Abstract
Caret software is widely used for analyzing and visualizing many types of fMRI data, often in conjunction with experimental data from other modalities. This article places Caret’s development in a historical context that spans three decades of brain mapping – from the early days of manually generated flat maps to the nascent field of human connectomics. It also highlights some of Caret’s distinctive capabilities. This includes the ease of visualizing data on surfaces and/or volumes and on atlases as well as individual subjects. Caret can display many types of experimental data using various combinations of overlays (e.g., fMRI activation maps, cortical parcellations, areal boundaries), and it has other features that facilitate the analysis and visualization of complex neuroimaging datasets.
Keywords: Surfaces, Cerebral Cortex, Atlases, Visualization, Connectivity, Human
Introduction
Caret software for brain mapping began in the early 1990’s and is thus of the same vintage as fMRI. As suggested by its full name (Computerized Anatomical Reconstruction and Editing Toolkit), Caret was originally designed to generate and visualize cortical surface reconstructions from postmortem brain sections – initially from the macaque monkey (Carman et al., 1995; Drury et al., 1996) and later from humans (Van Essen and Drury, 1997). Over the past decade Caret has evolved into a platform that is widely used for the analysis and visualization of structural and functional MRI data. This is often done in conjunction with other software applications that carry out initial stages of analysis on volumes and/or surfaces. Looking towards the future, Caret provides the foundation for the Connectome Workbench, which will be the primary platform for visualization and mining of structural and functional connectivity data obtained by the Human Connectome Project. Thus, the history of Caret’s development provides a unique perspective on how a software program evolved to meet the rapidly expanding needs associated with MR-based studies of brain structure, function, development, and connectivity.
Cortical surfaces and flat maps – the early days
Caret’s roots lie in an earlier era of brain mapping, in which manually generated cortical flat maps were the coin of the realm for coping with the complex and irregular pattern of cortical convolutions. My involvement (obsession is perhaps more accurate) with cortical surface representations started in 1975, when I began studying extrastriate visual cortex in the macaque while a postdoc at University College London. I found it highly frustrating to read one macaque connectivity study after another that displayed complex anatomical relationships using drawings or photographs of histological sections through the irregularly convoluted cortex. This format made it difficult to evaluate data contained in multiple sections within a single hemisphere and to compare results across different brains. An attractive way forward in principle was to generate cortical surface maps that restore the topological relationships between closely spaced sections. This had previously been done for area V1 by Daniel and Whitteridge (1961) using a 3-D physical model (cleverly generated out of clay!) and for other restricted cortical regions using ‘straight-line’ contour maps (Hubel and Wiesel, 1972; Jones and Burton, 1976). However, these methods were inadequate for the complex folding of macaque extrastriate cortex. Hence, I spent most of my postdoctoral year developing and using a manual ‘pencil and tracing paper’ method for generating cortical flat maps (Van Essen and Zeki, 1978; see Fig. 1A). Once in my own lab at Caltech, we showed that this method could be used to generate flat maps of the entire cerebral hemisphere (Van Essen and Maunsell, 1980) on which many types of experimental data could be displayed, such as the classic Brodmann architectonic map shown in Figure 1B.
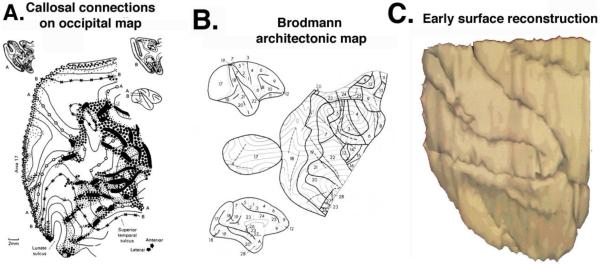
Figure 1. Early cortical surface maps.
A. The manual ‘pencil and tracing paper’ method for generating cortical flat maps was introduced and used to display interhemispheric connections of macaque occipital cortex. (Reproduced with permission from Van Essen and Zeki, 1978.) B. The first flat map of the entire cerebral neocortex, showing Brodmann’s architectonic areas. (Reproduced with permission from Van Essen and Maunsell, 1980). C. A computerized surface reconstruction of macaque occipital cortex. (D. Van Essen and G. McCann, unpublished results.)
It is fair to say that cortical flat maps played to mixed reviews. Some colleagues ‘got it’ and applauded the approach. Many others found flat maps confusing and not to their liking. For me, it was a lesson in perseverance. Had I followed the opinions of the majority of my colleagues, I would have abandoned the approach altogether.
