Abstract
The transcription factor CREB is a key regulator of many neuronal processes, including brain development, circadian rhythm, and long-term memory. Studies of CREB have focused on its phosphorylation, although the diversity of CREB functions in the brain suggests additional forms of regulation. Here we expand on a chemoenzymatic strategy for quantifying glycosylation stoichiometries to characterize the functional roles of CREB glycosylation in neurons. We show that CREB is dynamically O-GlcNAc-modified in response to neuronal activity and glycosylation represses CREB-dependent transcription by impairing its association with the co-activator CRTC/TORC. Blocking glycosylation of CREB altered cellular function and behavioral plasticity, enhancing both axonal and dendritic growth and long-term memory consolidation. Our findings demonstrate a new role for O-glycosylation in memory formation and provide a mechanistic understanding of how glycosylation contributes to critical neuronal functions. Moreover, we identify a previously unknown mechanism for the regulation of activity-dependent gene expression, neural development, and memory.
cAMP-response element binding protein (CREB) controls gene expression programs that underlie diverse neuronal processes, ranging from neural development and survival to complex adaptive behaviors such as long-term memory and drug addiction1–3. Extensive studies have focused on the importance of protein phosphorylation in regulating CREB activity in the nervous system4–8. Phosphorylation of Ser133, a key regulatory site in the protein, leads to recruitment of the co-activator CREB-binding protein (CBP) and activation of CREB-mediated transcription9. However, phosphorylation is not always sufficient to stimulate CREB-dependent transcription6,10,11, suggesting the existence of additional undiscovered mechanisms for the complex coordination of CREB activity.
O-Glycosylation of proteins by the monosaccharide β-N-acetyl-D-glucosamine (O-GlcNAc) is a dynamic, inducible post-translational modification with striking similarities to phosphorylation12–14. Attachment of this simple glycan to serine or threonine residues occurs on more than 1,000 proteins, including proteins involved in transcription and translation, signal transduction, cell cycle progression and synaptic plasticity12–16. The abundance of O-GlcNAc glycosylation in the brain13,15,16 and the fact that it shares many features with phosphorylation, a key regulator of cell signaling, synaptic plasticity, and learning and memory17, suggests critical roles for O-GlcNAc in the nervous system. Indeed, changes in overall O-GlcNAc levels have been shown to modulate long-term potentiation18, calcium influx via inositol phosphate channels19, and neurite branching20. Overall O-GlcNAc levels were also inversely related to phosphorylation levels on the protein tau21, implicating O-GlcNAc glycosylation in the pathogenesis of Alzheimer’s disease. However, a mechanistic understanding of how O-GlcNAc glycosylation contributes to neuronal processes and higher-order brain functions is unclear.
A major challenge toward understanding the biological roles of O-GlcNAc has been the difficulty of detecting and studying the modification. Like phosphorylation, O-GlcNAc glycosylation is chemically and enzymatically labile, often sub-stoichiometric, and subject to complex cellular regulation12–14. In addition, O-GlcNAc possesses several unique features that render it even more difficult to study than phosphorylation. For instance, phosphorylation has only three major forms (phosphorylated Ser, Thr, or Tyr), while O-GlcNAc glycosylation represents one of hundreds of different cellular glycans (e.g., mucin polysaccharides, glycosaminoglycans, etc.)22. Traditional methods for O-GlcNAc detection, such as radiolabeling with tritiated sugars, lack the sensitivity of 32P-labeling with ATP. Moreover, site-specific O-GlcNAc antibodies are rare and notoriously difficult to generate13, in contrast to the widespread use of phosphorylation-state specific antibodies. Finally, the presence of only one O-GlcNAc transferase (OGT) gene12,23 complicates genetic approaches toward elucidating the precise roles of the modification because knocking out or inhibiting OGT may have broad pleiotropic effects.
Recently, we developed a new chemoenzymatic method for detecting O-GlcNAc-modified proteins and quantifying in vivo glycosylation levels24. In this method, the O-GlcNAc residues on proteins are labeled with a polyethylene glycol (PEG) mass tag to shift the molecular weight of the glycosylated species. By immunoblotting the labeled cell lysate for proteins of interest, one can quantify glycosylation stoichiometries in vivo and establish whether proteins are mono-, di-, or multiply O-GlcNAc glycosylated. In addition, different post-translationally modified subpopulations can be rapidly visualized by immunoblotting with phospho- or other modification-specific antibodies. As such, the approach provides a direct read-out of whether phosphorylation and O-GlcNAc glycosylation are mutually exclusive on proteins of interest (i.e. yin-yang) or whether they co-exist on the same molecule. Here, we use this chemoenzymatic mass tagging method in combination with biochemical, molecular, cellular, and neurobiological approaches to understand the role of O-GlcNAc glycosylation in regulating the transcription factor CREB. We demonstrate that glycosylation at Ser40 inhibits both basal and activity-induced CREB-mediated transcription and serves to regulate important neuronal functions, including neurite outgrowth and memory consolidation.
RESULTS
CREB is highly glycosylated in neurons at Ser40
Previously, we demonstrated that CREB is O-GlcNAc-modified in mammalian cells and the rat forebrain24,25. To evaluate the role of O-glycosylation in regulating the function of CREB, we quantified the stoichiometry of CREB glycosylation in neurons using our chemoenzymatic approach24. Proteins with terminal GlcNAc sugars were selectively labeled with a 2,000-Da PEG mass tag24 and immunoblotted with an anti-CREB antibody to visualize the glycosylated species (Fig. 1a). We found that a large fraction of CREB (44–48%) was mono-glycosylated in both cultured cortical neurons and various brain regions of adult mice. Unlike other post-translational modifications on CREB, which are largely undetectable in unstimulated neurons, the stoichiometry of CREB glycosylation was high under basal conditions and was comparable to the levels of Ser133-phosphorylated CREB in activated PC12 cells26.
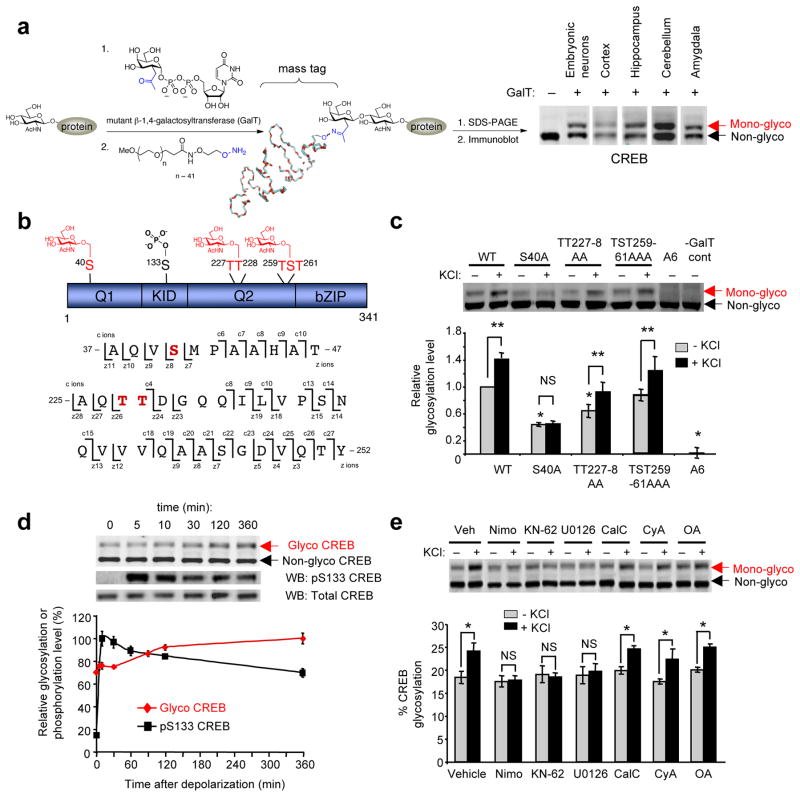
Figure 1.
