Abstract
MUC16/CA125 is a tumor marker currently used in clinics for the follow-up of patients with ovarian cancer. However, MUC16 expression is not entirely restricted to ovarian malignancies and has been reported in other cancers including breast cancer. Although it is well established as a biomarker, function of MUC16 in cancer remains to be elucidated. In the present study, we investigated the role of MUC16 in breast cancer and its underlying mechanisms. Interestingly, our results showed that MUC16 is overexpressed in breast cancer tissues whereas not expressed in non-neoplastic ducts. Further, stable knockdown of MUC16 in breast cancer cells (MDA MB 231 and HBL100) resulted in significant decrease in the rate of cell growth, tumorigenicity and increased apoptosis. In search of a mechanism for breast cancer cell proliferation we found that MUC16 interacts with the ezrin/radixin/moesin domain-containing protein of Janus kinase (JAK2) as demonstrated by the reciprocal immunoprecipitation method. These interactions mediate phosphorylation of STAT3 (Tyr705), which might be a potential mechanism for MUC16-induced proliferation of breast cancer cells by a subsequent co-transactivation of transcription factor c-Jun. Furthermore, silencing of MUC16 induced G2/M arrest in breast cancer cells through downregulation of Cyclin B1 and decreased phosphorylation of Aurora kinase A. This in turn led to enhanced apoptosis in the MUC16-knockdown breast cancer cells through Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated extrinsic apoptotic pathway with the help of c-Jun N-terminal kinase signaling. Collectively, our results suggest that MUC16 has a dual role in breast cancer cell proliferation by interacting with JAK2 and by inhibiting the apoptotic process through downregulation of TRAIL.
Keywords: MUC16, JNK, Aurora kinase, Cyclin B1, G2/M arrest and breast cancer
Introduction
Mucins are high molecular weight glycoproteins that are synthesized by epithelial cells and serve several functions that include lubrication, cell signaling and formation of chemical barriers. Aberrant expression of mucins has been reported to promote cancer development, and influence cellular growth, differentiation, transformation, adhesion, invasion and immune surveillance (Kufe, 2009). The functions of MUC1 and MUC4 membrane-bound mucins are well established in different cancers (Singh et al., 2007; Bafna et al., 2008; Chaturvedi et al., 2008; Munro et al., 2009; Ponnusamy et al., 2010). In breast cancer, many mucins such as MUC1, MUC3 and MUC4 are overexpressed (Rakha et al., 2005), but they do not have a strong relationship with patient outcome with the exception of MUC1.
Cancer antigen 125 (CA125/MUC16) is a blood biomarker routinely used to monitor the progression of epithelial ovarian cancers and is encoded by the MUC16 mucin gene (Bast et al., 1998; Yin et al., 2002). MUC16 expression is also observed in other cancers such as pancreatic (Wu et al., 2009) and breast cancer (Moritani et al., 2008). However, the functional role(s) of MUC16 in breast cancer progression are not well understood. MUC16 is a 20–25MDa molecule with 22 152 amino acids in its protein sequence (O’Brien et al., 2002). The N-terminus of MUC16 comprises of a heavily O-glycosylated, non-tandem repeat domain of ~12 000 amino acids. The tandem repeat region adjacent to the N-terminus is composed of 60 repeats of 156 amino acids and 56 sea urchin sperm protein, enterokinase and agrin domains, while the carboxyl terminus contains a 32-residue cytoplasmic tail (Hattrup and Gendler, 2008). Recent reports have demonstrated that a polybasic amino acid sequence (RRRKK) in the cytoplasmic tail of MUC16 interacts with the ezrin/radixin/moesin (ERM) family of proteins (Blalock et al., 2007). Janus kinases (JAKs) are non-receptor tyrosine kinases (Firmbach-Kraft et al., 1990) and their amino terminus contains an SH2-like domain (JH3–JH4) and a ERM (4.1/ezrin/radixin/moesin) domain (JH6–JH7). The ERM domain of JAK proteins has been shown to be important in mediating its interaction with transmembrane proteins (Velazquez et al., 1992; Huang et al., 2001). A report from a previous study has also demonstrated that the ERM domain of JAK2 is required for receptor binding and regulating kinase activity of JAK2 at different activation states for downstream STAT signaling pathways (Harrison et al., 1995; Chen et al., 1997; Zhao et al., 2010).
In the present study, we demonstrate that MUC16 expression is upregulated in breast cancer tissues and correlates with the stage of the disease. Furthermore, MUC16 interacts with JAK2 and mediates breast cancer cell proliferation through rapid G2/M transition process. In addition, MUC16 also suppresses Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through c-Jun N-terminal kinase (JNK) phosphorylation.