The manual method of generating flat maps was not for the faint of heart. It was tedious, demanded considerable expertise, and involved much trial-and-error--the eraser was as important as the pencil! An alternative method for making cortical flat maps using wire-frame models (Gattass and Gross 1981) was comparably tedious, technically demanding, and not amenable to full-hemisphere flattening. It was clear to me even in the 1970’s that computers would be better suited than humans for the nuts and bolts job of reconstructing cortical convolutions and manipulating their shape. However, the computers needed proper instruction on how to do the job. This was far easier said than done; little did I realize that it would take nearly two decades for the concept to become a practical reality. My first stab at the problem was in the late 1970’s, when I teamed up with Gilbert McCann’s group at Caltech to reconstruct a chunk of macaque occipital cortex (Fig. 1C). However, surface generation and visualization were painfully slow, even though we were using state-of-the-art surface rendering software and the latest hardware (McCann’s PDP11 computer). A decade later, my lab acquired a Silicon Graphics Inc. (SGI) computer that had much better surface rendering capabilities (to the tune of $80,000, a whopping price at the time!). George Carman, Dave Bilitch, and I implemented a first-generation brain-mapping program (called ‘anatomy’) and tackled the cortical flattening problem. While we made progress, we came to fully appreciate the computational challenges of generating accurate surface reconstructions and flat maps (Carman, 1990). Around the same time frame, other groups implemented alternative approaches to the computational flattening problem (Wolfson and Schwartz, 1989; Schwartz et al., 1989; Dale and Sereno, 1993).
Our brain-mapping efforts accelerated after I moved to Washington University in 1992 and recruited the talented Heather Drury to lead the design and implementation of what became Caret software. In collaboration with Charlie Anderson, we developed a multi-resolution approach to cortical flattening that was computationally efficient (Drury et al., 1996). In collaboration with Mike Miller, we developed methods for landmark-constrained surface registration, initially applied to flat maps despite the drawback of having to deal with artificial cuts in the cortical surface (Van Essen et al. 1998). In 2001, I was very fortunate to bring John Harwell and Donna Dierker on board; their contributions over the past decade have allowed Caret to progress along many fronts. Here, I illustrate functionality and features of Caret that facilitate analyses of fMRI data and integration with other data types. These include (i) surface reconstruction, visualization, and shape manipulation; (ii) atlases and surface-based registration; (iii) mapping parcellations and functional data onto atlas surfaces; and (iv) interspecies comparisons between macaque and human cortex. Additional comments are provided on a few general user-friendly features (e.g., ‘scenes’ and study metadata) that are distinct to Caret.
Surface reconstructions
The ease of obtaining high-quality structural MR images from T1-weighted scans has motivated many efforts to develop segmentation algorithms that capture the shape of cortical convolutions in individual subjects (e.g., Dale and Sereno, 1993; Teo et al., 1997; MacDonald et al., 2000; Kriegeskorte and Goebel, 2001; Fischl et al., 2001; Han et al., 2004). Our lab developed the SureFit cortical segmentation algorithm, which was initially free-standing (Van Essen et al., 2001) but was later incorporated into the Caret platform. A distinctive feature of SureFit is that it generates segmentation and surfaces running along the cortical midthickness (Fig. 2A, B). This gives a representation of cortical surface area that is roughly proportional to the associated volume of cortical gray matter. Our emphasis on midthickness surface contrasts with other segmentation algorithms, including the widely used FreeSurfer method, which generate surfaces running along the pial and/or white-matter boundaries. Fortunately, the midthickness surface can easily be obtained by averaging FreeSurfer white and pial surfaces once they are imported into Caret. Caret can also handle datasets generated from perinatal human infant brains, which are poorly myelinated and hence require customized segmentation software (e.g., the LIGASE method) to obtain high-quality surfaces from T2-weighted scans (Hill et al., 2010).
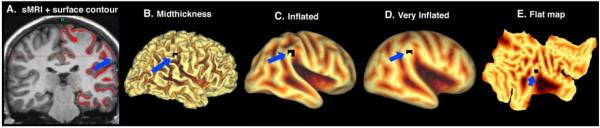
Figure 2. Visualization of volumes and surfaces in Caret.
A. Coronal volume slice with midthickness surface contour of right hemisphere. B. Lateral view of midthickness surface. C, D. Lateral view of inflated and very inflated surfaces with sulcal depth maps displayed. E. Flat map, with sulcal depth displayed. Crosshairs and blue arrow in panel A indicate the location of the highlighted node (black; blue arrows) in the surface views.
Flexible surface and volume representations
Surface inflation and flattening allow visualization of cortex that is buried in deep and irregular sulci. Inflated surfaces (e.g., Fig. 2C, D) retain a brain-like shape that aids localization. Cortical flat maps (Fig. 2E) display the entire hemisphere in one view, but require artificial cuts that disrupt the continuity between nearby cortical locations. Displaying maps of sulcal depth (distance to the ‘cerebral hull’) is an effective way to preserve a representation of underlying shape features on each of these smoother surfaces (Van Essen et al., 2001). Because no single surface or volume configuration serves all visualization purposes, it is often useful to load several surface configurations into Caret and to use multiple windows to view as many configurations as are helpful for whatever analyses are underway.
While the advantages of surface visualization are now reasonably obvious even to brain-mapping ‘newbies’, the road to widespread acceptance of surface-based approaches remained uphill for many years. The puzzlement and skepticism about manual flat maps that I experienced initially persisted among many investigators even when inflated surfaces could be twirled around effortlessly in Caret, FreeSurfer, and Brain Voyager. I attribute this to a ‘methodological inertia’ that can take years to overcome and as a result can hinder progress.
