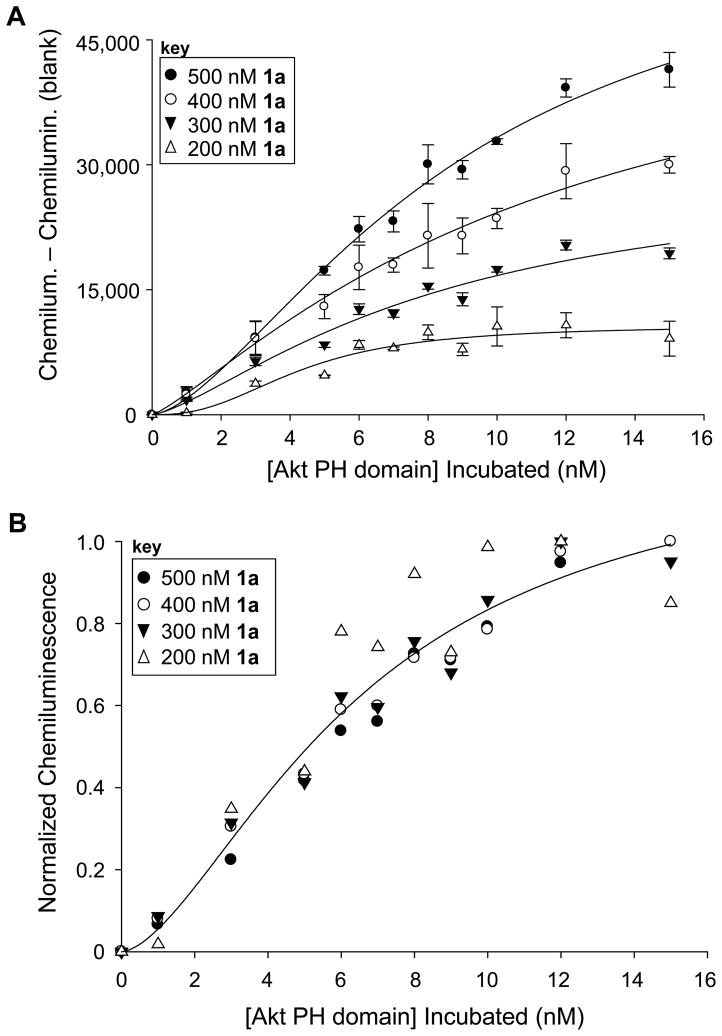
Figure 2.
Density-dependant studies to detect binding upon modulating the amount of ligand 1a on the surface. A. Raw binding curves at different concentrations show that increased ligand leads to increased protein recruitment. B. Normalized curves obtained by dividing by the maximal signal indicate that results overlay closely, indicating that surface density does not affect binding affinity.
2.3.2. Akt-PH glass slide binding analysis using PIPn-biotin conjugate 1a
The wash buffer employed for all glass slide experiments was 20 mM Tris buffer at pH 7.74, with 0.05% tween, and all washings were performed in petri dishes with agitation. For the uniform concentration grids (Figure 3A), 12 solutions of 1a at 500 μM were printed onto the slide in 20 replicates (2 grids of 12×10) and then incubated in a humidifying chamber overnight. For the varying concentration grids (Figure 3B), 12 solutions of 1a at 0, 15, 30, 45, 60, 75, 100, 200, 250, 300, 400, and 500 μM were printed onto the slide in 20 replicates (2 grids of 12×10) and incubated in a humidifying chamber overnight. The grids were outlined with a permanent marker, and the slide was washed 3 times in wash buffer in a Petri dish. The slide was then dried under a stream of nitrogen gas, after which 50 μg/mL of Akt-PH in wash buffer was added to the slide, such that the solution was retained held within the boundaries produced by the permanent marker. After incubation for 1 h in a humidifying chamber, the slide was then washed 3 times with wash buffer, dried under a stream of nitrogen, and a solution containing 0.2 μg/mL rabbit anti-GST and 0.1 μg/mL goat anti-rabbit–Cy3 in wash buffer was added and incubated in a humidifying chamber for 1 h. Following the incubation, the slide was washed 2 times with wash buffer, 3 times with deionized water, dried under a stream of nitrogen and then scanned for fluorescence using detection at 532 nm.
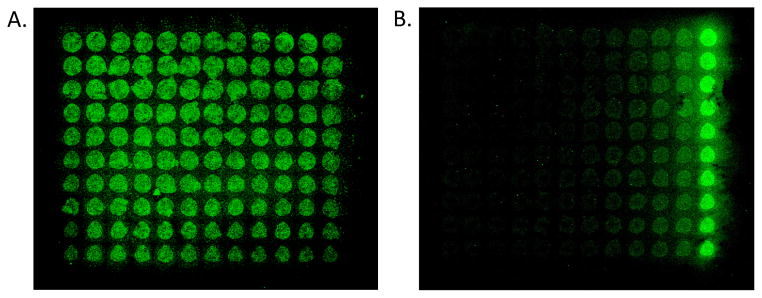
Figure 3.