Results
MUC16 is overexpressed in breast carcinoma
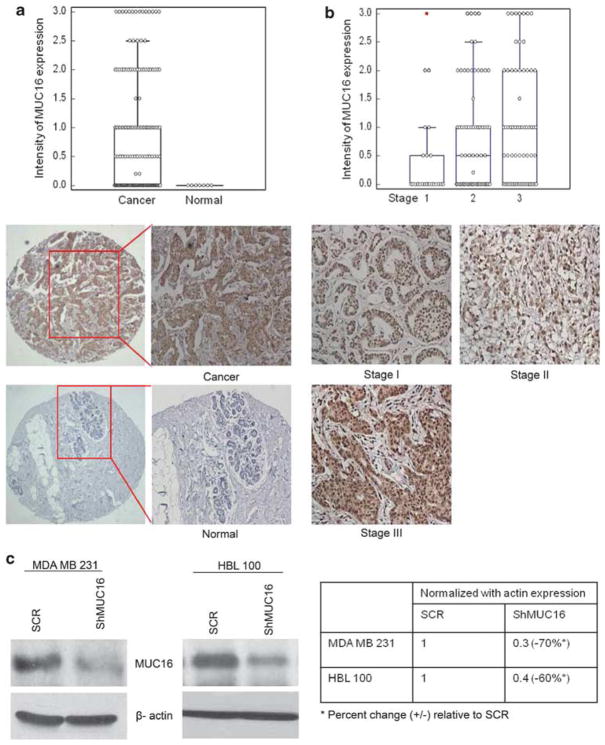
In order to investigate the expression of MUC16 during the progression of breast cancer, we analyzed MUC16 expression in breast cancer tissues by immunohistochemistry. A total of 178 tissue spots were examined, comprising 171 cases of invasive adenocarcinoma and 7 normal breast tissues. None of the normal tissues (0/7, 0%) expressed MUC16, whereas 93/171 (54%) ductal carcinomas were positive for MUC16. Thus, MUC16 expression was more common in breast cancer tissues compared with the normal ducts (P=0.0078 by χ2 test) (Figure 1a). Among the cases of invasive breast cancer, 23 were stage 1 (13.2%), 70 stage 2 (40%), 67 stage 3 (39%) and 2 stage 4 cases (1.2%). For nine cases, no information was available regarding the stage of the disease. There was a progressive and significant increase in MUC16 expression with advancing stage (mean MUC16 intensity being 0.5±0.1, 0.8±0.1 and 1.0±0.1 in stage 1, 2 and 3, respectively, P=0.03) (Figure 1b). Owing to the availability of only two cases of stage 4 disease, they were excluded from the analysis.
Figure 1.
Expression of MUC16 in breast cancer tissues and non-neoplastic breast tissues. Immunohistochemical analysis was carried out with CA125 antibody in breast cancer tissue. (a) Shows high immunoreactivity for MUC16 was observed in breast cancer tissues compared with non-neoplastic breast tissues. (b) Shows the comparison of MUC16 expression between various stages of breast carcinoma. Stable knockdown of MUC16 in two different breast cancer cell lines MDA MB 231 and HBL 100. Endogenously expressed MUC16 was stably knocked down using a MUC16 shRNA construct (pSUPER-Retro-MUC16-sh) in highly aggressive MDA MB 231 and HBL 100 breast cancer cells. (c) Western blot analysis confirms the decreased expression of MUC16 in MDA MB 231 and HBL 100 cells that were stably transfected with the MUC16 shRNA construct in comparison with empty-vector control cells.
MUC16 knockdown in aggressive breast cancer cells (MDA MB 231 and HBL100)
Having demonstrated an overexpression of MUC16 in breast cancer tissues, we next sought to determine the functional role of MUC16 in breast cancer cells. We screened a panel of breast cancer cells for MUC16 expression, in which we found strong expression of MUC16 in two highly aggressive cell lines such as MDA MB 231 and HBL 100 (Supplementary Figure 1A). In this study, MDA MB 231 and HBL 100 were chosen for MUC16 knockdown by RNAi method and it was confirmed by immunoblot and immunofluorescence analysis (Figure 1c & Supplementary Figure 1B). The results reveal a 70–80% knockdown of MUC16 in both the cell lines compared with the scrambled RNAi-transfected cells. These MUC16-knockdown and control cells (henceforth referred to as ShMUC16 and SCR, respectively) were subsequently used in functional assays to investigate the role of MUC16 in breast cancer.
