Summary
Copy number variations (CNVs) have been shown to contribute substantially to disease susceptibility in several inherited diseases including cancer. We conducted a genome-wide search for CNVs in blood-derived DNA from 79 individuals (62 melanoma patients and 17 spouse controls) of 30 high-risk melanoma-prone families without known segregating mutations using genome-wide comparative genomic hybridization (CGH) tiling arrays. We identified a duplicated region on chromosome 4q13 in germline DNA of all melanoma patients in a melanoma-prone family with three affected siblings. We confirmed the duplication using quantitative PCR and a custom-made CGH array design spanning the 4q13 region. The duplicated region contains 10 genes, most of which encode CXC chemokines. Among them, CXCL1 (melanoma growth-stimulating activity α) and IL8 (interleukin 8) have been shown to stimulate melanoma growth in vitro and in vivo. Our data suggests that the alteration of CXC chemokine genes may confer susceptibility to melanoma.
Keywords: Familial melanoma, Germline copy number variations, disease susceptibility, CXC chemokines, chromosome 4q13
Cutaneous malignant melanoma is an etiologically heterogeneous disease with genetic, host, and environmental factors, as well as their interactions contributing to its development (Tucker and Goldstein, 2003). Approximately 10% of melanoma cases occur in a familial setting (Goldstein and Tucker, 2001). To date, two high-risk melanoma susceptibility genes, CDKN2A on chromosome 9p21 and CDK4 on 12q14, have been identified. Germline mutations of these two genes account for only a small proportion of familial melanoma susceptibility, suggesting the existence of other high-risk genes. Recently, copy number variants (CNVs) have been shown to contribute substantially to disease susceptibility in several inherited diseases including cancer (Kuiper et al., 2010). Specifically, a recent genome-wide CNV mapping study reported the identification of a major susceptibility gene for a familial cancer, chordoma (Yang et al., 2009), suggesting that screening for complex genomic rearrangements that co-segregate with disease in families may provide a powerful alternative to traditional gene-mapping approaches.
We conducted a genome-wide search for CNVs in 30 high-risk melanoma-prone families without known segregating mutations using a whole-genome human array–comparative genomic hybridization (array-CGH) chip (Nimblegen 385K; average probe spacing, 7 kb). The families were from the United States and ascertained through health care professionals or self referrals. The families included at least two living first degree relatives with a history of invasive melanoma. All family members who were willing to participate in the study provided written informed consent under an NCI IRB-approved protocol. Each underwent a full-body skin examination and completed risk factor questionnaires for sun-related exposures. All diagnoses of melanoma were confirmed by histologic review of pathologic material or by pathology reports. We analyzed blood-derived genomic DNA from 79 individuals including 62 melanoma patients (1–4 patients per family) and 17 spouses. We used the Nexus Copy Number built-in Rank Segmentation algorithm to identify significant CNVs (P=1×10−6; number of probes per segment ≥10; log2 ratio>0.25 for gains and <−0.25 for losses). We focused on CNVs that were not present in the 17 spouses, did not overlap with previously reported CNVs, were located within gene regions, and occurred in all melanoma patients within a family.
We identified a duplicated region located on 4q13 in two melanoma patients (II-1 and II-3) in the family shown in Fig. 1. Three individuals (II-1, II-3, and II-4) had a confirmed history of melanoma at the time of initial examination (Fig. 1). Similar to other American melanoma-prone families, all of the melanoma patients had dysplastic nevi (DN), and relatively early onset of melanoma; one patient had multiple primary melanomas. The siblings’ mother (I-2) had confirmed squamous cell carcinomas of the skin and reportedly had lung cancer and melanoma; the maternal grandmother and a maternal cousin also were reported to have melanoma. None of these three melanomas could be confirmed.
Fig. 1.