CREB is O-GlcNAc glycosylated at Ser40 in response to neuronal activity. (a) Detection of O-GlcNAc glycosylated CREB in neurons by chemoenzymatic labeling with a 2000-Da mass tag and immunoblotting with an anti-CREB antibody. (b) Glycosylation sites on CREB mapped by ETD-MS. (c) Glycosylation levels of Flag-tagged WT CREB and various alanine mutants after expression in cultured cortical neurons, as detected by chemoenzymatic labeling and immunoblotting with an anti-Flag antibody. All levels were normalized to Flag-tagged WT CREB in untreated cells. Neurons were depolarized with KCl where indicated (n = 7 for WT and S40A CREB; n = 3–5 for other mutants; *P < 0.01 compared to WT, unstimulated cells; **P < 0.05; NS, not significant). (d) Kinetics of endogenous CREB glycosylation and Ser133 phosphorylation upon depolarization of cortical neurons with KCl. Levels of glycosylation or phoshorylation are plotted relative to the maximum signal for each modification. (n = 4–6). (e) Glycosylation levels of endogenous CREB in unstimulated or KCl-stimulated cortical neurons upon treatment with inhibitors of L-type calcium channels (nimodipine; nimo), CaMKs (KN-62), MAPK (U0126), protein kinase C (calphostin C; CalC), PP-2B (cyclosporin A; CyA), or PP-1/2A (okadaic acid; OA). (n = 3–6, *P < 0.02). Error bars, means, and standard errors of the mean (s.e.m). Full-length blots are presented in Supplementary Fig. 25.
To map the glycosylation sites, CREB was expressed in neuro2a cells, immunoprecipitated, and subjected to electron transfer dissociation-mass spectrometry (ETD-MS) analysis. In addition to the region of glycosylation identified previously (Thr259-Ser260-Thr261)25, O-GlcNAc glycosylation was mapped to Ser40 and Thr227 or Thr228 (Fig. 1b, Supplementary Fig. 1). To determine the major site of glycosylation, we expressed Flag-tagged WT CREB and various alanine mutants in cultured cortical neurons and measured their relative glycosylation levels under basal conditions using our chemoenzymatic approach. Mutation of Ser40 to alanine (S40A) led to a large reduction in CREB glycosylation levels compared to WT CREB (56.1 ± 3.1%; Fig. 1c), while mutation of both Thr227 and Thr228 (TT227-8AA) led to a smaller decrease in glycosylation (35.9 ± 9.6%). CREB glycosylation was also decreased by mutation of Thr259, Ser260, and Thr261 (TST259-61AAA; 13.6 ± 8.5%), and simultaneous mutation of all potential sites (A6) abolished the glycosylation of CREB. Our studies indicate that CREB is highly mono-glycosylated in neurons under basal conditions and is rarely glycosylated simultaneously at multiple sites. Additionally, we identify all major glycosylation sites on CREB and establish Ser40 as the predominant site of O-GlcNAc glycosylation in neurons.
Neuronal activity induces CREB glycosylation at Ser40
We next investigated whether CREB glycosylation is dynamically induced by neuronal stimuli. Although O-GlcNAc levels are modulated by glucose concentrations and cellular stress12,14, the signals and signaling pathways that regulate O-GlcNAc glycosylation in neurons are largely unknown. Two well-established methods for inducing neuronal activity include membrane depolarization using KCl and activation of N-methyl-D-aspartic acid (NMDA) receptors27. Notably, treatment of neurons with KCl or NMDA stimulated glycosylation of CREB (Fig. 1d, Supplementary Fig. 2). Upon membrane depolarization with KCl, CREB glycosylation levels increased steadily by 42.0 ± 4.8% over the course of 6 h (Fig. 1d). The kinetics of glycosylation was slower and more sustained than that of phosphorylation at Ser133, with glycosylation continuing to increase as phosphorylation levels declined. Mutation of Ser40 to alanine blocked depolarization-induced CREB glycosylation, while mutation of the other glycosylation sites had no effect (Fig. 1c). Treatment with the protein synthesis inhibitor cycloheximide did not block the increase in glycosylation (Supplementary Fig. 3), suggesting that glycosylation is activated directly by signal transduction pathways without requiring new protein synthesis. Inhibition of L-type calcium channels with nimodipine abolished the depolarization-induced glycosylation of CREB, indicating a requirement for voltage-sensitive calcium influx (Fig. 1e). Moreover, inhibition of Ca2+/calmodulin-dependent protein kinases (CaMKs) or mitogen-activated protein kinase (MAPK) blocked the increase in CREB glycosylation, while inhibitors of protein kinase C or protein phosphatases PP-2B or PP-1/2A had no effect (Fig. 1e). These results provide the first demonstration that neuronal activity triggers O-GlcNAc glycosylation in neurons and, specifically, leads to the calcium- and kinase-dependent glycosylation of CREB at Ser40.
Induction of Ser40 glycosylation requires phosphorylation
A variety of kinase pathways converge to phosphorylate CREB at Ser133 and activate CREB-mediated transcription in neurons, including the cAMP-induced protein kinase A (PKA), MAPK, and CaMKIV6–8,10,28. Previously, we found that CREB phosphorylation at Ser133 by PKA occurs independently of the glycosylation status of CREB in 293T cells24. However, the involvement of multiple different kinases in CREB Ser133 phosphorylation warrants the investigation of pathway-specific interactions between CREB glycosylation and phosphorylation. CaMKs and MAPK are known to phosphorylate CREB at Ser133 following neuronal depolarization28. As these kinases are also necessary for calcium-induced, activity-dependent glycosylation of CREB, we determined the interplay between Ser133 phosphorylation and CREB glycosylation. Importantly, mutation of Ser133 to Ala (S133A) blocked the depolarization-induced increase in CREB glycosylation (Fig. 2a). However, forskolin-mediated stimulation of Ser133 phosphorylation via the cAMP pathway had no effect on CREB glycosylation levels (Supplementary Fig. 4). These results provide evidence that phosphorylation is required but may not be sufficient by itself to activate CREB glycosylation.