Expression of MUC16 affects growth characteristics of breast cancer cells
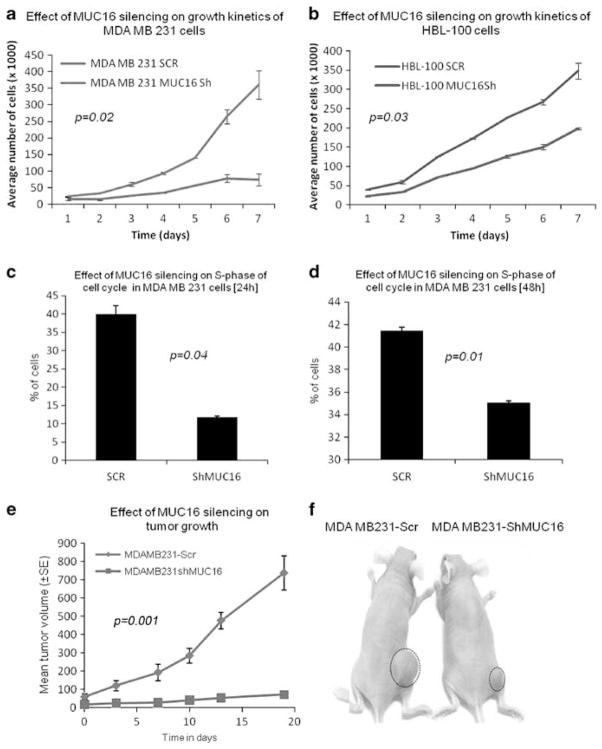
To investigate whether MUC16 influences the growth of breast cancer cells, MDA MB 231 and HBL 100–ShMUC16 and SCR cells were seeded at low density (1 × 104/well in a six-well plate) and growth rates determined by counting the number of viable cells for 7 days. The MDA MB 231 and HBL 100–ShMUC16 cells exhibited a significant decrease (P=0.02 and P=0.03, respectively) in the rate of proliferation compared to the SCR cells (Figures 2a and b). This suggests that MUC16 may have a significant role in the growth of breast cancer cells. In addition, cell cycle analysis (double thymidine block method) was performed to investigate the role of MUC16 in proliferation of breast cancer cells. The results show that there is a significant reduction in the percentage of cells in the S phase of MDA MB 231–ShMUC16 cells at 24 h (P=0.04) and 48 h (P=0.01) (Figures 2c and d) and at 48 h HBL 100 (P=0.02) (Supplementary Figure 1C) cells compared with SCR cells. These results suggest that MUC16 may have a crucial role in the S phase of cell cycle progression in breast cancer.
Figure 2.
MUC16 enhances breast cancer proliferation. Growth kinetic assays show that growth rate is significantly lower in MUC16-knockdown MDA MB 231 (P=0.02) (a) and HBL 100 (P=0.03) cells than in control cells (b). Effect of MUC16 on distribution of MDA MB 231 and HBL 100 cells in the S phase of the cell cycle. MUC16-knockdown and control cells of MDA MB 231 were stained with propidium iodide and analyzed by flow cytometry. The percentage of MUC16-knockdown cells is drastically decreased in the S phase of cell cycle than SCR cells at two different time point (c, d). (e, f) Shows that tumor formation is greatly decreased in MUC16-knockdown cells than in control cells (P=0.001).
MUC16 induces tumorigenicity in breast cancer cells
To assess the effect of MUC16 on the tumorigenicity of breast cancer cells, MUC16-knockdown MDA MB 231 and control cells were subcutaneously injected into immunodeficient female nude mice (2 × 106 cells/mouse). A palpable mass was first noticed 25 days post inoculation and monitored up to 45 days after inoculation. The tumor size was measured on alternate days. The mean tumor volume was significantly lower in mice injected with the MUC16-knockdown MDA MB 231 cells compared to mice injected with scramble-transfected cells (P=0.001) (Figure 2e and f). The results of this in vivo experiment strongly suggest that MUC16 enhances the tumorigenic potential of breast cancer cells.