The evolution of brain atlases
In order to compare results across individuals and across studies, it is important to represent data on a common target – an atlas – and to compensate for individual variability when registering individuals to the atlas. Brain atlases for human neuroimaging began with the classical Talairach atlas and stereotaxic space (Talairach and Szikla, 1967; Talairach and Tournoux, 1988), which were first promoted by Fox et al. (1985) as a spatial framework for reporting the centers of activation foci from PET and fMRI studies. Human brain atlases have evolved dramatically in the ensuing decades. Some MR-based volumetric atlases were based on individual subjects (Roland et al., 1994; Roland and Zilles, 1996; Holmes et al., 1998). However, the idiosyncratic pattern of cortical convolutions makes it problematic to rely on any single subject as an atlas target. Consequently, there has been a proliferation of population-average MR structural volumes, initially using linear (affine) transformations (Evans et al., 1993; Friston et al., 1995; Ojemann et al., 1997; Jenkinson and Smith, 2001) and more recently using high-dimensional nonlinear registration (see Klein et al., 2009). These atlases differ in overall brain dimensions and in the fidelity of intersubject alignment (see Devlin and Poldrack, 2007). However, even high-dimensional nonlinear registration is inherently limited in its alignment fidelity, unless it is constrained to respect the topology of the cortical sheet.
Surface-based and volume-based atlases are inherently complementary for visualization as well as analysis purposes. Providing support for atlases has been a major emphasis of Caret since its inception. Our laboratory introduced surface-based atlases that were initially based on physically sectioned brains – ‘Case 79-0’ for the macaque (Drury et al., 1996) and the Visible Man cryosectioned brain for human (Van Essen and Drury, 1997). A subsequent generation of surface-based atlases was based on high quality MR scans of individual subjects. This includes the ‘Colin’ human atlas (Fig. 3A; Drury and Van Essen, 1997; Van Essen, 2002a, 2004) and the macaque ‘F99’ atlas (Van Essen, 2004). Importantly, both the Colin and F99 atlases include cerebellar as well as cerebral cortical surface reconstructions along with the associated MR volumes, making them valuable substrates for analyses that utilize whole-brain coverage (Fig 3A). Obtaining accurate cerebellar surfaces was a special challenge, because automated algorithms are unable to segment its very thin cerebellar gray and white matter domains. Instead, generating these ‘one-of-a-kind’ cerebellar surfaces entailed hundreds of hours of tedious manual segmentation (Van Essen, 2002b).
Figure 3. Human surface-based atlases.
A. The single-subject ‘colin’ surface-based atlas of cerebral and cerebellar cortex (Van Essen, 2002a,b). B. The PALS-B12 average midthickness surfaces, with three highlighted nodes representing corresponding points in the left and right hemisphere (Van Essen, 2005). C. The FreeSurfer-generated fsaverage midthickness surfaces, which shows more detailed features than the PALS-B12 atlas, but lacks correspondence between left and right hemispheres (highlighted blue node ‘1’ differs in location in the left vs right hemisphere). D. The ‘fs_LR’ atlas midthickness surfaces, which are essentially identical in shape to the fsaverage surfaces, but are in precise geographic correspondence (Van Essen et al., 2011b).
Population-average atlases are preferable for surfaces just as for volumes, in order to have a target that is not biased by the shape features of an individual subject. Several human surface-based atlases have emerged in recent years. The PALS-B12 atlas (Van Essen, 2005) was generated from SureFit-segmented surfaces of 12 subjects, registered to a population-average using landmark-constrained registration (‘Landmark-SBR’) and a set of ‘Core 6’ landmarks that can be consistently delineated in essentially all subjects. The PALS-B12 average and midthickness surfaces (Fig. 3B) have the left and right hemispheres in register with one another. FreeSurfer’s ‘fsaverage’ atlas is based on an energy-based registration method (Energy-SBR) that aligned the ‘average convexity’ (similar to sulcal depth) of 40 individuals to a population-average average convexity map (Desikan et al., 2006). The fsaverage midthickness surface (Fig. 3C) has a finer-grained pattern of structural features in each hemisphere than can be discerned in the PALS-B12 surfaces. However, it has the drawback that the left and right hemisphere surface meshes are not in geographic correspondence. To address this limitation, we used Landmark-SBR (with 55 landmarks!) to generate a hybrid ‘fs_LR’ atlas that preserves the shape characteristics of the fsaverage left and right hemispheres but are in precise geographic correspondence (Fig. 3D; Van Essen et al., 2011b).
Mapping data onto atlas surfaces
Surface-based atlases provide a spatial framework for comparing many types of experimental data within and across studies. One important data type involves parcellation schemes that represent the mosaic of distinct cortical areas delineated using architectonic, retinotopic, or other types of experimental data. Caret has been used extensively for this purpose ever since the Felleman and Van Essen (1991) parcellation was transferred from a manual flat map onto the macaque 79-0 atlas (Drury et al., 1996). Many additional macaque parcellations have subsequently been registered to the F99 atlas (Van Essen et al., 2001, 2004, 2011a), including probabilistic architectonic maps derived from a population of hemispheres.