Fluorescence scans of microarray slides indicate high-throughput detection of Protein–PI(3,4,5)P3 binding using glass slides decorated with PI(3,4,5)P3-TEG-biotin 1a. A. Results from a grid of 1a printed at uniform concentration. B. Results from grid of 1a printed at varying concentration (each column includes 10 replicates of each concentration). In each case, 1a was immobilized through robotic pin printing, followed by incubation with GST-tagged Akt-PH and detection of bound protein using rabbit anti-GST and Cy3-labeled goat anti-rabbit IgG.
3. Results
When considering an appropriate platform for surface-based analysis of protein–lipid binding interactions, an important choice is the identity of the ligand, as these compounds exist within membrane bilayers in the biological context. This is particularly noteworthy due to the current understanding of the structural requirements for protein–lipid recognition, which appears to vary distinctly among different binding domains. Certain proteins are known to bind with high affinity to truncated lipid motifs outside of the membrane context, particularly with the PIPns due to their highly charged headgroups.(Ferguson et al., 1995; Garcia et al., 1995; Hirose et al., 1999; Kavran et al., 1998; Lemmon et al., 1995) However, other peripheral proteins require the presence of the membrane bilayer to interact, typically due to the insertion of hydrophobic residues into the membrane core.(Bravo et al., 2001; Kutateladze, 2006) As a result of this variation in binding mode, we have devised separate assays to detect protein–lipid binding employing lipid motifs both within and outside the context of the membrane environment.(Best et al., 2011)
To analyze binding in a bilayer environment, we employed immobilized whole liposomes to detect lipid recognition in a format that mimics cellular membranes, which we first used to analyze Protein Kinase C recognition.(Losey et al., 2009) Alternatively, we have devised an assay via the immobilization of simplified PIPn headgroup motifs for rapid characterization of effectors that bind with high affinity to these moieties outside of the membrane context. Our initial report of this latter approach utilized a synthetic PI(4,5)P2 analog linked via a tetraethylene glycol (TEG) spacer to biotin in order to detect binding of the PH domain of β-spectrin.(Gong et al., 2009b) This assay was shown to be effective for sensitive detection of the binding and inhibition of PIPn-binding proteins. Due to the diversity of PIPn targets exhibited by proteins, an important application of this approach involves the efficient delineation of the PIPn-binding selectivity of effectors. Herein, we describe the utility of this assay for characterizing PIPn specificity through studies focusing on the PH domain of the prominent lipid-binding protein Akt.
Probe design and synthesis
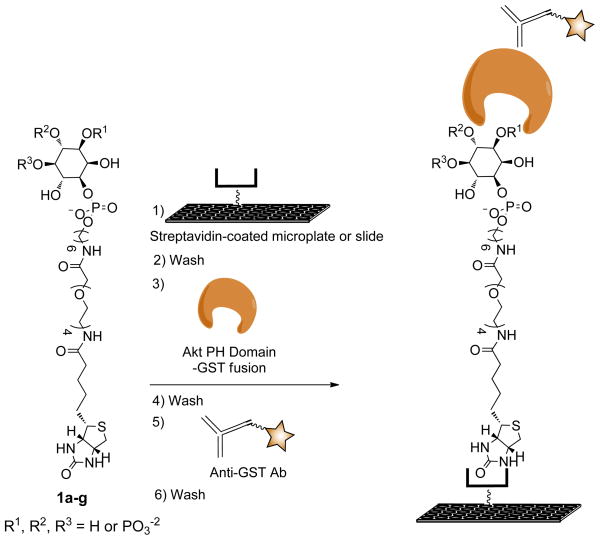
The devised PIPn assay employs PIPn-TEG-biotin conjugates as the recognition motif for binding studies, based upon our initial study that successfully employed synthetic PI(4,5)P2–TEG–biotin structure 1c (Scheme 1). The ligand structures include the appropriate headgroup with a phosphodiester moiety located at the same position as the parent PIPn lipids (P1). The phosphodiester linkage connects to a six-carbon hydrophobic region that is designed to, in part, mimic the hydrophobicity of the natural glycerolipid backbone, which is excluded in these structures. Next, the TEG linker is used to space the headgroup binding motif from the microplate surface. Recently, Iyer and co-workers showed that a long tether is critical in studies employing the coating of streptavidin-coated microplates, as the systematic shortening of this linker led to drastic reductions in binding.(Lewallen et al., 2009) Finally, the biotin moiety of 1c is employed for high-affinity non-covalent immobilization of these ligands onto streptavidin-coated microplates in an effort to avoid non-specific binding interactions.