Interaction of MUC16 with JAK2 mediates STAT3 phosphorylation
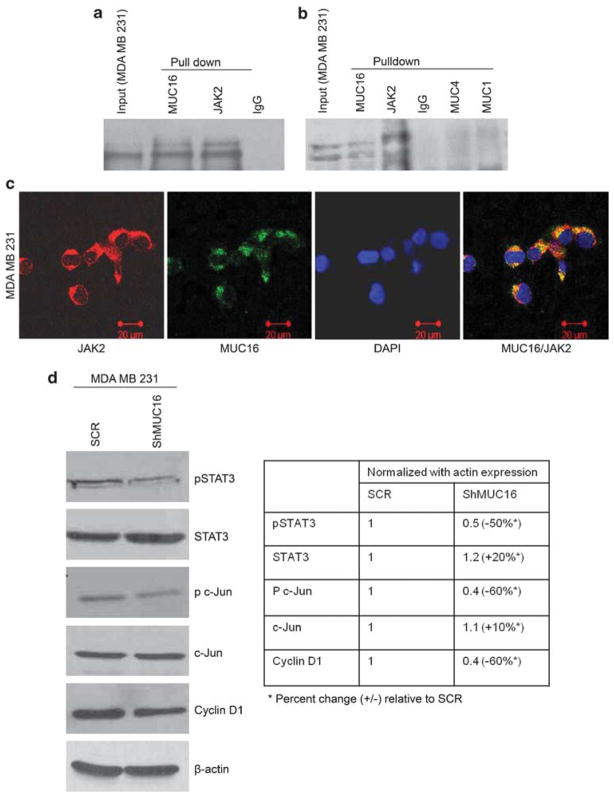
Having observed decreased proliferation in MUC16-knockdown cells, we next sought to determine the mechanism underlying MUC16-mediated cell proliferation. MUC16 contains a 32-amino-acid cytoplasmic tail with several tyrosine, serine and threonine residues that could serve as potential sites for phosphorylation (O’Brien et al., 2002). Recent reports have demonstrated that MUC16 interacts with the ERM family of proteins to interlink the actin cytoskeleton (Blalock et al., 2007). ERM-containing proteins include Band 4.1, PTP NE, FAK and JAK. JAKs are non-receptor tyrosine kinases, which contain a ERM domain required for mediating interactions with transmembrane proteins (Velazquez et al., 1992; Huang et al., 2001). Based on this rationale, we hypothesized that MUC16 interacts with JAK2 to mediate breast cancer cell proliferation. We have analyzed MUC16 interaction with JAK2 in breast cancer cells by reciprocal co-immunoprecipitation assay (Figure 3a). In addition, MUC16 also colocalized with JAK2 in MDA MB 231 cells (Figure 3c), suggesting that MUC16 interacts with JAK2 in breast cancer cells (possibly through ERM domain). Besides, we also checked whether JAK2 interacts with other mucins such as MUC1 and MUC4. The result shows that JAK2 does not interact with either MUC1 or MUC4, which indicates that JAK2 potentially interacts with MUC16 in breast cancer cells (Figure 3b). STAT3 is one of the downstream effectors of the JAK2 signaling pathway. Hence, we aimed to compare the degree of STAT3 activation in MUC16-expressing and -knock-down cells. The activated form of STAT3 (Tyr705) was significantly decreased in the MUC16-knock-down cells, MDA MB 231 (Figure 3d) and HBL 100 (Supplementary Figure 2A) when compared with the control cells. Taken together, our results suggest that MUC16 interacts with JAK2, which mediates phosphorylation and activation of STAT3, thus inducing the downstream signaling of MUC16. The activation of STAT is primarily regulated by JAK2 kinase, which is induced by cell surface receptor (Watson, 2001; Levy and Darnell, 2002; Calo et al., 2003). Usually, STAT molecules participate in various biological processes namely differentiation, proliferation, survival and apoptosis, in addition, STAT3 is implicated in breast cancer development (Watson, 2001; Clevenger, 2004).
Figure 3.
MUC16 interacts with JAK2 in breast cancer cells. On performing co-immunoprecipitation, a strong interaction between MUC16 and JAK2 in MDA MB 231 cells was observed (a, b). Co-immunolocalization was also performed and a strong binding affinity between MUC16 and JAK2 was observed (c). MUC16 interacts with JAK2 and mediates breast cancer proliferation via STAT3, c-Jun and Cyclin D1. Western blot analysis confirms that MUC16 interacts with JAK2 and mediates phosphorylation of STAT3 and c-Jun, which enables proliferation of breast cancer cells via Cylin D1 activation (d).