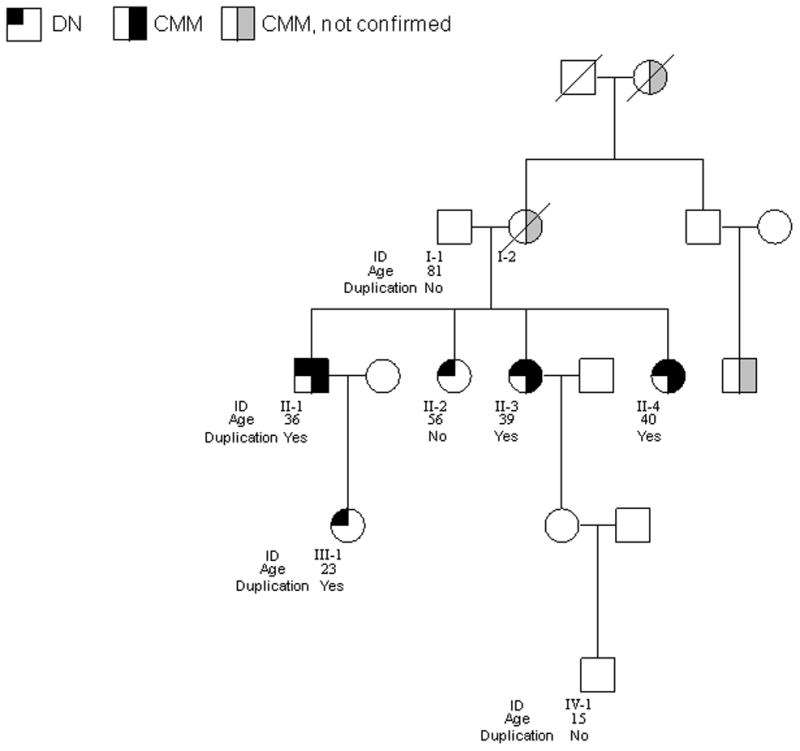
The pedigree of the melanoma family with the 4q13 duplication. Age as shown is age at diagnosis for melanoma patients and age at evaluation for unaffected individuals. II-1 and II-3 were analyzed by whole-genome array-CGH; I-1, II-1, II-2, II-3, II-4, III-1, and IV-1 were analyzed by qPCR; II-1, II-2, II-3, II-4, and III-1 were analyzed by custom-made 4q13 tiling arrays.
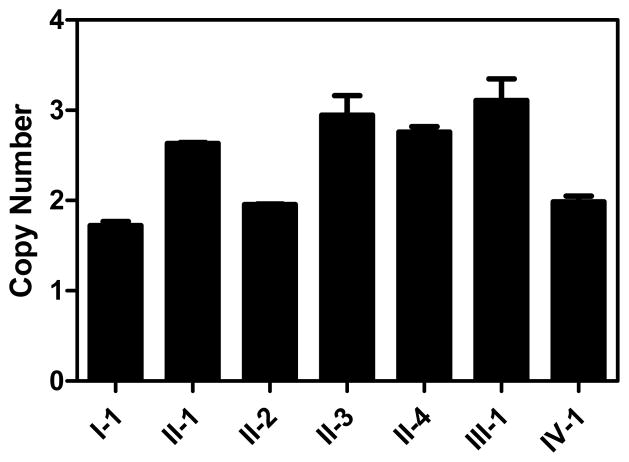
To validate the duplication and to examine the co-segregation of the duplication with melanoma status, we developed 2 quantitative PCR (qPCR) assays targeting two genes in this region, CXCL3 exon 4 and CXCL6 exon 4, respectively, and analyzed all 7 individuals with DNA available in this family. qPCR analyses confirmed the duplication in all three affected siblings, II-1, II-3, and II-4 (Fig. 2a). The unaffected father (I-1), the unaffected sibling (II-2), and an unaffected grandson (IV-1), did not have the duplication. The unaffected offspring (III-1) of one of the affected patients (II-1) also had the duplication. However, she was only 23 years old at ascertainment, had extensive number of nevi including DN, and had used sun protection for most of her life.
Fig. 2.