Scheme 1.
Synthesis of biotin conjugates of all seven naturally occurring phosphoinositide headgroup isomers.
To develop an assay for characterizing binding specificity, we set out to extend our previous approach to produce the corresponding analogs of all seven naturally occurring PIPn isomers (1a–g). With regard to synthesis, we have taken advantage of previously successful strategies for inositol polyphosphate and PIPn probe development using modular scaffolds bearing a reactive moiety that can be derivatized in the final step of synthesis for convenient access to diverse structures.(Dorman et al., 1995; Henne et al., 1988; Inoue et al., 1999; Marecek et al., 1992; Nakanishi et al., 2002; Olszewski et al., 1995; Prestwich et al., 1991; Schafer et al., 1990; Tegge and Ballou, 1992) Using this approach, we recently described the synthesis of amino-conjugates of all seven PIPn headgroup isomers (2a–g), and efficient subsequent derivatization to produce different reporter-tagged analogs.(Gong et al., 2009a) In the synthesis of enantiomerically pure headgroup analogs, we followed previously devised procedures for myo-inositol desymmetrization that were developed by Bruzik and co-worker(Kubiak and Bruzik, 2003), and Holmes and co-workers(Conway et al., 2010; Painter et al., 1999). To synthesize ligands for binding analysis using all isomers, 2a–g were allowed to react with synthetic biotin–TEG–succinimidyl ester reagent 3 to generate PIPn–TEG–biotin conjugates 1a–g for studies.
Microplate binding studies employing PIPn headgroup-biotin conjugates
Following the synthesis of 1a–g, we sought to apply these compounds for side-by-side microplate binding analysis in order to directly compare all seven PIPn isomers and thus assess protein binding specificity. For studies, we employed the PH domain of Akt (Akt-PH) due to the key role this protein plays in the cell cycle and since few binding studies have been reported using all PIPn isomers. GST-tagged Akt-PH was expressed and purified as described previously.(Manna et al., 2007) In the assay, compounds 1a–g were immobilized in separate rows of streptavidin-coated 96-well microplates by incubating with solutions with optimized concentrations, typically 500 nM (Scheme 2). Each isomer was then treated with varying concentrations of GST-tagged Akt-PH, followed by detection via chemiluminescence-based ELISA signal transduction using an HRP-tagged anti-GST antibody. Each step of the assay was followed by washes to remove any unbound species.
Scheme 2.
Assay for high-throughput detection of Akt-PH binding to immobilized synthetic analogs of all seven PIPn isomers.
The results from this approach are shown in Figure 1. Here, the analogs corresponding to PI(3,4,5)P3 (1a) and PI(3,4)P2 (1b) yielded strong dose-dependant responses when incubating with relatively low concentrations of Akt-PH (0–20 nM). The other isomers yielded minimal signal and thus essentially doubled as negative controls for the experiment. The data shown in Figure 1 represent the averages of at least three analyses with error bars showing the standard error produced by multiple runs. This indicates the reproducibility of the assay, despite the multiple components in the analysis and the potential variations for surface derivatization in the microplate. The results obtained from these are consistent with previous reports implicating PI(3,4,5)P3 and PI(3,4)P2 as the primary ligands for Akt binding, highlighting the importance of PI 3-K. While the other PIPn headgroups do not bind in this particular case due to the headgroup binding specificity of Akt, related studies using synthetic headgroup analogs indicate that these compounds are effective for studying other target binding domains.(Conway et al., 2010) Results similar to those shown in Figure 1 have been reported through studies using full PIPn analogs including SPR, vesicle pelleting, and tryptophan quenching assays.(Frech et al., 1997; James et al., 1996; Manna et al., 2007) In the latter study, it was observed that full PIPn structures bound with significantly higher affinity than the corresponding soluble inositol phosphates (InsPs), which contain a phosphate group at P-1 instead of the phosphodiester-linked glycerolipid backbone. In the current study we observe strong binding to headgroup analogs bearing the traditional phosphodiester but lacking the full glycerolipid scaffold. However, these compounds also contain a short hydrocarbon spacer linked to the phosphodiester that may mimic the hydrophobic nature of the lipid backbone. Thus, this provides further evidence that simplified headgroup analogs containing the phosphodiester linkage and a short hydrophobic region are effective for binding.