MUC16 promotes breast cancer cell proliferation by interacting with JAK2 and activating STAT3 and c-Jun
The transcription factor c-Jun acts along with STAT3 to induce tumorigenesis in many different types of cancer cells (Johnson et al., 1996). We observed a significant decrease in the phosphorylation of c-Jun in MDA MB 231–ShMUC16 (Figure 3d) and HBL 100–ShMUC16 (Supplementary Figure 2A) cells compared with the SCR cells, while the levels of total c-Jun protein remained unchanged. c-Jun has also been suggested to drive the cell cycle directly by transcriptionally upregulating Cyclin D1 through the transcription factor AP-1 (Li et al., 2003; Zenz et al., 2003). In our results, Cyclin D1 expression was significantly decreased in the MDA MB 231–ShMUC16 (Figure 3d) and HBL 100–ShMUC16 (Supplementary Figure 2A) cells compared with SCR cells. This suggests that MUC16 is involved in the phosphorylation of c-Jun to stimulate the proliferation of breast cancer cells by upregulating of Cyclin D1. Our confocal microscopy also revealed a significant downregulation of Cyclin D1, Cyclin A and Cyclin E in the MDA MB 231–ShMUC16 cells than the SCR cells (Supplementary Figure 2B). In addition, we observed that p21 is highly localized in the cytoplasmic region of MUC16-expressing (that is, SCR) MDA MB 231 cells as compared with the MDA MB 231–ShMUC16 cells (Supplementary Figure 2B). p21 is a cell cycle inhibitor, which blocks the kinase activity of the cyclin/cyclin-dependent kinase complex in response to DNA damage (Coqueret, 2003), and an elevated level of p21 in the cytoplasm has been shown to promote cellular transformation and tumor progression (Tanaka et al., 2002; Lee and Helfman, 2004). Thus, these results suggest that MUC16 also regulates p21 activity during breast cancer cell proliferation.
MUC16 promotes breast cancer cell proliferation via G2/M transition process
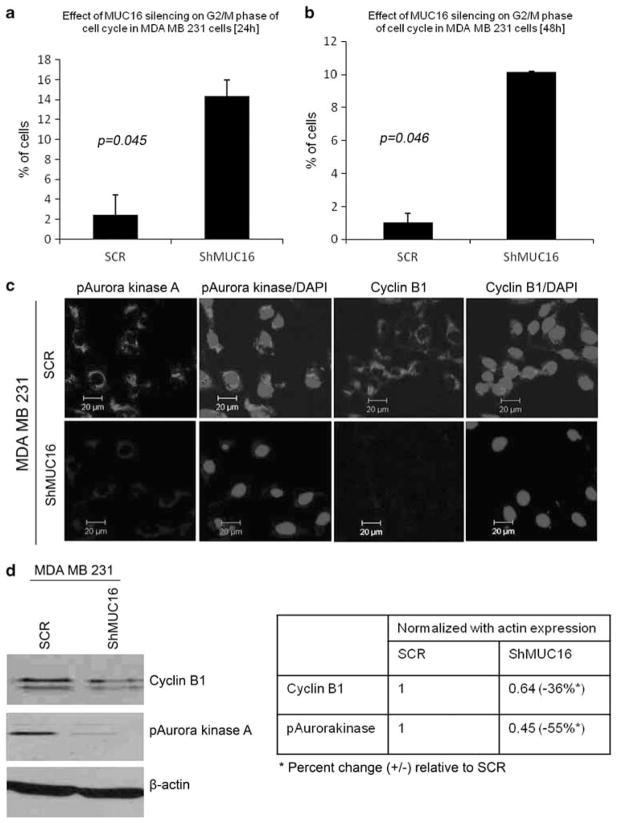
Cell cycle data showed that a large number of MUC16-knockdown cells were accumulated in G2/M phase of the cell cycle at two different time points such as 24 h (P=0.045) and 48 h of MDA MB 231 (P=0.046) (Figures 4a and b) and at 48 h of HBL 100 (P=0.04) (Supplementary Figure 3A). This indicates that the expression of MUC16 promotes rapid G2/M transition and thus drives breast cancer cell proliferation. This conclusion is further supported by the decrease in Cyclin B1 expression and Aurora kinase A phosphorylation (key G2/M checkpoint regulatory proteins) in ShMUC16 compared with the SCR cells by both confocal (Figure 4c) and western blot analysis (Figure 4d and Supplementary Figure 3B).
Figure 4.
Involvement of MUC16 in G2/M transition of breast cancer cells. The cell cycle analysis demonstrated that rapid G2/M transition was observed in MUC16-expressing cells than MUC16-knockdown MDA MB 231 (a, b). Cyclin B1 and phospho-Aurora kinase A are highly distributed in MUC16-expressing MDA MB 231 cells in comparison with MUC16-knockdown cells as observed by confocal analysis (c). Furthermore, western blot results show that levels of Cyclin B1 and phospho-Aurora kinase A are significantly reduced in MUC16-knockdown cells compared with control cells (d).