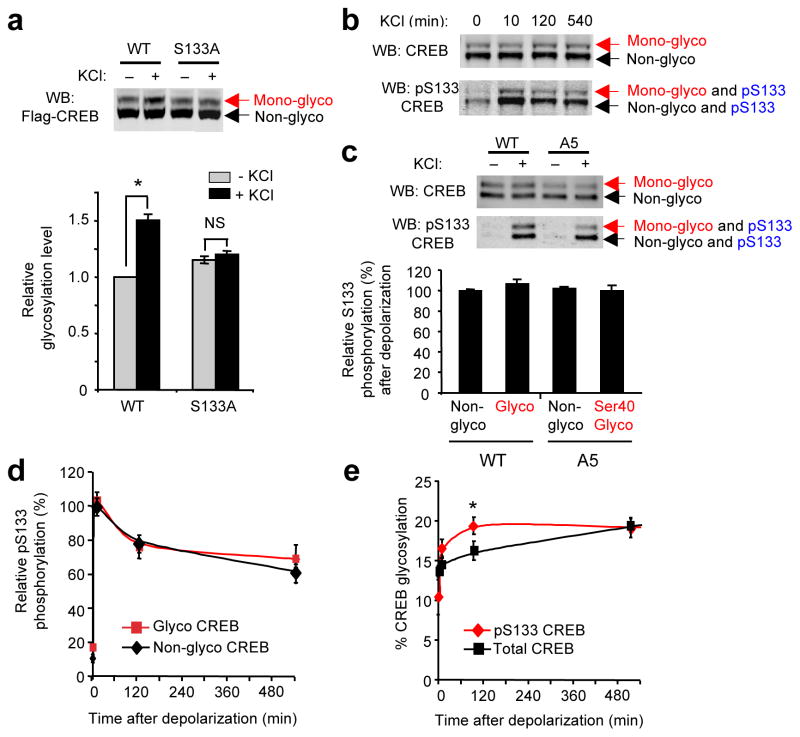
Figure 2.
Neuronal activity induces CREB glycosylation preferentially on the Ser133-phosphorylated subpopulation. (a) Glycosylation is not induced on S133A CREB. Glycosylation levels were analyzed on Flag-tagged WT or S133A CREB expressed in cortical neurons. (n = 22, *P < 0.001). (b) Chemoenzymatic labeling of endogenous CREB for visualizing phosphorylation and glycosylation within the same protein molecule and for quantifying the levels of each modification within distinct post-translationally modified subpopulations. (c) Quantification of pSer133 levels on the nonglycosylated and glycosylated subpopulations of Flag-tagged WT or A5 mutant CREB following 10-min depolarization of neurons. All levels were normalized relative to nonglycosylated, Flag-tagged WT CREB. (n = 3). (d and e) Kinetics of Ser133 phosphorylation (d) and glycosylation (e) for specific post-translationally modified subpopulations of endogenous CREB. Relative S133 phosphorylation and glycosylation levels were calculated as described in Methods (n = 4, *P < 0.03). Error bars, means, and s.e.m. Full-length blots are presented in Supplementary Fig. 25.
We next examined the interdependence of Ser133 phosphorylation and Ser40 glycosylation on CREB. Cortical neuronal lysates were labeled with a 2,000-Da mass tag and immunoblotted with a phospho-Ser133-specific or total CREB antibody to enable visualization of four distinct subpopulations (Fig. 2b)24. We found that both O-GlcNAc glycosylation and Ser133 phosphorylation can occur concomitantly on the same protein molecule (Fig. 2b). To assess further the role of glycosylation specifically at Ser40, we constructed a mutant CREB (A5) in which all glycosylation sites except Ser40 were mutated to alanine. Both the Ser40 glycosylated and nonglycosylated subpopulations of the A5 mutant were phosphorylated at Ser133 following 10 min of neuronal depolarization (Fig. 2c). Furthermore, phospho-Ser133 levels were similar across all glycosylated and nonglycosylated subpopulations of WT and A5 CREB. Thus, the same molecule of CREB can be modified simultaneously by O-GlcNAc at Ser40 and O-phosphate at Ser133, and glycosylation at Ser40 does not affect Ser133 phosphorylation levels in response to neuronal depolarization. To determine the kinetics of glycosylation and phosphorylation within each post-translationally modified subpopulation, we monitored the time course of induction. The kinetics of Ser133 phosphorylation, which included both a rapid and slow phase as reported previously28, were similar for both the glycosylated and nonglycosylated subpopulations of endogenous CREB upon KCl depolarization (Fig. 2b, d), confirming further that Ser133 phosphorylation is independent of the glycosylation state of CREB. Importantly, glycosylation was more rapidly induced on the Ser133-phosphorylated subpopulation compared to the total CREB population (Fig. 2b, e). Together, the results strongly suggest that phosphorylation and glycosylation work cooperatively to regulate CREB activity, with activity-dependent glycosylation induced preferentially on the phosphorylated subpopulation. The close coupling of these two post-translational modifications may allow for graded suppression of CREB following its activation.
CREB glycosylation represses CRTC-dependent transcription
We next determined whether glycosylation modulates the transcriptional activity of CREB. As Ser40 is the major glycosylation site and the only site responsive to neuronal activity, we compared the ability of wild-type (WT) and S40A mutant CREB to regulate CRE-dependent gene expression. A short hairpin RNA (shRNA) was used to knockdown endogenous CREB in neuro2a cells, and shRNA-resistant WT or S40A mutant CREB was overexpressed (Supplementary Fig. 5). Replacement of endogenous CREB with the S40A mutant resulted in increased CRE-luciferase activity, suggesting that glycosylation functions to repress CREB activity (Fig. 3a). The S40A substitution also upregulated the expression of endogenous CREB target genes involved in cell cycle arrest, cell survival, and mitochondrial function, including CDKN1A, NR4A2, and OPA3 (Supplementary Fig. 6). To confirm that the effects of mutating Ser40 are likely due to loss of an O-GlcNAc moiety at Ser40, we overexpressed the β-N-acetylglucosaminidase enzyme (O-GlcNAcase or OGA) to decrease O-GlcNAc glycosylation levels in cells and assessed the activity of WT and S40A CREB. As expected, OGA overexpression enhanced CRE-luciferase activity in cells expressing WT CREB, mimicking the effects of the S40A mutation (Supplementary Fig. 7). Notably, cells expressing S40A CREB did not undergo any further increase in CRE-dependent transcription upon OGA overexpression, suggesting that OGA overexpression enhances CREB activity by decreasing glycosylation at Ser40. Together, these data suggest that Ser40 glycosylation represses the transcriptional activity of CREB.