As we observed in our previous report, binding studies employing microplate surfaces coated with ligand yielded somewhat higher protein binding affinities than membrane- and solution-phase analysis. In the current studies, apparent dissociation constants for protein–surface association, commonly termed “Kd, surf”,(Liang et al., 2007) were determined to be approximately 5 nM for both 1a and 1b. While these values are essentially identical to those from other protein-PIPn binding studies employing immobilized biotinylated PIPn-motifs,(Gong et al., 2009b; Mehrotra et al., 2000) they represent higher affinities than those previously observed for the binding of PI(3,4,5)P3 (234 – 590 nM) and PI(3,4)P2 (350 – 570 nM) to Akt in membrane contexts.(Frech et al., 1997; James et al., 1996; Manna et al., 2007) However, variations in observed binding strengths are not unexpected due to the different conditions employed in the various assays. The primary differences in the current study include the truncated structures of the PIPn ligands and the use of surfaces coated with the ligand motif as the platform for analysis. In order to probe the latter characteristic, we next sought to evaluate the effect of ligand density on protein binding.
One of the benefits of surface-based analysis is that the density of the ligand on the surface can be controlled via the concentration of the solution employed for immobilization. This approach has previously been used to measure variations in binding affinity resulting from the modulation of ligand density.(Liang et al., 2008; Oyelaran and Gildersleeve, 2009) Since the current studies employ an individual binding domain, it was expected that protein affinity would not be affected by ligand density, which we set out to evaluate. Here, the binding assay was pursued using solutions with different concentrations of PI(3,4,5)P3-TEG-biotin ligand 1a ranging from 200 – 500 nM. The raw data from these studies, shown in Figure 2A, indicated that the amount of Akt-PH recruited to the microplate surface decreased upon the immobilization of decreased ligand, as judged by the amplitude of the binding curves. This effect was only observed when varying the amount of ligand on the surface, indicating that it does not result from non-selective binding. Further evidence of this can be seen in Figure 1 where high concentrations of other PIPn headgroups did not lead to protein binding.
While an increase in amplitude was observed, the alteration of ligand density did not affect protein binding affinity, which is best seen through the overlay of normalized binding curves at each concentration, obtained by dividing by the maximal signal for each case (Figure 2B). In the resulting plot, normalized binding curves obtained at different surface densities were essentially identical. Thus, increasing ligand deposition led to enhanced protein recruitment but did not affect the binding affinity in this case. This provides evidence against a “surface effect” affecting binding through non-specific interactions caused by the accumulation of charge on the microplate surface. Nevertheless, the primary difference between liposomal binding studies and those employing individual lipid motifs immobilized onto surfaces involves the presentation of the ligand, which likely leads to the variations that have been observed in binding affinity between these methods. In liposomal studies, signaling lipids such as PI(3,4,5)P3 are typically incorporated at low percentages and are fluid within the bilayer, whereas surface studies would lead to higher concentrations and the constraining of lipid mobility. It is possible that immobilization of PI(3,4,5)P3 onto the surface at higher effective concentrations inherently passes a threshold in which a slight clustering effect is observed to boost binding affinity. Formation of additional contacts with the protein would be in line with previous work since Akt-PH has been found to possess a separate binding domain for PS (Huang et al., 2011) and since reports have indicated that Akt membrane recruitment may be affected by the presence of lipid rafts (Lasserre et al., 2008). Despite this possibility for enhanced binding due to surface presentation, the assay is effective for determining PIPn binding specificity, as depicted by the data in Figure 1 in which only the 3-phosphorylated targets of Akt–PH result in protein binding.
Microarray analysis of Akt-PH binding using PI(3,4,5)P3 analogs printed on glass slides
While the previously described microplate binding assay allows for rapid detection of protein–PIPn binding interactions, such studies would further benefit from true microarray analysis, which generally employs glass slides as the platform for high-throughput detection in a miniaturized format. To pursue this goal for the detection of Akt-PI(3,4,5)P3 binding, we modified the prior microplate assay by immobilizing PI(3,4,5)P3-TEG-biotin conjugate 1a onto streptavidin-coated glass slides. For immobilization, a robotic pin printer was used to deposit ~0.6 femtoliters of solutions of 1a at each location, resulting in spot sizes of ~50 microns. This minimizes the amount of compound required for analysis and allows for thousands of experiments to be performed on a single 25mm × 75mm glass slide. In the prior microplate binding studies, chemiluminescence-based detection was exploited due to the beneficial enhancement in sensitivity. However, this is not effective for microarray analysis on slides due to the mobility of the soluble luminal substrate. As such, we amended our detection scheme to analyze bound protein via fluorescence by switching to an anti-GST antibody followed by a secondary goat anti-rabbit antibody labeled with a cyanine-3 (Cy3) fluorophore. Protein bound to the PI(3,4,5)P3-functionalized glass slide was then detected using a fluorescence slide scanner. Blocking and washing steps were also probed to optimize detection. We also pursued binding analysis via the immobilization of PI(3,4,5)P3-amine conjugate 2a onto slides bearing epoxysilane groups as reactive electrophiles. However, we generally found this latter approach to yield significantly increased background signal compared to deposition via the biotin-avidin interaction.