One might hope that by now neuroscientists would have converged on a consensus parcellation for macaque cortical organization. No such luck - the problem remains very hard but hopefully not intractable! Of the numerous parcellations in widespread use, none can convincingly claim to represent ‘ground truth’, though some are on more solid ground than others. Caret provides several ways to view these parcellations and to compare different schemes. For example, Figure 4A shows a composite parcellation for the macaque that contains 129 distinct areas covering 90% of the neocortical surface (Van Essen et al., 2011a). Figure 4B shows the same areal boundaries, but with the Felleman and Van Essen (1991) parcellation painted on the surface. Many other parcellations concurrently loaded in this dataset can be interrogated by clicking on any surface node of interest. For example, selecting the highlighted node in Figure 4B yields the list of cortical areas and parcellation schemes shown in Figure 4C (TEa_m_LV00, PIDd_FV91, TEM_PHT00, etc.). Links to the publications associated with each of these areas can be specified using ‘study files’ that include the PubMedID plus other relevant study metadata.
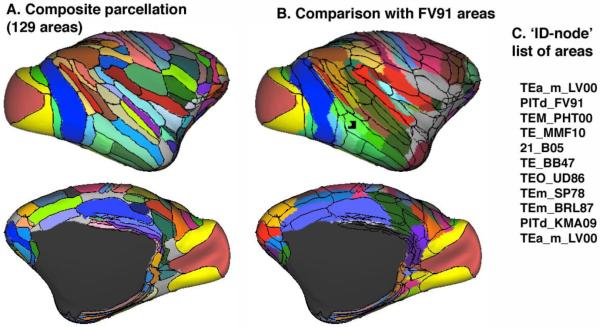
Figure 4. Macaque cortical parcellations.
A. A composite parcellation based on the Lewis & Van Essen (2000; LVE00), Paxinos et al. (2000; PHT00) and Ferry et al. (2000; FOA00) parcellations. B. The Felleman & Van Essen (1991; FV91) parcellation, with an overlay of the areal boundaries from panel A. C. A list of cortical areas associated with the highlighted node (arrow, TBD) from 12 concurrently loaded parcellations in Caret.
For human cortex, the classical Brodmann (1909) architectonic map, initially mapped to the Visible Man atlas surface (Drury et al., 1999), was subsequently transferred to the PALS-B12 and fs_LR atlases (Fig. 5A). Because many studies still refer to Brodmann areas, this parcellation remains useful even though it is inaccurate in a number of respects. More accurate parcellations for portions of human cortex are now available (Van Essen et al., 2011b), including a map of 52 areas based on architectonic and retinotopic analyses that were surface-registered from individuals to the fs_LR atlas (Fig. 5B). Additional options for displaying cortical parcellations include (i) probabilistic area 2 mapped via SBR and via VBR (Fig. 5C) and the centers of gravity of probabilistic areas computed in the volume and projected to the surface as ‘foci’ based on their stereotaxic coordinates (Fig. 5D). The recent demonstration that cortical ‘myelin maps’ can be obtained using the ratio of T1-weighted and T2-weighted images (Glasser and Van Essen, 2011) provides another useful modality for cortical parcelation (Fig. 5E).
Figure 5. Human cortical parcellations mapped to the fs_LR atlas (Van Essen et al., 2011b).
A. Brodmann (1909) map. B. Composite parcellation of cytoarchitectonic areas mapped by Fischl et al. (2008) and then to fs_LR. C. Probabilistic area 2 mapped by SBR (top) and VBR (bottom). D. Additional areas mapped using the centers of gravity of VBR-based parcellations. E. Myelin maps from a population average (Glasser and Van Essen, 2011).
Mapping fMRI data to atlas surfaces enables a variety of informative analyses, including comparisons across different fMRI studies as well as comparisons with the cortical parcellations illustrated in the preceding figures. Caret supports three distinct ways to map fMRI results to atlas surfaces: (i) projecting stereotaxic coordinates of activation foci; (ii) mapping volume-averaged activations (statistical parametric maps) to the atlas surface; and (iii) SBR from individual subjects to atlas surfaces. Each approach has its own advantages and limitations.