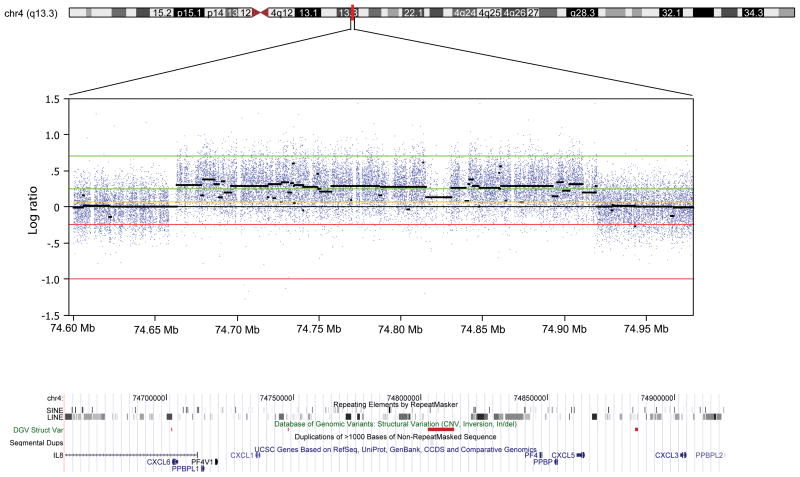
The 4q13 duplication identified in the melanoma-prone family. Panel a. Quantitative PCR (qPCR) of CXCL3 in genomic DNA of the melanoma family. Each qPCR assay was performed in duplicate. Results of qPCR assays for each individual are shown as a point estimate and 1 SD interval expressed as fold-difference compared to a reference sample. Data for CXCL6 were similar (not shown). Panel b. The duplication in melanoma patient II-3 by 4q13-focused array-CGH. Previously-reported CNVs listed in the Database of Genomic Variations are shown as red bars and none were located within genes. The duplicated region contains short and long interspersed repeat elements (SINE and LINE). Panel c. Breakpoint junction of the duplication showing a head-to-tail orientation. Tel-REF: telomeric reference sequence; Cen-REF: centromeric reference sequence. Matched sequences between reference and the melanoma patient are shown in colors (blue for telomeric and red for centromeric).
To further confirm the 4q13 duplication and to better define the breakpoints of the amplicons, we analyzed genomic DNA from fifteen individuals (individuals II-1, II-2, II-3, II-4, and III-1 in the family with the 4q13 duplication, 5 melanoma patients from families without the 4q13 duplication, and 5 unaffected controls) using a Nimblegen custom-made fine-tiling CGH array spanning the 4q13 region (average probe spacing, 15 bp). We confirmed the duplication in all three affected siblings and in individual III-1. As expected, the duplications in these four individuals were identical in size and location. In contrast, no duplication was observed in II-2, or in five other melanoma patients from five families that did not carry the duplication, or in five controls. We subsequently PCR amplified and sequenced the junction fragments from individuals II-1 and III-1 and determined that the duplication was 257kb (74663132 to 74919990 bp, hg19) with a head-to-tail tandem orientation (Fig. 2b, 2c). Bioinformatic analysis revealed that the breakpoints were located at or near repetitive short and long interspersed repeat (SINE and LINE) elements (Fig. 2b). In contrast, no junction fragment was amplified from individual II-2 who did not carry the duplication.
The duplicated region was not present in the other individuals evaluated by array-CGH. In addition, the 4q13 duplication was not observed in 318 control chromosomes (159 control subjects) by qPCR, suggesting that the duplication is unlikely a common polymorphism. Furthermore, the duplication was not observed in index patients from additional CDKN2A/CDK4 mutation-negative melanoma-prone families, including 16 American, 182 Italian, 170 Spanish, and 96 Australian families using either qPCR or array-CGH.
The duplicated region affected 10 genes, including IL8 (interleukin 8), CXCL6 (granulocyte chemotactic protein-2), PPBPL1 (pro-platelet basic protein-like 1), PF4V1 (platelet factor-4 variant 1), CXCL1 (melanoma growth-stimulating activity α), PF4 (platelet factor-4), PPBP (pro-platelet basic protein), CXCL5 (chemokine (C-X-C motif) ligand 5), CXCL3 (melanoma growth-stimulating activity γ), and PPBPL2 (pro-platelet basic protein-like 2). IL8 and PPBPL2 were partially affected and the remaining eight genes were completely contained in the duplicated region (Fig. 2b). Most of these genes belong to a family of CXC chemokines, which are characterized by one amino acid between the first and second cysteine residues. Among them, CXCL1, CXCL3, CXCL5, CXCL6, PPBP, and IL8 contain a three amino acid ELR motif (glutamine–leucine–arginine; ELR+) between the N-terminus and the first cysteine, and function as potent promoters of angiogenesis, therefore promoting tumorigenesis and metastasis (Payne and Cornelius, 2002).