Results from microarray studies using this assay are depicted in Figure 3. In an initial experiment, we printed a 12×10 grid of spots consisting of PI(3,4,5)P3-TEG-biotin analog 1a at a constant concentration of 500 μM. Following protein binding analysis, the fluorescence image depicted in Figure 3A was obtained. Here, green spots were present corresponding to the locations of 1a on the slide surface, and minimal background fluorescence was observed. One note is that variations in spot intensities were seen in this grid, despite the fact that the ligand was immobilized at a constant concentration. This is a common technical challenge in microarray analysis, which can be attributed to issues including differences in the density of reactive functional groups on the slides, the numerous steps that are required for the assay (ligand immobilization, blocking, washing, binding and detection), and variations in the efficacy of pin printing at different locations. The latter affect seems to be a factor in these studies, since spot intensity appears to decrease at the right and bottom of the grid, which correlates to the spots that were printed the latest in the pin printer sequence.
Following these initial results, we set out to vary the concentration of 1a immobilized on the surface to view the effect on protein binding. In this case, a 12×10 grid was produced with 12 different concentrations (0, 15, 30, 45, 60, 75, 100, 200, 250, 300, 400, and 500 μM) printed that increased to the right along each row. Ten replicates were then printed for each concentration in each column of the grid. Following protein binding analysis, the fluorescence slide scan depicted in Figure 3B was obtained. In this case, the fluorescence intensities were seen to increase to the right of the slide, correlating with increasing concentration of 1a. Additionally, this experiment led to improved homogeneity of signal among replicates when compared to the prior data shown in Figure 3A. Thus, these results indicate successful initial microarray analysis through printing of synthetic PI(3,4,5)P3 ligands. Due to the high-throughput nature of this technique and the minimal amounts of materials that are necessary for analysis, this approach could provide a valuable tool for characterizing the biological activities of PIPns and their target proteins.
4. Summary
This report details two assays for the detection and characterization of protein–PIPn binding interactions employing microplates and glass slides as the platforms. The microplate-based binding platform was used to characterize protein–PIPn binding interactions using synthetic headgroup ligands corresponding to all seven naturally occurring PIPn isomers. This approach was applied for rapid side-by-side analysis of the binding of Akt-PH to the seven PIPn headgroup isomers. The assay allowed for highly sensitive detection of binding, and high-affinity interactions of the protein with its primary PIPn headgroup targets were observed, exhibiting dissociation constants in the low nanomolar range. Results showed the headgroup analogs corresponding to PI(3,4,5)P3 and PI(3,4)P2 to be the primary ligand targets, with little binding observed from the other isomers, in agreement with previous reports. Studies in which the density of the ligand on the microplate surface was modulated indicated that increasing ligand enhanced the amount of protein recruited to the surface, but did not affect the affinity of the interaction. These results also provided further information regarding the structural features required for Akt binding, as immobilized headgroups bearing the phosphodiester linkage and short hydrocarbon spacers exhibited high-affinity protein binding outside of a membrane environment. Additionally, microarray analysis was achieved in a miniaturized format through robotic pin printing of PI(3,4,5)P3 ligands onto glass slides. Subsequently, sensitive fluorescence-based detection of binding was achieved, and the assay benefits from the minimal amounts of materials that are required.
The described assays will be beneficial for probing and perturbing protein–PIPn binding for numerous reasons. First, these platforms entail high-throughput screening to maximize the efficiency of analysis. In addition, the surface-based ligand display involved in these assays can be used to mimic the presentation of lipid motifs on the surfaces of cell membranes, thus providing an effective model system for biological recognition. Furthermore, surface deposition through the use of a high affinity non-covalent interaction, particularly biotin-streptavidin, allows for the ligand density to be controlled, enabling the investigation of potential ligand clustering effects. Finally, while traditional binding assays are limited to in vitro binding studies using protein and ligand motifs in solution, antibody-based signal transduction allows for the detection of a specific protein target, and could thus be used for target detection in a more complex biological context, such as a cell extract. We are now pursuing these avenues to apply this platform to address important issues in protein–PIPn binding interactions.
Supplementary Material
Highlights.
We have developed assays employing microarray analysis using 96-well microplates and robotic pin printing onto glass slides for high-throughput analysis of protein-phosphoinositide binding interactions.