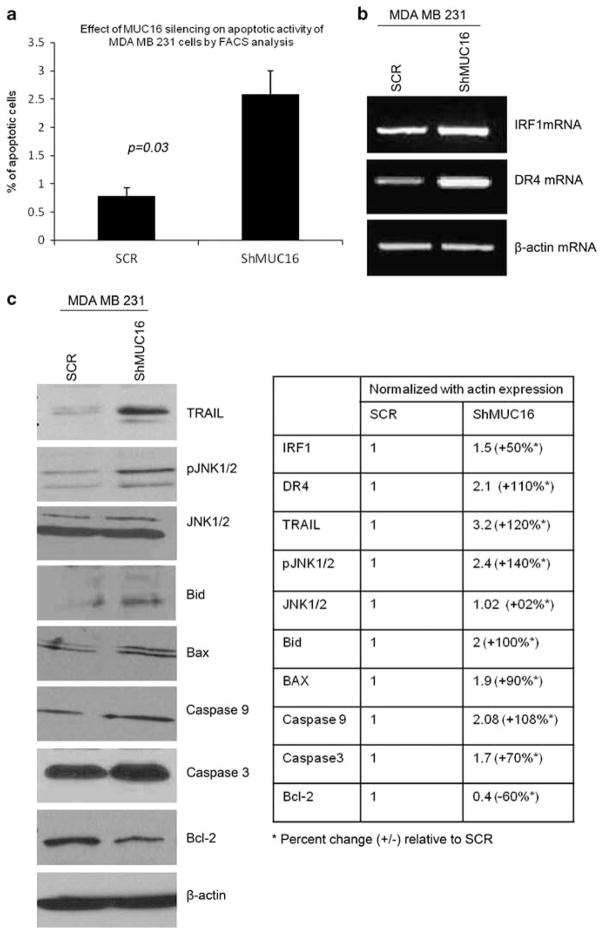
MUC16 suppresses TRAIL-mediated apoptosis in breast cancer cells
Our previous result showed that silencing of MUC16 led to an accumulation of breast cancer cells in the G2/M phase. Cell cycle control is the major mechanism to regulate cell growth (Hartwell and Weinert, 1989; Sherr, 1996) and cell cycle arrest at G2/M phase leads to apoptosis of the cells (Orren et al., 1997; Fujimoto et al., 1999; Gamet-Payrastre et al., 2000). We also observed a significant increase in the percentage of apoptotic cells in the MUC16-knockdown MDA MB 231 (P=0.03) and HBL 100 (P=0.04)) compared with SCR cells (Figure 5a and Supplementary Figure 3C). Micro array data have shown that TRAIL was significantly upregulated (3.1-fold, P=0.001) in the MDA MB 231–ShMUC16 vs SCR cells. TRAIL (or Apo2 ligand) is a member of the TNF cytokine family whose members induce apoptosis. Interestingly, TRAIL induces apoptosis in a variety of cancer cells but has no cytotoxic effect in normal cells (Chinnaiyan et al., 2000; LeBlanc and Ashkenazi, 2003). One of the recent reports suggested that TRAIL preferentially induced apoptosis in triple negative breast cancer cells (Rahman et al., 2009). We observed a significant upregulation of TRAIL expression in the MDA MB 231–ShMUC16 cells (Figure 5c). The expression of TRAIL is positively regulated by the transcription factor interferon regulatory factor 1 (Huang et al., 2009). We also observed a significant upregulation in interferon regulatory factor 1 mRNA levels in the MDA MB 231–ShMUC16 cells as compared with the SCR cells (Figure 5b). Taken together, these results indicate that MUC16 mediates suppression of TRAIL gene expression through interferon regulatory factor 1, which is the underlying mechanism for the pro-survival function of MUC16 in breast cancer cells.
Figure 5.
Enhanced apoptosis in MUC16-knockdown breast cancer cells. Fluorescence-activated cell sorting analysis reveals that the rate of apoptosis is significantly increased in MUC16-knockdown breast cancer cells compared with the control cells (a). (b) shows that when MUC16 is low, interferon regulatory factor 1 and Death receptor 4 mRNA expression increased for execution of extrinsic apoptotic activity in breast cancer cells. (c) A significant increase in TRAIL, pro-apoptotic molecules such as Bid and Bax upregulated in MDA MB 231–ShMUC16 than SCR cells, and furthermore, phosphorylation of JNK1/2 and activities of caspase 3 and 9 in MUC16-knockdown breast cancer cells, as observed by western blot analysis. Levels of anti-apoptotic molecule Bcl-2 are decreased in MUC16-knockdown breast cancer cells in comparison with the control cells (c).