The foci-based approach was originally introduced in conjunction with the Visible Man atlas (Van Essen and Drury, 1997). Its attractiveness is based on the fact that many thousands of studies have reported the centers of fMRI activations or deactivations in published tables of stereotaxic coordinates. In recent years, a growing number of these coordinates have been systematically stored, along with key metadata, in databases. The BrainMap database (http://brainmap.org/), pioneered by Peter Fox and colleagues in San Antonio, currently contains 81,000 foci from 2100 studies. The SumsDB (‘Surface Management System) database (http://sumsdb.wustl.edu/sums/) was developed in my laboratory by James Dickson (Dickson et al., 2001) to provide a flexible and searchable repository for a wide variety of surface and volume neuroimaging data. More recently, it has been expanded in its data management and visualization capabilities by Ping Gu. It includes a stereotaxic Foci Library (http://sumsdb.wustl.edu/sums/searchload.do?dispatch=celldata) that currently contains more than 52,254 from 1,636 studies. Eleven foci in the Foci Library are tagged according to the stereotaxic space in which the data were originally analyzed, and each was projected to a version of the PALS-B12 atlas in the appropriate stereotaxic space (Van Essen, 2005; Van Essen and Dierker, 2007). This effectively brings all foci into a common surface-based spatial framework. Coordinate data in SumsDB can be searched online and viewed using WebCaret visualization interface, which has many of Caret’s capabilities but can be used without downloading software or experimental data. Search results of interest (or the entire Quick-Search Foci Library, as in Fig. 6A) can be downloaded for offline analysis in Caret. For example, Figure 6B shows foci related to processing of color, faces, and houses, extracted from the Foci Library and analyzed in Caret. To date, the time-consuming aspects of manual data entry have been rate-limiting for rapid growth of the Foci Library. The emergence of automated text mining tools that can extract stereotaxic coordinates and some associated metadata (Yarkoni et al., 2011) may help to alleviate this bottleneck.
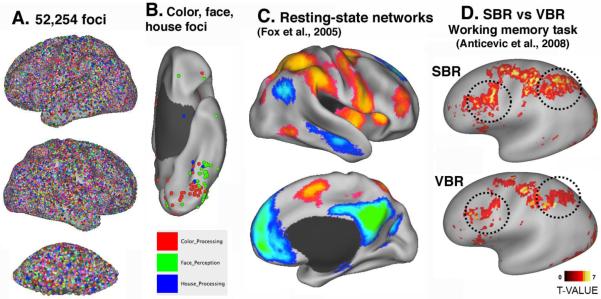
Figure 6. Multiple ways to map fMRI activations to PALS-B12 atlas surfaces in Caret.
A. 52,254 stereotaxic foci from 1,636 studies from the SumsDB Foci Library mapped to the inflated cerebral and cerebellar atlas surfaces. B. Foci specifically involved in processing of color (red), faces (green), or houses (blue) on a ventral view of the left hemisphere. C. Resting-state networks mapped to the atlas surface using average fiducial mapping (Fox et al., 2005). D. Working memory activations from 29 schizophrenia patients analyzed by Landmark-SBR (top) vs affine-registered VBR (adapted, with permission from Anticevic et al., 2008).
Figure 6C illustrates results from the first publication (Fox et al., 2005) that used the PALS-B12 atlas to visualize results from volume-to-surface mapping of fMRI data. It is also one of the most widely cited, in part because it was the first resting-state fMRI study to examine the default mode network using surface visualization. Mapping from population-average volumes to atlas surfaces has the advantage of providing much greater detail about the magnitude and spatial pattern of fMRI activations and other complex data types. However, the fidelity of the representation remains limited by whatever spatial smoothing and/or intersubject registration was done in the volume domain. Also, the results depend upon the whether data are mapped directly to the average midthickness (fiducial) atlas surface or indirectly, using midthickness surfaces of the 12 individual hemispheres contributing to the atlas as intermediaries that capture data from a larger volume (Van Essen, 2005).
Compensation for individual variability can be achieved more accurately using surface-based instead of volume-based registration (SBR vs. VBR), because VBR does not respect the topology of the cortical surface (Fischl et al. 2008; Anticevic et al., 2008). For example, Figure 6D shows results for a verbal working memory task in schizophrenia patients, where the group-average activations are more prominent in dorsolateral prefrontal and parietal cortex when analyzed by SBR (top panel) vs VBR (bottom panel). Registration of individuals to the PALS-B12 atlas originally required manual generation of landmarks, which was an impediment to high-throughput analyses. This limitation is now circumvented by an automated landmark identification (ALI) method (Anticevic et al., 2011) that is freely available.
It is often desirable to view single-subject data (e.g., fMRI activations) on both an atlas surface and on the individual hemisphere from which the data were acquired. Caret supports this capability using ‘standard-mesh’ surfaces that establish geographic correspondences between the atlas surface and the individual-subject surfaces (Saad et al., 2004), as does SUMA software (Saad manuscript, this issue of NeuroImage). Standard-mesh representations of an individual hemisphere (e.g., white, pial, and midthickness, and inflated configurations) look virtually identical to the native-mesh version, but they encode the point-to-point geographic correspondences established by the registration algorithm.
Interspecies comparisons
Human cortex is 9-fold greater in surface area than the macaque, but its evolutionary expansion has been far from uniform. Early sensory and motor regions have a similar functional organization in both species and have expanded only modestly. Regions of lateral temporal, parietal, and frontal cortex have expanded much more dramatically. Efforts to compare cortical organization objectively and quantitatively must deal not only with these large regional differences in cortical expansion but also major differences in the location of cortical areas relative to the convolutions. Landmark-SBR provides a powerful approach to monkey-human comparisons because it allows objective specification of putative homologies, using landmarks along the boundaries of corresponding areas or functional ROIs. Our early efforts along these lines (Denys et al., 2004; Orban et al., 2004; Van Essen and Dierker, 2007) used up to 23 landmarks based mainly on putative homologies of cortical areas identified by architectonics or other criteria. We also used landmarks delineated by fMRI results obtained using equivalent tasks in both species in order to constrain the registration in parietal and temporal cortex. Future interspecies comparisons will benefit from improvements in the Landmark-SBR algorithm based on the LVD (Landmark Vector Difference) method (Van Essen et al., 2011a) and in the availability of additional landmarks, such as those present in myelin maps (Glasser et al., 2011), that are likely to reflect genuine homologies.