For these assays, we designed and synthesized biotin-conjugates corresponding to the headgroups of all seven naturally occurring phosphoinositide isomers.
Binding analysis was performed by immobilizing synthetic phosphoinositide-conjugates onto surfaces followed by antibody-based detection of protein binding.
These platforms were applied to study the phosphoinositide-binding specificity of the PH domain of Akt and understand the effects of ligand surface density on protein binding.
Acknowledgments
This work was made possible by funding from the National Science Foundation (CHE-0954297 for MDB) and the National Institutes of Health (GM68849 for WC). We also acknowledge Dr. Sujata Agarwal at the UTIA genomics hub facility for assistance with microarray experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Anderson RJ, Osborne SL, Meunier FA, Painter GF. Regioselective Approach to Phosphatidylinositol 3,5-Bisphosphates: Syntheses of the Native Phospholipid and Biotinylated Short-Chain Derivative. J Org Chem. 2010;75:3541–3551. doi: 10.1021/jo100393c. [DOI] [PubMed] [Google Scholar]
- Assinder SJ, Dong QH, Kovacevic Z, Richardson DR. The TGF-beta, PI3K/Akt and PTEN pathways: established and proposed biochemical integration in prostate cancer. Biochem J. 2009;417:411–421. doi: 10.1042/BJ20081610. [DOI] [PubMed] [Google Scholar]
- Best MD, Rowland MM, Bostic HE. Exploiting bioorthogonal chemistry to elucidate protein–lipid binding interactions and other biological roles of phospholipids. Acc Chem Res. 2011;44:686–698. doi: 10.1021/ar200060y. [DOI] [PubMed] [Google Scholar]
- Best MD, Zhang HL, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat Prod Rep. doi: 10.1039/B923844C. In Press. [DOI] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJG, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Catimel B, Schleber C, Condron M, Patsiouras H, Connolly L, Catimel J, Nice EC, Burgess AW, Holmes AB. The PI(3,5)P2 and PI(4,5)P2 Interactomes. J Proteome Res. 2008;7:5295–5313. doi: 10.1021/pr800540h. [DOI] [PubMed] [Google Scholar]
- Catimel B, Yin MX, Schieber C, Condron M, Patsiouras H, Catimel J, Robinson D, Wong LSM, Nice EC, Holmes AB, Burgess AW. PI(3,4,5)P3 Interactome. J Proteome Res. 2009;8:3712–3726. doi: 10.1021/pr900320a. [DOI] [PubMed] [Google Scholar]
- Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Ann Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Cho WW, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- Chow LML, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Connors KA. Binding constants: The measurement of molecular complex stability. John Wiley & Sons; New York: 1987. [Google Scholar]
- Conway SJ, Gardiner J, Grove SJA, Johns MK, Lim ZY, Painter GF, Robinson DEJE, Schieber C, Thuring JW, Wong LSM, Yin MX, Burgess AW, Catimel B, Hawkins PT, Ktistakis NT, Stephens L, Holmes AB. Synthesis and biological investigation of phosphatidylinositol phosphate affinity probes. Org Biomol Chem. 2010;8:66–76. doi: 10.1039/b913399b. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Miller GJ. Biology-enabling inositol phosphates, phosphatidylinositol phosphates and derivatives. Nat Prod Rep. 2007;24:687–707. doi: 10.1039/b407701f. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dorman G, Chen J, Prestwich GD. Synthesis of D-myo-P-1-(O-aminopropyl)-inositol-1,4,5-trisphosphate affinity probes from alpha d-glucose. Tetrahedron Lett. 1995;36:8719–8722. [Google Scholar]
- Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415:333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Ferguson CG, James RD, Bigman CS, Shepard DA, Abdiche Y, Katsamba PS, Myszka DG, Prestwich GD. Phosphoinositide-containing polymerized liposomes: Stable membrane-mimetic vesicles for protein-lipid binding analysis. Bioconj Chem. 2005;16:1475–1483. doi: 10.1021/bc050197q. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the High-Affinity Complex of Inositol Trisphosphate with a Phospholipase-C Pleckstrin Homology Domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High affinity binding of inositol phosphates and phosphoinositides to the Pleckstrin homology domain of RAC protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ. The pleckstrin homology domain of phospholipase C-delta(1) binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- Gong D, Bostic HE, Smith MD, Best MD. Synthesis of modular headgroup conjugates corresponding to all seven phosphatidylinositol polyphosphate isomers for convenient probe generation. Eur J Org Chem. 2009a:4170–4179. [Google Scholar]