Absence of MUC16 induces TRAIL-mediated apoptosis in breast cancer cells
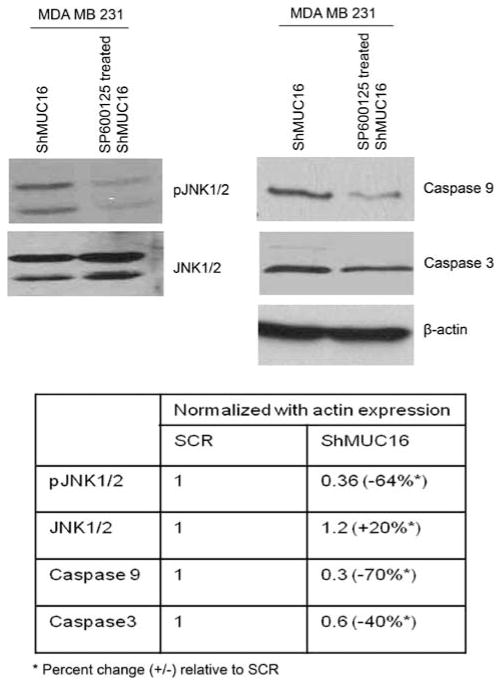
In addition to the TRAIL gene upregulation, the MUC16 knockdown also upregulated various apoptosis-inducing molecules such as Bid (Figure 5c) and death receptor 4 (DR4) (Figure 5b). These findings suggest that downregulation of MUC16 activates the extrinsic apoptotic pathway (Kischkel et al., 2000). We also observed an upregulation of the pro-apoptotic Bcl-2 family member Bax and downregulation of anti-apoptotic molecule such as Bcl-2 in the MUC16-knockdown cells (Figure 5c). The upregulation of these proteins suggests that TRAIL mediates the activation of the extrinsic apoptotic pathway of apoptosis. Proapoptotic molecules of Bid serve to link the extrinsic (mediated by the DR) and intrinsic apoptotic (by mitochondrial) pathways (Wang et al., 1996), which then activates Bax to translocate to the mitochondria followed by release of cytochrome C to execute the apoptotic process (Eskes et al., 2000; Munoz-Pinedo et al., 2006). These results indicate that in the absence of MUC16, TRAIL gets upregulated, which in turn induces apoptosis by binding to DR4 in breast cancer cells (which is facilitated through Bid activation). Furthermore, we observed an increased activity of JNK, initiator caspase (caspase 9) and effector caspase (caspase 3) in the MDA MB 231–ShMUC16 cells compared with the SCR cells (Figure 5c). To investigate whether the JNK pathway is also required for apoptosis in MDA MB 231–ShMUC16 cells, we measured apoptosis (through caspase 3 and 9 expression) in the absence or presence of the JNK1/2 inhibitor SP600125 (Bennett et al., 2001). Treatment with the inhibitor led to a significant decrease in phosphorylation of JNK1/2, which is associated with a significant reduction in caspase 3 and 9 expression (Figure 6). These results suggest that MUC16 suppresses apoptosis in MDA MB 231 cells through inhibition of JNK signaling. These results also suggest that JNK signaling is required for TRAIL-mediated apoptosis in breast cancer cells in the absence of MUC16.
Figure 6.
Inhibition of JNK promotes apoptosis in breast cancer cells. The cells were treated with pharmacological JNK1/2 inhibitor SP600125 (40 nm), which resulted in a significant decrease of caspase 3 and 9 activities.
Discussion
MUC16 is overexpressed in epithelial ovarian cancer and its expression is also observed in several other malignancies including breast cancer (Moritani et al., 2008). MUC16, originally isolated from ovarian tumor cells, was known as the ovarian tumor cell marker CA125 before its cloning and protein characterization (Perez and Gipson, 2008). CA125/MUC16 is considered to be one of the best available serum markers for epithelial ovarian cancer; however, the functional role of MUC16/CA125 in cancer progression remains largely unknown. In this study, we observed that the expression of MUC16 was significantly higher in breast cancer tissues when compared to non-neoplastic breast tissues. The results of the immunohistochemical analysis suggest that MUC16 is overexpressed in breast cancer and may have a potential role in the development and/or progression of breast cancer. An interesting finding showed that stable silencing of endogenous MUC16 in breast cancer cells resulted in a significant decrease in their growth rates. Further analyses revealed that majority of the MUC16-knockdown cells were arrested in the G2/M phase of the cell cycle. Thus, suggesting that MUC16 has an important role in promoting proliferation in breast cancer cells.