A ‘scenic tour’
Many analyses carried out in Caret involve complex combinations of data overlaid on surfaces and volumes. It can take considerable time and effort to set up such a display when carrying out an analysis or when generating figures for a publication or a presentation. It is inherently inefficient to start from scratch the next time an investigator wants to regenerate the same or a similar display. One of Caret’s distinctive innovations is the introduction of ‘scenes’ that capture all of the metadata needed to recreate any given display, no matter how complex it may be. For example, the data for Figures 2 - 6 in this article can be effortlessly regenerated using the SumsDB URL in each figure legend (or in the SumsDB directory: http://sumsdb.wustl.edu/sums/directory.do?id=8287748&dir_name=CARET_NeuroImage12 and opening the scene of interest in WebCaret, or in Caret after downloading the relevant archive. More generally, scenes save time and facilitate communication between investigators.
Caret and Connectome Workbench
The emerging field of connectomics opens many new vistas for brain mapping. Diffusion imaging (dMRI) and tractography can provide estimates of structural connectivity from each gray matter location to every other gray matter location via fiber fascicles running through the white matter. Resting-state fMRI (R-fMRI) takes advantage of temporal correlations of fluctuations in the BOLD signal to generate maps of functional connectivity. However, the resultant connectivity-related datasets can be far larger than standard functional imaging datasets (e.g., maps of fMRI task-activations). For example, a ‘dense connectome’ file that represents connectivity between each gray matter location and all other locations at a spatial resolution of 3 mm may be ~10 GB in size. Effective utilization of such datasets requires novel types of analysis and visualization.
When Matt Glasser joined my lab in 2008 with interest and expertise in tractography, we began designing improvements to Caret for visualizing connectivity data. These efforts accelerated in 2009, when Matt, along with Tim Laumann and Alex Cohen, took part in (and emerged as co-winners of) the Pittsburgh Brain Connectivity Competition sponsored by Walter Schneider at OHBM. In mid-2009, NIH announced an RFA for the Human Connectome Project (HCP). Washington University teamed up with the University of Minnesota and seven other institutions to generate a proposal. For the grant submission, John Harwell implemented a prototype ‘point-and-click’ viewer of dense connectivity datasets.
Our ‘WU-Minn HCP Consortium’ was thrilled to receive NIH funding (http://humanconnectome.org/). This enabled us to recruit Jon Schindler and Tim Coalson to the programming team and to accelerate development of the Connectome Workbench visualization platform, which is based on Caret (Marcus et al., 2011). Connectome Workbench will include many new features, especially ones customized for connectivity analyses. One type of data visualization already implemented is a capability to view connectivity maps concurrently on surfaces and on subcortical gray matter using a new CIFTI data format (http://www.nitrc.org/projects/cifti/) that combines surface vertices and volume voxels in a unified file format. For example, Figure 7 illustrates resting-state functional connectivity maps for two nearby seed locations in parietal cortex of a single subject, displayed on a prototype of the Connectome Workbench.
Figure 7. Resting-state functional connectivity maps from an individual subject.
Upper panels show a functional connectivity map for seed location 1 in medial parietal cortex, computed by cross-correlation of cortical surface vertices and subcortical/cerebellar gray-matter voxels from a 20-minute resting-state scan. Data were stored using the CIFTI format that includes surface and gray-matter volume data in a single file and were displayed using a prototype of Connectome Workbench. Bottom panels show the functional connectivity map from a seed location (2) that is more lateral in parietal cortex and has a dramatically different pattern of functional connectivity. Unpublished data, courtesy M. Glasser.
Many additional capabilities are envisioned for Connectome Workbench, including visualization of probabilistic white matter fiber trajectories; maps of ‘parcellated’ as well as ‘dense’ connectome datasets’; and cross-modal visualization of data derived from structural connectivity, functional connectivity, task-fMRI, and MEG/EEG recordings in individuals or in population averages. Also, new features are planned for the Connectome Workbench user interface to enhance its user friendliness. Finally, the Workbench will be closely linked to the ConnectomeDB database to enable data mining and visualization of query results that relate connectivity to behavioral and other phenotypes obtained by the HCP.
Concluding remarks
Remarkable advances in brain mapping software have occurred over the past two decades, and a multitude of powerful platforms, including Caret, are widely used. Because there are many complementarities, it is increasingly important for investigators to be competent and comfortable in migrating data and analyses across platforms. Capabilities that are nowadays considered routine were beyond the scope of this author’s imagination not so many years ago. If the next two decades are anything like the previous two, our scientific grandchildren will operate in a brain mapping environment that makes our current capabilities seem distinctly quaint, much as today’s generation of Google-map and GPS navigators are bemused by the bulky paper maps that 20th century navigators relied on so heavily.