- Gong D, Smith MD, Manna D, Bostic HE, Cho W, Best MD. Microplate-based characterization of protein-phosphoinositide binding interactions using a synthetic biotinylated headgroup analogue. Bioconjugate Chem. 2009b;20:310–316. doi: 10.1021/bc8004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne V, Mayr GW, Grabowski B, Koppitz B, Soling HD. Semisynthetic derivatives of inositol 1,4,5-trisphosphate substituted at the 1-phosphate group - Effects on calium release from permeabilized guinea-pig parotid acinar cells and comparison with binding to alolase-A. Eur J Biochem. 1988;174:95–101. doi: 10.1111/j.1432-1033.1988.tb14067.x. [DOI] [PubMed] [Google Scholar]
- Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M. Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science. 1999;284:1527–1530. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- Huang BX, Akbar M, Kevala K, Kim HY. Phosphatidylserine is a critical modulator for Akt activation. J Cell Biol. 2011;192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Thayer DA, Chang AY, Best MD, Hoffmann J, Head S, Wong CH. Carbohydrate microarray for profiling the antibodies interacting with Globo H tumor antigen. Proc Natl Acad Sci U S A. 2006;103:15–20. doi: 10.1073/pnas.0509693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. Membrane binding domains. Biochim Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Kikuchi K, Hirose K, Iino M, Nagano T. Synthesis and evaluation of 1-position-modified inositol 1,4,5-trisphosphate analogs. Bioorg Med Chem Lett. 1999;9:1697–1702. doi: 10.1016/s0960-894x(99)00256-5. [DOI] [PubMed] [Google Scholar]
- James SR, Downes CP, Gigg R, Grove SJA, Holmes AB, Alessi DR. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: Implications for development, immunity, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by Pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, Davidson K, Eguinoa A, Ellson CD, Lipp P, Manifava M, Ktistakis N, Painter G, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Dove SK, Michell RH, Grewal A, Nazarian A, Erdjument-Bromage H, Tempst P, Stephens LR, Hawkins PT. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Kubiak RJ, Bruzik KS. Comprehensive and uniform synthesis of all naturally occurring phosphorylated phosphatidylinositols. J Org Chem. 2003;68:960–968. doi: 10.1021/jo0206418. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta. 2006;1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre R, Guo XJ, Conchonaud F, Hamon Y, Hawchar O, Bernard AM, Soudja SM, Lenne PF, Rigneault H, Olive D, Bismuth G, Nunes JA, Payrastre B, Marguet D, He HT. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. In: Wakelam MJO, editor. Cell Biology of Inositol Lipids and Phosphates. Portland Press Ltd; London: 2007. pp. 81–93. [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Obrien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewallen DM, Siler D, Iyer SS. Factors affecting protein-glycan specificity: Effect of spacers and incubation time. Chembiochem. 2009;10:1486–1489. doi: 10.1002/cbic.200900211. [DOI] [PubMed] [Google Scholar]
- Liang PH, Wang SK, Wong CH. Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: Determination of surface and solution dissociation constants. J Am Chem Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- Liang PH, Wu CY, Greenberg WA, Wong CH. Glycan arrays: Biological and medical applications. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ZY, Thuring JW, Holmes AB, Manifava M, Ktistakis NT. Synthesis and biological evaluation of a PtdIns(4,5)P-2 and a phosphatidic acid affinity matrix. J Chem Soc, Perkin Trans. 2002;1:1067–1075. [Google Scholar]
- Losey EA, Smith MD, Meng M, Best MD. Microplate-based analysis of protein-membrane interactions via immobilization of whole liposomes containing a biotinylated anchor. Bioconjugate Chem. 2009;20:376–383. doi: 10.1021/bc800414k. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Manifava M, Thuring J, Lim ZY, Packman L, Holmes AB, Ktistakis NT. Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)-bisphosphate-coupled affinity reagents. J Biol Chem. 2001;276:8987–8994. doi: 10.1074/jbc.M010308200. [DOI] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marecek JF, Estevez VA, Prestwich GD. New tetherable derivatives of myo-inositol 2,4,5-trisphosphates and 1,3,4-trisphosphates. Carbohyd Res. 1992;234:65–73. doi: 10.1016/0008-6215(92)85039-3. [DOI] [PubMed] [Google Scholar]