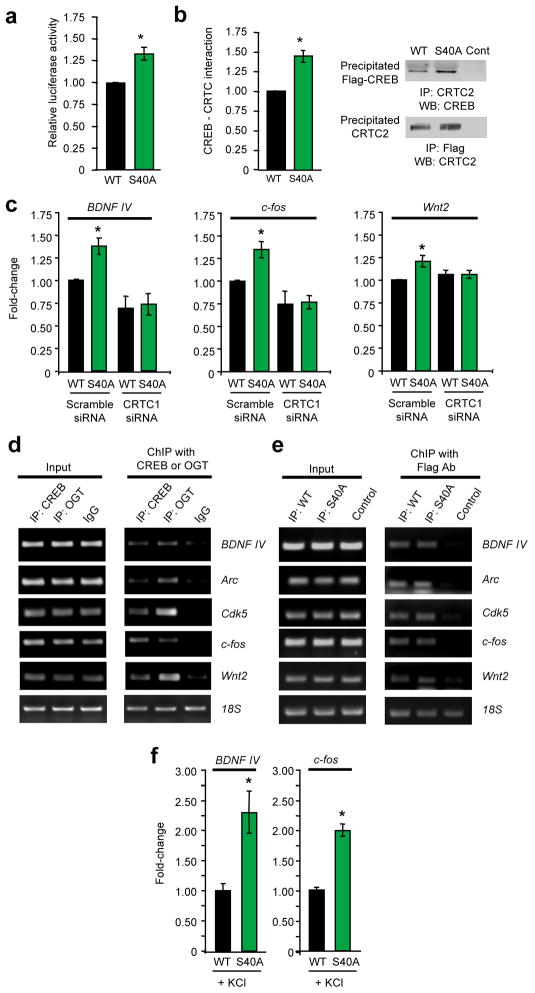
Figure 3.
Glycosylation at Ser40 represses CREB activity via a CRTC-dependent mechanism. (a) CRE-luciferase activity in neuro2a cells expressing WT or S40A CREB. (n = 11, *P < 0.01). (b) Reciprocal co-immunoprecipitation of the CREB-CRTC2 complex from neuro2a cells expressing WT or S40A CREB. The bar graph represents the average of all co-immunoprecipitation experiments (n = 8). (c) qPCR analysis of BDNF exon IV, c-fos, and Wnt2 expression in cultured cortical neurons electroporated with the indicated siRNAs or expression vectors using RPL3 as an internal control. Fold-change is plotted relative to neurons electroporated with WT CREB and scramble siRNA. (n = 4–9, *P < 0.01). (d) Chromatin immunoprecipitation with an anti-CREB, anti-OGT, or IgG antibody was followed by PCR for the indicated promoters. (n = 3). (e) Chromatin immunoprecipitation with an anti-Flag antibody after electroporation of neurons with Flag-tagged WT CREB, Flag-tagged S40A CREB, or no vector as a control. PCR was performed for the indicated promoters. (n = 3). (f) qPCR analysis of BDNF exon IV and c-fos expression after membrane depolarization of cultured cortical neurons expressing WT or S40A CREB. (n = 10, *P < 0.01). Error bars, means, and s.e.m. Full-length blots are presented in Supplementary Fig. 25.
To investigate the mechanism, we evaluated whether glycosylation affects the ability of CREB to associate with DNA or transcriptional co-activators. Binding of CREB to the CRE promoter was unaffected by the S40A mutation in an electrophoretic mobility shift assay (Supplementary Fig. 8). CREB interacts with two co-activators, the Ser133 phosphorylation-dependent CREB-binding protein (CBP) and the Ser133 phosphorylation-independent CREB-regulated transcriptional co-activator (CRTC/TORC)9,11. As the S40A mutant increased CREB activity when Ser133 phosphorylation was not activated (Fig. 3a, Supplementary Fig. 6), we examined whether glycosylation affects the CREB-CRTC interaction. Binding of CREB to CRTC was significantly enhanced by the S40A mutation in reciprocal co-immunoprecipitation assays (Fig. 3b, Supplementary Fig. 9). Furthermore, knockdown of CRTC2 expression in neuro2a cells abolished the observed increases in CDKN1A, NR4A2, and OPA3 transcript levels for S40A CREB compared to WT CREB (Supplementary Fig. 6 and 10). Together, these findings indicate that glycosylation impairs the ability of CREB to activate transcription by disrupting the CREB-CRTC interaction.
We next determined whether glycosylation at Ser40 regulates gene expression in neurons. In particular, we focused on well-characterized, neuronal CREB target genes important for brain development and memory consolidation, including BDNF exon IV, Arc, Cdk5, c-fos, and Wnt229–33. Relative to WT CREB, expression of S40A CREB in cortical neurons increased the levels of BDNF exon IV, Arc, Cdk5, c-fos, and Wnt2 transcripts (Fig. 3c, Supplementary Fig. 11). The observed increases likely underestimate the contribution of O-GlcNAc glycosylation to CREB activity given the moderate transfection efficiency of primary neurons (~30–40%) and the contribution of other transcription factors to the regulation of those genes. Therefore, we used CREB siRNA to obtain a rough approximation of the contribution of CREB to the expression of each gene. Assuming equal transfection efficiency of the cDNA plasmids and siRNA, the observed increases correspond to approximately 2.5–3.6-fold inductions in CREB activity (Supplementary Fig. 12). Consistent with a mechanism involving direct regulation of these genes, both CREB and O-GlcNAc transferase (OGT) were bound to the promoters of each gene (Fig. 3d), and WT and S40A CREB showed comparable levels of promoter occupancy in chromatin immunoprecipitation assays (Fig. 3e). Moreover, siRNA-mediated knockdown of CRTC1 reversed the effects of S40A CREB on neuronal gene expression (Fig. 3c, Supplementary Fig. 11 and 13). Thus, glycosylation of CREB at Ser40 modulates the constitutive expression levels of genes important for neuronal development, survival, and synaptic plasticity via a CRTC-dependent mechanism.
To investigate whether glycosylation also contributes to activity-induced gene expression, neurons expressing WT or S40A CREB were depolarized with KCl. Blocking glycosylation of CREB at Ser40 increased the levels of BDNF exon IV and c-fos transcripts to a greater extent in membrane-depolarized neurons compared to unstimulated neurons (Fig. 3f). These results, together with the observation that neuronal activity enhances glycosylation of the Ser133-phosphorylated CREB subpopulation, suggest dual functions for CREB glycosylation: first, to repress basal transcript levels and second, to attenuate activity-dependent CREB-induced gene expression.
CREB glycosylation at Ser40 regulates neuronal growth
As CREB has critical roles in axon growth, activity-dependent dendrite development, and synaptogenesis1,31,34, the ability of O-glycosylation to regulate CREB-mediated gene expression could have important consequences for neuronal development. To assess the functional consequences of Ser40 glycosylation on neuronal growth, we assayed axonal and dendritic extension in cortical neurons expressing WT or S40A mutant CREB (Supplementary Fig. 14). Dendrites of neurons expressing WT CREB or a GFP control exhibited similar lengths, and as expected, their growth was stimulated by membrane depolarization (Fig. 4a). In contrast, neurons expressing S40A CREB had significantly longer dendrites than WT CREB-expressing neurons (2.77-fold increase), displaying lengths comparable to those of depolarized neurons, and their dendrites showed no further elongation upon membrane depolarization (Fig. 4a). Additionally, neurons expressing S40A CREB had significantly longer axons compared to WT CREB-and GFP-expressing controls (Supplementary Fig. 15). Thus, blocking glycosylation of CREB accelerates the rate of both dendrite and axon elongation and leads to dysregulation of basal and activity-induced dendritic growth.