Acknowledgments
I thank Matt Glasser for comments on the manuscript and for sharing unpublished results, Susan Danker for assistance in ms preparation, E. Reid for technical assistance, J. Harwell, D. Dierker, P. Gu, T. Coalson, and J. Schindler for excellent software development. Supported by NIMH grant R01 MH 60974 and by the Human Connectome Project (1U54MH091657-01) from the 16 NIH Institutes and Centers that Support the NIH Blueprint for Neuroscience Research and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For Special Issue of NeuroImage (‘Twenty years of fMRI’)
References
- Anticevic A, Dierker DL, Gillespie SK, Repovs G, Csernansky JG, Van Essen DC, Barch DM. Comparing surface-based and volume-based analyses of functional neuroimaging data in patients with schizophrenia. Neuroimage. 2008;41:835–848. doi: 10.1016/j.neuroimage.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A, Repovs G, Dierker D, Harwell J, Coalson T, Barch DM, Van Essen DC. Automated landmark identification for human cortical surface-based registration. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.08.093. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. J.A. Barth; Leipzig: 1909. [Google Scholar]
- Carman GJ. Thesis. California Institute of Technology; 1990. Mappings of the cerebral cortex. [Google Scholar]
- Carman GJ, Drury HA, Van Essen D. Computational methods for reconstructing and unfolding the cerebral cortex. Cereb Cortex. 1995:5. doi: 10.1093/cercor/5.6.506. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno M.i. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: A linear approach. J Cogn Neurosci. 1993;5:162–170. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Daniel PM, Whitteridge D. The representation of the visual field on the cerebral cortex in monkeys. J Physiol. 1961;159:203–221. doi: 10.1113/jphysiol.1961.sp006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denys K, Vanduffel W, Fize D, Nelissen K, Sawamura H, Georgieva S, Vogels R, Van Essen D, Orban GA. Visual activation in prefrontal cortex is stronger in monkeys than in humans. J Cogn Neurosci. 2004;16:1505–1516. doi: 10.1162/0898929042568505. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Devlin JT, Poldrack RA. In praise of tedious anatomy. Neuroimage. 2007;37:1033–1041. doi: 10.1016/j.neuroimage.2006.09.055. discussion 1050-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson J, Drury H, Van Essen DC. ‘The surface management system’ (SuMS) database: a surface-based database to aid cortical surface reconstruction, visualization and analysis. Philos Trans R Soc Lond B Biol Sci. 2001;356:1277–1292. doi: 10.1098/rstb.2001.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury H, Van Essen D, Corbetta M, Snyder AZ. Surface-based analyses of the human cerebral cortex. In: Warping Brain, Toga A, et al., editors. Academic Press; 1999. pp. 337–363. [Google Scholar]
- Drury HA, Van Essen DC. Functional specializations in human cerebral cortex analyzed using the Visible Man surface-based atlas. Hum Brain Mapp. 1997;5:233–237. [PubMed] [Google Scholar]
- Drury HA, Van Essen DC, Anderson CH, Lee CW, Coogan TA, Lewis JW. Computerized mappings of the cerebral cortex: a multiresolution flattening method and a surface-based coordinate system. J Cogn Neurosci. 1996;8:1–28. doi: 10.1162/jocn.1996.8.1.1. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peterse TM. 3D statistical neuroanatomical models from 305 MRI volumes; Proc IEEE-Nuclear Science Symp and Med Imag Conf; 1993. [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Ferry AT, Ongur D, An X, Price JL. Prefrontal cortical projections to the striatum in macaque monkeys: evidence for an organization related to prefrontal networks. J Comp Neurol. 2000;425:447–470. doi: 10.1002/1096-9861(20000925)425:3<447::aid-cne9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BT, Mohlberg H, Amunts K, Zilles K. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Perlmutter JS, Raichle ME. A stereotactic method of anatomical localization for positron emission tomography. J Comput Assist Tomogr. 1985;9:141–153. doi: 10.1097/00004728-198501000-00025. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes CJ, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: A general linear approach. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- Gattass R, Gross CG. Visual topography of striate projection zone (MT) in posterior superior temporal sulcus of the macaque. J Neurophysiol. 1981;46:621–638. doi: 10.1152/jn.1981.46.3.621. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neuroscience. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Preuss TM, Snyder LH, Nair G, Rilling JR, Zhang X, Li L, Van Essen DC. Comparative mapping of cortical myelin content in humans, chimpanzees, and Macaques using T1-weighted and T2-weighted MRI. Soc. Neurosci. Abstract. 2011 in press. [Google Scholar]