CXCL1 has been shown to be up-regulated in melanoma cells and play an important role in melanoma pathogenesis (Dhawan and Richmond, 2002), with secretion of CXCL1 in melanoma cell lines 6- to 16-fold higher than in normal melanocytes (Norgauer et al., 1996). Overexpression of CXCL1 in melanocytes has been associated with enhanced growth and tumor formation in nude mice (Balentien et al., 1991). Blocking antibodies to either CXCL1 or its receptor, chemokine (C-X-C motif) receptor 2 (CXCR2), inhibited melanoma cell growth and reduced angiogenic activity (Haghnegahdar et al., 2000). Similarly, IL8 was the first chemokine reported to induce melanoma cell chemotactic and haptotactic migration (Wang et al., 1990). Neutralizing antibodies to the IL8 receptors CXCR1 and CXCR2, as well as to IL8 itself, inhibited melanoma cell proliferation and invasive potential (Varney et al., 2003), suggesting the potential for these chemokines and their receptors in melanoma treatment. In a study identifying gene expression signatures in melanoma progression, Haqq et al. found that elevated expression of CXCL1 not only distinguished primary tumors from moles but also was useful in identifying metastases (Haqq et al., 2005). Similarly, data from molecular profiling analyses also found upregulation of CXCL3 and IL8 in melanoma tumors and metastases (Lukk et al., 2010) (Mauerer et al., 2011) (Bertucci et al., 2007). In addition, recent data suggest that ciglitazone, an oral thiazolidinedione anti-diabetic drug, inhibited growth and survival of melanoma cells through repressing the expression and secretion of CXCL1, while the recombinant CXCL1 abrogated the apoptotic effect induced by ciglitazone and inhibition of CXCL1 production mimicked the effects of ciglitazone on growth inhibition (Botton et al., 2011). Furthermore, CXCL1 inhibition by ciglitazone was mediated by the decreased expression of microphthalmia-associated transcription factor (MITF), the master gene of melanocyte differentiation. These results further highlight the importance of CXCL1 in melanoma tumorigenicity and its potential as a therapeutic target.
In summary, we identified a germline duplication that may explain the melanoma susceptibility in a melanoma-prone family. The duplicated region contains genes that are known to play important roles in melanoma development and progression. Although the duplication is very rare, other types of variants such as point mutations and small insertions/deletions in these genes may explain susceptibility in additional melanoma families and therefore evaluations of variations within this novel duplication are needed to further determine the role of these genes in melanoma susceptibility.
Significance.
Several CXC chemokines are known to play important roles in melanoma development and progression. Here we show for the first time that CXC chemokine genes may confer major susceptibility to familial melanoma. Variations in these genes, including CNVs and single-nucleotide mutations, should be screened in melanoma-prone families without a defined etiology. Our results further support the role of rare, inherited copy number variations in cancer susceptibility.
Acknowledgments
We are indebted to the participating families, whose generosity and cooperation have made this study possible. We thank SeqWright, Roche Nimblegen, and Biodiscovery for their molecular and informatic services. We also acknowledge the contributions to this work that were made by Virginia Pichler, Deborah Zametkin, Mary Fraser, and Barbara Rogers. This research was supported by the Intramural Research Program of the NIH, NCI, DCEG. The research at the Melanoma Unit in Barcelona is partially funded by Grants 06/0265 and 09/1393 from Fondo de Investigaciones Sanitarias, Spain; by the CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain; by the AGAUR 2009 SGR 1337 of the Catalan Government, Spain; by the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and by the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115) and a personal grant to Paula Aguilera from Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Rio Hortega 10/00120. Genoa team research has been supported by : Fondazione CARIGE 2010, IMI and ACM 2011, Italian Ministry of Health DGRST.4/4235-P1.9.A.B. NKH is supported by a Senior Principal Research Fellowship from the National Health and Medical Research Council of Australia.