- Mehrotra B, Myszka DG, Prestwich GD. Binding kinetics and ligand specificity for the interactions of the C2B domain of synaptogmin II with inositol polyphosphates and phosphoinositides. Biochemistry. 2000;39:9679–9686. doi: 10.1021/bi000487o. [DOI] [PubMed] [Google Scholar]
- Nakanishi W, Kikuchi K, Inoue T, Hirose K, Iino M, Nagano T. Hydrophobic modifications at 1-phosphate of inositol 1,4,5-trisphosphate analogues enhance receptor binding. Bioorg Med Chem Lett. 2002;12:911–913. doi: 10.1016/s0960-894x(02)00044-6. [DOI] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski JD, Dorman G, Elliott JT, Hong Y, Ahern DG, Prestwich GD. Tethered benzophenone reagents for the synthesis of photoactivatable ligands. Bioconjugate Chem. 1995;6:395–400. doi: 10.1021/bc00034a009. [DOI] [PubMed] [Google Scholar]
- Ooms LM, Horan KA, Rahman P, Seaton G, Gurung R, Kethesparan DS, Mitchell CA. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem J. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- Osborne SL, Wallis TP, Jimenez JL, Gorman JJ, Meunier FA. Identification of secretory granule phosphatidylinositol 4,5-bisphosphateinteracting proteins using an affinity pulldown strategy. Mol Cell Prot. 2007;6:1158–1169. doi: 10.1074/mcp.M600430-MCP200. [DOI] [PubMed] [Google Scholar]
- Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter GF, Grove SJA, Gilbert IH, Holmes AB, Raithby PR, Hill ML, Hawkins PT, Stephens L. General synthesis of 3-phosphorylated myo-inositol phospholipids and derivatives. J Chem Soc, Perkin Trans. 1999;1:923–935. [Google Scholar]
- Painter GF, Thuring JW, Lim ZY, Holmes AB, Hawkins PT, Stephens LR. Synthesis and biological evaluation of a PtdIns(3,4,5)P-3 affinity matrix. Chem Commun. 2001:645–646. [Google Scholar]
- Pasquali C, Bertschy-Meier D, Chabert C, Curchod ML, Arod C, Booth R, Mechtler K, Vilbois F, Xenarios I, Ferguson CG, Prestwich GD, Camps M, Rommel C. A chemical proteomics approach to phosphatidylinositol 3-kinase signaling in macrophages. Mol Cell Proteom. 2007;6:1829–1841. doi: 10.1074/mcp.T600066-MCP200. [DOI] [PubMed] [Google Scholar]
- Pendaries C, Tronchere H, Plantavid M, Payrastre B. Phosphoinositide signaling disorders in human diseases. FEBS Lett. 2003;546:25–31. doi: 10.1016/s0014-5793(03)00437-x. [DOI] [PubMed] [Google Scholar]
- Prestwich GD, Marecek JF, Mourey RJ, Theibert AB, Ferris CD, Danoff SK, Snyder SH. Tethered Ip3 - Synthesis and Biochemical Applications of the 1-O-(3-Aminopropyl) Ester of Inositol 1,4,5-Trisphosphate. Journal of the American Chemical Society. 1991;113:1822–1825. [Google Scholar]
- Richer SM, Stewart NK, Tomaszewski JW, Stone MJ, Oakley MG. NMR Investigation of the Binding between Human Profilin I and Inositol 1,4,5-Triphosphate, the Soluble Headgroup of Phosphatidylinositol 4,5-Bisphosphate. Biochemistry. 2008;47:13455–13462. doi: 10.1021/bi801535f. [DOI] [PubMed] [Google Scholar]
- Richer SM, Stewart NK, Webb SA, Tomaszewski JW, Oakley MG. High Affinity Binding to Profilin by a Covalently Constrained, Soluble Mimic of Phosphatidylinositot-4,5-bisphosphate Micelles. ACS Chem Biol. 2009;4:733–739. doi: 10.1021/cb900121r. [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang ZH, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell DM, Riggins GJ, Willson JKV, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554–554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schafer R, Nehlssahabandu M, Grabowsky B, Dehlingerkremer M, Schulz I, Mayr GW. Synthesis and application of photoaffinity analogs of inositol 1,4,5-trisphosphate selectively substituted at the 1-phosphate group. Biochem J. 1990;272:817–825. doi: 10.1042/bj2720817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong H, van der Sluijs P, van Meer G. How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol. 2001;2:698–698. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- Tegge W, Ballou CE. Syntheses of D-myo-Inositol 1,4,5-trisphosphate affinity ligands. Carbohyd Res. 1992;230:63–77. doi: 10.1016/s0008-6215(00)90513-5. [DOI] [PubMed] [Google Scholar]
- Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci. 2008;65:2833–2841. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SA, Stewart NK, Belcher LJ, Mechref Y, Tomaszewski JW, Wu G, Novotny MV, Oakley MG. Synthesis and characterization of covalent mimics of phosphatidylinositol-4,5-bisphosphate micelles. Biomacromolecules. 2007;8:1790–1793. doi: 10.1021/bm061166g. [DOI] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.