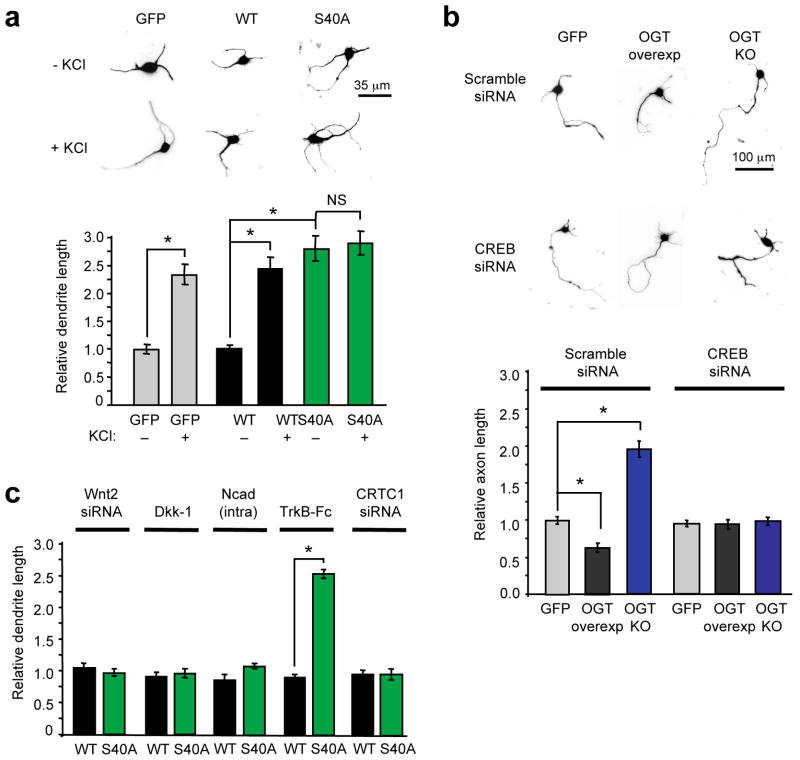
Figure 4.
CREB glycosylation at Ser40 represses dendritic and axonal growth. (a) Relative total dendrite lengths of cortical neurons expressing GFP, WT CREB or S40A CREB. (For each condition, thirty dendrites were measured in each of three independent experiments, *P < 0.001; NS, not significant). (b) Relative axon lengths of OGT floxed cortical neurons expressing GFP, Flag-tagged OGT, or Cre recombinase (OGT KO). Neurons were also transfected with scramble or CREB siRNA as indicated. (For each condition, thirty axons were measured in each of three independent experiments, *P < 0.001). (c) Relative dendrite lengths of unstimulated neurons transfected with WT or S40A CREB, Wnt2 signaling inhibitors (Wnt2 siRNA, Dickkopf-1 (Dkk-1), Ncad(intra)) or CRTC1 siRNA as indicated. The BDNF signaling inhibitor TrkB-Fc was added in solution to neurons. (For each condition, thirty dendrites were measured in each of three independent experiments, *P < 0.001). Lengths are shown relative to unstimulated GFP-expressing neurons in (A). Error bars, means, and s.e.m.
To confirm that these effects result from direct O-glycosylation of CREB, we performed OGT gain- and loss-of-function experiments. OGT-null neurons were generated from OGT floxed-mice23 by expression of CRE recombinase in cultured cortical neurons (Supplementary Fig. 16). Knocking out OGT stimulated axonal growth, whereas OGT overexpression (Supplementary Fig. 17) attenuated axonal growth (Fig. 4b). In both cases, siRNA-mediated knockdown of endogenous CREB reversed the effects of OGT knockout or overexpression and restored axon lengths to those of GFP-expressing neurons (Fig. 4b), indicating that O-GlcNAc glycosylation regulates axonal growth through a CREB-dependent mechanism.
To investigate further the underlying molecular mechanisms, we considered known mediators of dendrite and axon elongation. Activation of CREB drives the expression of the secreted mitogen Wnt2 to regulate activity-dependent dendritic growth, whereas application of the neurotrophin BDNF leads to axon elongation31,35. Earlier, we showed that both Wnt2 and BDNF transcript levels were significantly increased in cortical neurons expressing S40A CREB as compared to WT CREB (Fig. 3c). To examine whether CREB glycosylation modulates dendritic growth via the Wnt2 pathway, we knocked down Wnt2, overexpressed the Wnt2 antagonist Dickkopf-1, or overexpressed the β-catenin sequestrant Ncad(intra). All three treatments blocked the enhancement of dendritic growth following neuronal depolarization (Supplementary Fig. 18), demonstrating the efficacy of the Wnt2 blockers and consistent with the importance of Wnt2 in dendritic growth31. Most importantly, the same treatments reversed the stimulatory effects of S40A CREB on dendritic growth (Fig. 4c, Supplementary Fig. 19).
To probe the mechanism of glycosylation-mediated axonal growth, we treated neurons with the BDNF/NT-4/5 scavenger TrkB-Fc. Addition of TrkB-Fc to the cells blocked the effects of S40A CREB specifically on axonal, but not dendritic, growth (Fig. 4c, Supplementary Fig. 19 and 20). Furthermore, knockdown of CRTC1 abolished the S40A CREB-dependent increases in both Wnt2 and BDNF gene expression (Fig. 3c), dendritic growth (Fig. 4c, Supplementary Fig. 19), and axonal growth (Supplementary Fig. 20). Together, the results demonstrate that CREB glycosylation modulates dendrite and axon elongation via the CRTC-dependent downregulation of Wnt2 and BDNF signaling, respectively. These findings provide strong evidence that O-glycosylation of CREB has a large, chronic repressive effect on multiple developmental pathways and functions as a critical regulator of neuronal growth. As the directed growth of axons and dendrites in response to synaptic activation, depolarization, and trophic factors is critical for the formation and maturation of neuronal circuits36, these results also implicate O-GlcNAc in regulating such processes.
CREB glycosylation at Ser40 modulates long-term memory
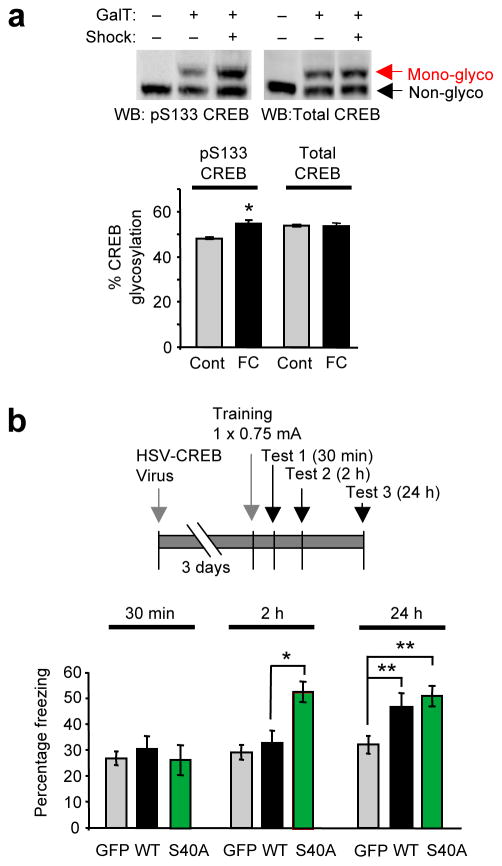
Having shown that CREB glycosylation affects important cellular processes, we next examined whether glycosylation impacts higher-order brain functions in vivo. Extensive studies have shown that CREB is not only a requirement but also a critical driving force for the consolidation of long-term conditioned fear memories3,37–41. We first investigated whether glycosylation is induced on endogenous CREB in response to auditory fear conditioning in mice. Specifically, the glycosylation levels of CREB were compared in the lateral amygdala of fear-conditioned mice and tone-only trained controls. CREB phosphorylation was induced in the amygdala, consistent with previous studies (Supplementary Fig. 21)39. An increase in glycosylation (13.6 ± 0.3%) was detected within the activated subpopulation (i.e., CREB phosphorylated at Ser133; Fig. 5a) but not the total CREB population. Therefore, glycosylation is specifically induced upon physiologic activation of the amygdala in vivo to modify Ser133-phosphorylated CREB. This induction of CREB glycosylation was comparable to the induction observed in neuronal cultures after 10-min depolarization with KCl (Fig. 1d). The chemoenzymatic mass tagging approach also revealed that more than half of the phosphorylated CREB subpopulation (54.7 ± 1.4%, 10 min after fear conditioning) was glycosylated, suggesting that CREB glycosylation may contribute significantly to amygdala function.