- Han X, Pham DL, Tosun D, Rettmann ME, Xu C, Prince JL. CRUISE: cortical reconstruction using implicit surface evolution. Neuroimage. 2004;23:997–1012. doi: 10.1016/j.neuroimage.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Hill J, Dierker D, Neil J, Inder T, Knutsen A, Harwell J, Coalson T, Van Essen D. A surface-based analysis of hemispheric asymmetries and folding of cerebral cortex in term-born human infants. J Neurosci. 30:2268–2276. doi: 10.1523/JNEUROSCI.4682-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN. Laminar and columnar distribution of geniculo-cortical fibers in the macaque monkey. J Comp Neurol. 1972;146:421–450. doi: 10.1002/cne.901460402. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jones EG, Burton H. Areal differences in the laminar distribution of thalamic afferents in cortical fields of the insular, parietal and temporal regions of primates. J Comp Neurol. 1976;168:197–247. doi: 10.1002/cne.901680203. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R. An efficient algorithm for topologically correct segmentation of the cortical sheet in anatomical mr volumes. Neuroimage. 2001;14:329–346. doi: 10.1006/nimg.2001.0831. [DOI] [PubMed] [Google Scholar]
- Lewis JW, Van Essen DC. Mapping of architectonic subdivisions in the macaque monkey, with emphasis on parieto-occipital cortex. J Comp Neurol. 2000;428:79–111. doi: 10.1002/1096-9861(20001204)428:1<79::aid-cne7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Kabani N, Avis D, Evans AC. Automated 3-D extraction of inner and outer surfaces of cerebral cortex from MRI. Neuroimage. 2000;12:340–356. doi: 10.1006/nimg.1999.0534. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Harwell J, Olsen T, Hodge M, Glasser MF, Prior F, Jenkinson M, Laumann T, Curtiss SW, Van Essen DC. Informatics and data mining tools and strategies for the human connectome project. Front Neuroinform. 2011;5:4. doi: 10.3389/fninf.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. Neuroimage. 1997;6:156–167. doi: 10.1006/nimg.1997.0289. [DOI] [PubMed] [Google Scholar]
- Orban GA, Van Essen D, Vanduffel W. Comparative mapping of higher visual areas in monkeys and humans. Trends Cogn Sci. 2004;8:315–324. doi: 10.1016/j.tics.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Academic Press; 2000. [Google Scholar]
- Roland PE, Graufelds CJ, Wahlin J, Ingelman L, Andersson M, Ledberg A, Pedersen J, Akerman S, Dabringhaus A, Zilles K. Human Brain Atlas: For high resolution functional and anatomical mapping. Hum Brain Mapp. 1994;1:173–184. doi: 10.1002/hbm.460010303. [DOI] [PubMed] [Google Scholar]
- Roland PE, Zilles K. The developing European computerized human brain database for al imaging modalities. Neuroimage. 1996;4:S39–S47. doi: 10.1006/nimg.1996.0050. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall RC, Japee S, Cox RW. SUMA: an interface for surface-based intra- and inter-subject analysis with AFNI. Proc IEEE Int Symp Biomed Imag. 2004:1510–1513. [Google Scholar]
- Schwartz EL, Shaw A, Wolfson E. A numerical solution to the generalized mapmaker’s problem: Flattening nonconvex polyhedral surfaces. IEEE Trans Pattern Analysis Machine Intelligence. 1989;11:1005–1008. [Google Scholar]
- Talairach J, Szikla G. Atlas d’anatomie stereotaxicque du telencephale: etudes anatomo-radiologiques. Masson and Cie; Paris: 1967. [Google Scholar]
- Talairach J, Tournoux P. Coplanar Stereotaxic Atlas of the Human Brain. Thieme Medical; New York: 1988. [Google Scholar]
- Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Trans Med Imaging. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Surface-based atlases of cerebellar cortex in the human, macaque, and mouse. Ann N Y Acad Sci. 2002;978:468–479. doi: 10.1111/j.1749-6632.2002.tb07588.x. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr Opin Neurobiol. 2002a;12:574–579. doi: 10.1016/s0959-4388(02)00361-6. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Surface-based atlases of cerebellar cortex in the human, macaque, and mouse. Ann N Y Acad Sci. 2002b;978:468–479. doi: 10.1111/j.1749-6632.2002.tb07588.x. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Organization of visual areas in macaque and human cerebral cortex. In: Chalupa L, Werner J, editors. The Visual Neurosciences. 2004. pp. 507–521. [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA. Structural and functional analyses of human cerebral cortex using a surface-based atlas. J Neurosci. 1997;17:7079–7102. doi: 10.1523/JNEUROSCI.17-18-07079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Joshi S, Miller MI. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. Proc Natl Acad Sci U S A. 1998;95:788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker D, Harwell J. Cortical parcellations of the Macaque monkey analyzed on surface-based atlases. Cereb Cortex. 2011a doi: 10.1093/cercor/bhr290. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker D, Harwell J, Coalson T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex. 2011 doi: 10.1093/cercor/bhr291. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Maunsell JH. Two-dimensional maps of the cerebral cortex. J Comp Neurol. 1980;191:255–281. doi: 10.1002/cne.901910208. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Zeki SM. The topographic organization of rhesus monkey prestriate cortex. J Physiol. 1978;277:193–226. doi: 10.1113/jphysiol.1978.sp012269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson E, Schwartz EL. Computing minimal distances on polyhedral surfaces. IEEE Trans Pattern Analysis Machine Intelligence. 1989;11:1005–1008. [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]