References
- Balentien E, Mufson BE, Shattuck RL, Derynck R, Richmond A. Effects of MGSA/GRO alpha on melanocyte transformation. Oncogene. 1991;6:1115–24. [PubMed] [Google Scholar]
- Bertucci F, Pages C, Finetti P, Rochaix P, Lamant L, Devilard E, Nguyen C, Houlgatte R, Birnbaum D, Xerri L, et al. Gene expression profiling of human melanoma cell lines with distinct metastatic potential identifies new progression markers. Anticancer Res. 2007;27:3441–9. [PubMed] [Google Scholar]
- Botton T, Puissant A, Cheli Y, Tomic T, Giuliano S, Fajas L, Deckert M, Ortonne JP, Bertolotto C, Tartare-Deckert S, et al. Ciglitazone negatively regulates CXCL1 signaling through MITF to suppress melanoma growth. Cell Death Differ. 2011;18:109–21. doi: 10.1038/cdd.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan P, Richmond A. Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol. 2002;72:9–18. [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Tucker MA. Genetic epidemiology of cutaneous melanoma: a global perspective. Arch Dermatol. 2001;137:1493–6. doi: 10.1001/archderm.137.11.1493. [DOI] [PubMed] [Google Scholar]
- Haghnegahdar H, Du J, Wang D, Strieter RM, Burdick MD, Nanney LB, Cardwell N, Luan J, Shattuck-Brandt R, Richmond A. The tumorigenic and angiogenic effects of MGSA/GRO proteins inmelanoma. J Leukoc Biol. 2000;67:53–62. doi: 10.1002/jlb.67.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, 3rd, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–7. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Ligtenberg MJ, Hoogerbrugge N, Geurts Van Kessel A. Germline copy number variation and cancer risk. Curr Opin Genet Dev. 2010;20:282–9. doi: 10.1016/j.gde.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–4. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauerer A, Roesch A, Hafner C, Stempfl T, Wild P, Meyer S, Landthaler M, Vogt T. Identification of new genes associated with melanoma. Exp Dermatol. 2011;20:502–7. doi: 10.1111/j.1600-0625.2011.01254.x. [DOI] [PubMed] [Google Scholar]
- Norgauer J, Metzner B, Schraufstatter I. Expression and growth-promoting function of the IL-8 receptor beta in human melanoma cells. J Immunol. 1996;156:1132–37. [PubMed] [Google Scholar]
- Payne AS, Cornelius LA. The role of chemokines in melanoma tumor growth and metastasis. J Invest Dermatol. 2002;118:915–22. doi: 10.1046/j.1523-1747.2002.01725.x. [DOI] [PubMed] [Google Scholar]
- Tucker MA, Goldstein AM. Melanoma etiology: where are we? Oncogene. 2003;22:3042–52. doi: 10.1038/sj.onc.1206444. [DOI] [PubMed] [Google Scholar]
- Varney ML, Li A, Dave BJ, Bucana CD, Johansson SL, Singh RK. Expression of CXCR1 and CXCR2 receptors in malignant melanoma with different metastatic potential and their role in interleukin-8 (CXCL-8)-mediated modulation of metastatic phenotype. Clin Exp Metastasis. 2003;20:723–31. doi: 10.1023/b:clin.0000006814.48627.bd. [DOI] [PubMed] [Google Scholar]
- Wang JM, Taraboletti G, Matsushima K, Van Damme J, Mantovani A. Induction of haptotactic migration of melanoma cells by neutrophil activating protein/interleukin-8. Biochem Biophys Res Commun. 1990;169:165–70. doi: 10.1016/0006-291x(90)91449-3. [DOI] [PubMed] [Google Scholar]
- Yang XHR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li SF, Goldstein AM, Parry DM, Kelley MJ. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nature Genetics. 2009;41:1176–1178. doi: 10.1038/ng.454. [DOI] [PMC free article] [PubMed] [Google Scholar]