Figure 5.
CREB glycosylation at Ser40 modulates long-term conditioned fear memory. (a) Glycosylation levels of activated Ser133-phosphorylated CREB and total CREB in the amygdala 15 min after auditory fear conditioning. (n = 3, *P < 0.01). (b) Freezing behavior after auditory fear conditioning of mice infused with HSV vectors expressing GFP, WT or S40A CREB. (n = 11 for GFP, n = 16 for WT, and n = 20 for S40A at 2 h and 24 h, n = 6 for all vectors at 30 min. *P < 0.05, **P < 0.005). Error bars, means, and s.e.m. Full-length blots are presented in Supplementary Fig. 25.
To determine whether CREB glycosylation affects memory formation in vivo, replication-defective herpes simplex viral (HSV) vectors expressing WT CREB and GFP, S40A CREB and GFP, or GFP alone were bilaterally microinjected into the lateral amygdala of mice before fear conditioning (Supplementary Fig. 22), and memory was assessed 30 min, 2 h, and 24 h after training. Similar to previous experiments39,41, mice infused with WT CREB vector had enhanced memory compared to GFP vector-infused mice after 24 h, but not after 30 min or 2 h (Fig. 5b; F1,25 = 4.34, P = 0.048), indicating that CREB overexpression increases long-term memory. Notably, mice infused with S40A CREB vector exhibited significant memory enhancement by 2 h after training compared to mice infused with WT CREB or GFP (F2,45 = 9.70, P = 0.0003), and the enhanced memory persisted at 24 h (Fig. 5b). The mice exhibited the same short-term memory at 30 min independent of the genotype, suggesting that the enhancements at 2 h and 24 h likely represent changes in long-term memory and are not due to other non-specific events such as differences in pain sensation or cell death. Taken together, the data suggest that expression of S40A CREB in the amygdala may promote more rapid long-term memory consolidation compared to WT CREB.
We next tested further whether the effect of S40A CREB represents enhanced long-term memory formation, a CREB-dependent process that requires de novo mRNA and protein synthesis3,37. Mice were injected with anisomycin at various points after training and then memory was assessed. Anisomycin is a commonly used inhibitor of protein synthesis3,37,42, although it may also have secondary effects such as the activation of p38 MAPK43. Previous reports have shown that anisomycin injection disrupts protein synthesis-dependent long-term memory consolidation, but not protein synthesis-independent short-term memory42. Anisomycin injection immediately after training blocked the memory enhancement of S40A CREB at 2 h (Supplementary Fig. 23; F1,12 = 24.57, P = 0.0003), while anisomycin injection 2 h after training failed to block the memory enhancement at 24 h (Supplementary Fig. 24; F1,13 = 0.23, P = 0.64). These results provide further evidence that mice expressing S40A CREB have enhanced, consolidated long-term memory at 2 h. Collectively, our studies show that disinhibition of CREB activity through a single glycosylation site mutation can modulate the formation of long-term memory, and they demonstrate a new role for O-GlcNAc glycosylation in memory processing.
DISCUSSION
Since the discovery of O-GlcNAc over twenty years ago, the O-GlcNAc modification has been shown to play key roles in many cellular processes, ranging from glucose homeostasis and the stress response to insulin signaling and transcription12–14,44,45. Despite tantalizing evidence linking O-GlcNAc to neuronal signaling and neurodegeneration13,15,16,18–21, a mechanistic understanding of how this modification contributes to important neuronal functions has been lacking. In this work, we demonstrate that O-GlcNAc glycosylation regulates CREB, a central transcription factor in the brain. Our studies show that O-GlcNAc glycosylation of CREB at a specific site, Ser40, has important effects on neuronal gene expression, axonal and dendritic growth, and long-term memory.
In contrast with previous studies linking changes in O-GlcNAc to cell stress and metabolism12,44,45, we found that O-GlcNAc glycosylation was dynamically induced by neuronal activity, both in vitro and in vivo, and upon activation of MAP and CaM kinase pathways. As these same pathways regulate the phosphorylation of many proteins, our results suggest a strong coupling of glycosylation to phosphorylation in neurons. Moreover, the sustained levels of both basal and induced CREB glycosylation suggest that glycosylation may affect CREB activity over longer time periods compared to phosphorylation and may be important for longer lasting alterations in cellular function.
Our chemoenzymatic strategy combined with site-directed mutagenesis provided a powerful method to study the complex interplay between O-GlcNAc and phosphorylation and to dissect the function of the modification at individual sites. The complex interrelationship between the two modifications on CREB expands a simple yin-yang model of O-GlcNAc and phosphorylation as opposing, independent, or synergizing with one another. Furthermore, we found that specific sites of O-GlcNAc glycosylation can be activated independently and uncoupled from one another, depending on the stimulus. We showed previously that CREB is glycosylated within Thr259-Ser260-Thr261, which is located in the binding domain of TAFII130, a component of the TFIID transcriptional complex25. Hyperglycosylation of CREB disrupts the interaction of CREB with TAFII130 in vitro. Here we demonstrate a distinct function for Ser40 glycosylation, and together, our results highlight the capacity for O-GlcNAc glycosylation to exert multiple site-specific functions in a protein. As CREB has been shown to be SUMOylated, acetylated, and ubiquitinated46,47, these studies raise the exciting possibility that CREB activity may be controlled through the complex, combinatorial effects of various post-translational modifications. The continued development of new chemical methods to dissect such intricate interrelationships will be essential for understanding the regulation of CREB and numerous other proteins.
The ability of O-GlcNAc to hinder the binding of transcription factors to transcriptional co-activators such as CRTC represents a novel function for the O-GlcNAc modification. Overall nuclear CRTC levels have been proposed to limit the pool of active CREB11. Thus, glycosylation adds the capacity to control a defined subpopulation of CREB and may provide an elegant mechanism for the regulation of specific subsets of genes. Supporting this notion, both CREB and OGT were localized to the promoters of glycosylation-sensitive genes. As OGT has been shown to associate with several transcriptional regulatory complexes, including the mSin3a-HDAC complex and Polycomb repressor complex48,49, OGT may have the potential to glycosylate CREB locally while it is bound to specific gene promoters. This would enable even a small cellular pool of glycosylated CREB to effect significant changes in gene expression and allow for the differential regulation of a subset of CREB-mediated genes. Future studies will examine the effects of CREB glycosylation on a genome-wide scale.
Our results suggest that O-GlcNAc glycosylation functions as a constant repressor of CREB activity, thereby controlling the basal expression levels of CREB-mediated genes such as BDNF, Wnt2, and c-fos. By keeping basal transcript levels low, O-GlcNAc glycosylation would provide a larger dynamic range for gene induction to enable neurons to respond properly upon activation. Consistent with this notion, blocking glycosylation of CREB led to dysregulation of KCl-induced dendritic growth. As both CREB repression (via glycosylation) and activation (via phosphorylation) regulate a common Wnt2-dependent pathway, the increase in Wnt2 transcripts observed upon expression of S40A CREB may saturate the Wnt2/beta-catenin pathway and desensitize the cell to further increases in Wnt2 expression following depolarization. Thus, O-GlcNAc glycosylation of CREB may contribute to the proper establishment of neural connections through its ability to repress the expression of trophic factors in the absence of their developmental cues.
In addition to modulating constitutive transcription, CREB glycosylation also limited activity-dependent transcription. These results suggest that the O-GlcNAc modification may regulate the kinetics of CREB deactivation in response to neuronal depolarization and other stimuli. The close coupling of CREB’s activation to its inhibition could be an important strategy for balancing the expression of inducible genes and restoring neuronal homeostasis.
Consistent with its roles in regulating gene expression and dendritic/axonal growth, glycosylation of CREB had a significant impact on long-term memory consolidation. One hypothesis is that expression of S40A CREB enhances the levels of plasticity-related transcripts before and after associative learning. Such changes in gene expression would be expected to facilitate the more rapid accumulation of transcripts necessary for synaptic remodeling and memory consolidation. Although further studies are needed to address the underlying molecular mechanisms of long-term memory, our results provide the first demonstration that the addition of a GlcNAc sugar to a single site within a protein – a seemingly minor chemical perturbation – has important functional consequences in neurons and participates in long-term memory formation. These findings also point to the potential for O-GlcNAc glycosylation to contribute to complex, higher-order brain functions.
Our study expands the scope of cellular regulation by O-GlcNAc glycosylation to the brain and demonstrates that it has important functions in the nervous system. We provide mechanistic insights into how protein O-glycosylation adds a new layer of regulation to phosphorylation-dependent neural processes. Furthermore, our studies identify a previously unknown mechanism for balancing basal gene expression with activity-induced gene expression in neurons. These results, combined with the observation that many transcription factors and synaptic proteins are O-GlcNAc-modified13,15,16, demonstrate the functional breadth and potential of O-GlcNAc glycosylation as a critical regulator of neuronal function.
METHODS
Quantification of O-GlcNAc glycosylation and Ser133 phosphorylation levels on CREB
Cultured neurons or dissected brain tissues were lysed and chemoenzymatically labeled with a PEG mass tag as described previously24, with the modifications noted in the Supplementary Methods. The lysates were subjected to 4–12% SDS-PAGE and immunoblotted. Anti-CREB (Chemicon) and anti-phospho-Ser133 CREB (Affinity BioReagents) antibodies were used to quantify the percentage of glycosylation on endogenous CREB (Figs. 1a, 1d, 1e, 2b, 2d, 2e, Supplementary Fig. 3 and 4). An anti-Flag (M2, Sigma) antibody was used to quantify the percentage of glycosylation on exogenously expressed CREB mutants (Figs. 1c, 2a, 2c). An anti-phospho-Ser133 CREB antibody was also used to measure phospho-Ser133 levels relative to total CREB levels. All Western blots were visualized and quantified using an Odyssey Infrared Imaging System and software (Li-Cor, Version 2.1).
To quantify O-GlcNAc stoichiometries, the intensities of the PEG-shifted band (glycosylated protein fraction) and the unshifted band (nonglycosylated protein fraction) were measured using Odyssey imaging software. The resulting values were corrected for local background, and the PEG-shifted bands were further corrected against the region of corresponding molecular weight in the unlabeled negative control lane. The percentage of glycosylation was calculated from the signal intensities as the percentage of the glycosylated protein fraction over the total protein fraction (the sum of the glycosylated and non-glycosylated fractions). For Figs. 1c and 2a, this value was further normalized to the percentage of glycosylation for Flag-tagged WT CREB in untreated neurons, and averaged across multiple independent sample sets. For kinetic studies, the percentage of glycosylation was further normalized relative to the glycosylation stoichiometry of the basal sample for each sample set, and averaged across multiple independent sample sets.
Fig. 2d was calculated from Fig. 2b by measuring the relative levels of S133 phosphorylation in the non-glycosylated or mono-glycosylated CREB population. The pS133 signals were corrected for total CREB levels in each fraction and then normalized with respect to the basal pS133 phosphorylation level in each case. Fig. 2e was calculated from Fig. 2b by measuring the glycosylation stoichiometries from the CREB and pS133 CREB immunoblots as described above.
Cell treatments
For experiments using exogenously expressed CREB mutants (Figs. 1c, 2a, 2c), WT or mutant pLenti CREB constructs were electroporated into neurons. Neurons were treated with KCl (55 mM, 2 h for Figs. 1c, 1e, 2a, and Supplementary Fig. 3; 10 min for Fig. 2c; 10 min–9 h for Figs. 1d, 2b, 2d, 2e) or forskolin (10 μM, 2 h) after 4–6 DIV and NMDA (25 μM, 5 and 10 min) after 13 DIV. Prior to KCl treatments, both treated and control neurons were silenced overnight with tetrodotoxin (TTX, 1 μM; Tocris Biosciences). For NMDA treatments, cells were transferred to a 37 °C warming plate, and the media was exchanged with warm HCSS (20 mM HEPES, 55 mM glucose, 5 mM KCl, 0.8 mM MgCl·6H2O, 120 mM NaCl, 16.2 mM CaCl2, pH 7.4). Where indicated, cells were treated with the following inhibitors for 30 min prior to the addition of KCl: nimodipine (5 μM), KN-62 (5 μM), U0126 (10 μM), calphostin C (2.5 μM), cyclosporin A (5 μM), okadaic acid (50 nM), cycloheximide (0.3 mg ml−1) or vehicle (water, EtOH, or DMSO). All drugs except KCl and TTX were from Axxora Alexis.
Neurite outgrowth
Neurons were electroporated with WT or S40A pLEMPRA, pMaxGFP, pcDNA3-Dkk-1-Flag (provided by X. Yu), or Ncad(intra) (provided by X. Yu) vectors and scramble, CRTC1, or Wnt-2 siRNA (Santa Cruz) as indicated and then plated at a density of 25,000 neurons cm−2. Neurons from B6.129-Ogttm1Gwh/J mice (Jackson Laboratories) were electroporated with pMaxGFP, the CRE recombinase pBOB-CAG-iCRE-SD (Addgene), or pA2UCOE-OGT vector, along with scramble or CREB siRNA as indicated. One day prior to imaging for dendrites, neurons were depolarized with KCl (50 mM) where indicated. After 1 DIV, neurons were treated with TrkB-Fc (R&D Biosystems; 0.7 μg ml−1) where indicated. After 4–5 DIV, all neurons were fixed with 4% paraformaldehyde for 20 min at room temperature, washed twice with PBS, once with H2O, and mounted onto glass slides. Transfected GFP-expressing cells were imaged using a Nikon Eclipse TE2000-S inverted microscope equipped with Metamorph software. Neurite lengths were quantified with NeuronStudio (Version 0.9.92)50. The longest neurite was assigned as the axon, and the remaining neurites were assigned as dendrites. Staining of representative cultures with the dendrite specific marker MAP2 confirmed the results. Lysates from neurons electroporated with WT or S40A CREB were immunoblotted for Flag-CREB to confirm equal levels of CREB expression. Neurons from B6.129-Ogttm1Gwh/J or C57BL/6 mice were immunostained with an anti-OGT antibody DM17 (Sigma) to confirm the effects of OGT knockdown. Flag-tagged OGT overexpression was confirmed by immunoprecipitation using anti-Flag M2 affinity resin (Sigma) per the manufacturer’s protocol, followed by Western blotting for OGT using the anti-OGT antibody DM17.
Statistics
P values were calculated from Student’s paired t-test when comparing within groups and from Student’s unpaired t-test when comparing between groups. ANOVA was used to analyze in vivo data. All calculations were performed using the program Excel.
Additional methods
Information on DNA plasmids and virus construction, cell culture and transfection, glycosylation site mapping and quantification, glycosylation/phosphorylation interaction studies, luciferase reporter assays, EMSA, quantitative RT-PCR, auditory fear conditioning and amygdala biochemistry, co-immunoprecipitation and ChIP experiments can be found in the Supplementary Methods.
All animal experiments were approved by the Institutional Animal Care and Use Committee at Caltech.
Supplementary Material
Acknowledgments
We thank M. Antoniou (King’s College London School of Medicine) for the pA2UCOE-EGFP construct, M. Greenberg (Harvard University) for the pLEMPRA-GOI and pLLX-shRNA constructs, G. Hart (The John Hopkins University School of Medicine) for the OGT antibody, S. Josselyn (University of Toronto) for the p1005+:CREB construct, R. Lansford (California Institute of Technology) for the pLenti PGK:H2B:mCherry construct, R. Malenka (Stanford University) and X. Yu (Shanghai Institutes for Biological Sciences) for the pcDNA3-Dkk-1-Flag and Ncad(intra) constructs, P. Qasba (US National Cancer Institute) for the Y289L GalT construct, and L. Wells (University of Georgia) for the pDEST-HA-OGA construct. We thank S.-H. Yu for synthesizing the UDP-ketogalactose substrate and D. Anderson for providing the fear conditioning apparatus. We thank A. Silva for a critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (R01 GM084724 to L.C.H.-W., F31 NS056525-02 to J.E.R., and NRSA Training Grant 5T32 GM07737 to P.M.C.).
Footnotes
Author contributions
L.C.H.-W. designed, directed and coordinated the project. P.M.C. and J.E.R. designed and performed the experiments except where otherwise noted. D.E.M. and E.C.P. performed the mass spectrometry analyses; R.L.N. prepared the HSV. P.M.C., J.E.R., and L.C.H.-W. wrote the manuscript, and all authors participated in editing it.
Competing financial interest
The authors declare no competing financial interests.
Additional Information
Supplementary information and chemical information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 2.Carlezon WA, Jr, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 3.Kida S, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5:348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- 4.Kornhauser JM, et al. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 5.Gau D, et al. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 6.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 7.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 8.Barco A, Jancic D, Kandel ER. CREB-Dependent Transcription and Synaptic Plasticity. In: Dudek SM, editor. Transcriptional Regulation by Neuronal Activity. Springer US; 2008. pp. 127–154. [Google Scholar]
- 9.Chrivia JC, et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Deisseroth K, Tsien RW. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron. 2002;34:179–182. doi: 10.1016/s0896-6273(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 11.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 13.Rexach JE, Clark PM, Hsieh-Wilson LC. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 15.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vosseller K, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Greengard P. The neurobiology of slow synaptic transmission. Science. 2001;294:1024–1030. doi: 10.1126/science.294.5544.1024. [DOI] [PubMed] [Google Scholar]
- 18.Tallent MK, et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J Biol Chem. 2009;284:174–181. doi: 10.1074/jbc.M807431200. [DOI] [PubMed] [Google Scholar]
- 19.Rengifo J, Gibson CJ, Winkler E, Collin T, Ehrlich BE. Regulation of the inositol 1,4,5-trisphosphate receptor type I by O-GlcNAc glycosylation. J Neurosci. 2007;27:13813–13821. doi: 10.1523/JNEUROSCI.2069-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francisco H, et al. O-GlcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev Neurobiol. 2009;69:162–173. doi: 10.1002/dneu.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rexach JE, et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamarre-Vincent N, Hsieh-Wilson LC. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J Am Chem Soc. 2003;125:6612–6613. doi: 10.1021/ja028200t. [DOI] [PubMed] [Google Scholar]
- 26.Hagiwara M, et al. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 31.Wayman GA, et al. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Ohshima T, et al. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleischmann A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguado F, et al. The CREB/CREM transcription factors negatively regulate early synaptogenesis and spontaneous network activity. J Neurosci. 2009;29:328–333. doi: 10.1523/JNEUROSCI.5252-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tucker KL, Meyer M, Barde YA. Neurotrophins are required for nerve growth during development. Nat Neurosci. 2001;4:29–37. doi: 10.1038/82868. [DOI] [PubMed] [Google Scholar]
- 36.Zhang LI, Poo MM. Electrical activity and development of neural circuits. Nat Neurosci. 2001;4 (Suppl):1207–1214. doi: 10.1038/nn753. [DOI] [PubMed] [Google Scholar]
- 37.Viosca J, Lopez de Armentia M, Jancic D, Barco A. Enhanced CREB-dependent gene expression increases the excitability of neurons in the basal amygdala and primes the consolidation of contextual and cued fear memory. Learn Mem. 2009;16:193–197. doi: 10.1101/lm.1254209. [DOI] [PubMed] [Google Scholar]
- 38.Bourtchuladze R, et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 39.Han JH, et al. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 40.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci. 2009;12:1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourtchouladze R, et al. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong W, et al. Anisomycin activates p38 MAP kinase to induce LTD in mouse primary visual cortex. Brain Res. 2006;1085:68–76. doi: 10.1016/j.brainres.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 44.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 45.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 46.Comerford KM, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci USA. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Q, Hutchins AE, Doyle CM, Lundblad JR, Kwok RP. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J Biol Chem. 2003;278:15727–15734. doi: 10.1074/jbc.M300546200. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 49.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 50.Wearne SL, et al. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.