We then examined the mechanism by which MUC16 regulates breast cancer cell proliferation. The cytoplasmic tail of MUC16 has several potential phosphorylation sites including three tyrosines and one serine residue (Fendrick et al., 1997; O’Brien et al., 2002). JAKs are non-receptor tyrosine kinases containing the ERM domain, which mediates the interaction of JAKs with transmembrane proteins. One of the important observations of the present study is the interaction of MUC16 with JAK2 in breast cancer cells. JAK2 leads to phosphorylation of STAT proteins, specifically STAT3; the active form of STAT3 has been identified in many human cancers and appears to be required for continued cellular growth (Yu et al., 2009). STAT3 phosphorylation induces its homodimerization, leading to its nuclear translocation, DNA binding and subsequent activation of gene transcription (Yu et al., 2009; Yue and Turkson, 2009), and also coiled-coil domain of STAT3 has been demonstrated to interact with the transcription factor c-Jun. c-Jun in turn is required for the transcription of STAT3 target genes (Ginsberg et al., 2007), which is required for cellular growth. In our study, the expression of active c-Jun was significantly reduced in the MUC16-knockdown cells. We also observed that Cyclin D1, target for c-Jun, was significantly downregulated in MDA MB 231 and HBL 100–ShMUC16 cells, further strengthening the proposed mechanism. Thus these results suggest that, MUC16 interacts with JAK2 to induce phosphorylation of STAT3, which in turn transactivates c-Jun to induce Cyclin D1 expression and thereby promotes breast cancer cell proliferation.
Moreover, p21 is a cell cycle inhibitor, which blocks the kinase activity of the cyclin/cyclin-dependent kinase complexes in response to DNA damage (Coqueret, 2003). An elevated level of p21 in the cytoplasm has been shown to promote cellular transformation and tumor progression (Tanaka et al., 2002; Lee and Helfman, 2004). We observed a decrease in cytosolic p21 expression in the MUC16-knockdown cells suggesting an additional mechanism for breast cancer cell proliferation.
Cell cycle analysis revealed that MUC16-knockdown cells were being arrested in the G2/M phase of the cell cycle. Concomitantly, we also observed a downregulation of Cyclin B1 and the active form of Aurora kinase A in these cells. Aurora kinase A is a member of the Ser/Thr kinase family, and serves to promote G2/M transition and is therefore committed to mitosis (Marumoto et al., 2002; Hirota et al., 2003). Similarly, activity of Cyclin B1 is highly observed in G2/M phase and has an essential role in G2/M transition process (Tong and Pollard, 1999; Korgun et al., 2006). As the G2/M transition is controlled by the activities of Aurora kinase A (Ouchi et al., 2004; He et al., 2008) and Cyclin B1 (Jin et al., 1998), our findings suggest this as an additional mechanism by which MUC16 regulates cell proliferation by promoting rapid G2/M transition. Cell cycle checkpoints are interconnected with the apoptotic process. Here, we observed a significant increase in the percentage of apoptotic cells upon knockdown of MUC16. This together with the G2/M arrest in the ShMUC16 cells suggest that expression of MUC16 promotes breast cancer cell growth by a dual effect of increasing proliferation and decreasing apoptosis. There are two types of apoptosis signaling pathways such as the extrinsic (mediated via DR) and the intrinsic pathway (mediated via the mitochondria). TRAIL is a DR ligand, which on binding to DR4 or DR5 stimulates their trimerization and induces subsequent changes in the conformation of the intracellular death domain of the receptor. This leads to the downstream signaling pathway ultimately leading to apoptosis (Kischkel et al., 2000). When MUC16 levels are low, there is an elevated expression of TRAIL. This enables it to bind with DR4, which then induces apoptosis via activation of Bid. In support of this mechanism, we also noted a concomitant decrease in the levels of the anti-apoptotic protein Bcl-2 and an increase in the levels of Bax, caspase 9, caspase 3 and the phosphorylated form of JNK. JNK activation is required for either cell survival or cell death, which is dependent on the cell type (Bode and Dong, 2007; Lin et al., 2007). When we blocked JNK signaling in MUC16-knockdown cells, expression levels of caspase 3 and caspase 9 were significantly reduced, suggesting that JNK signaling is required for MUC16-mediated apoptosis in breast cancer cells, particularly TRAIL-mediated apoptosis in MUC16-knockdown cells.
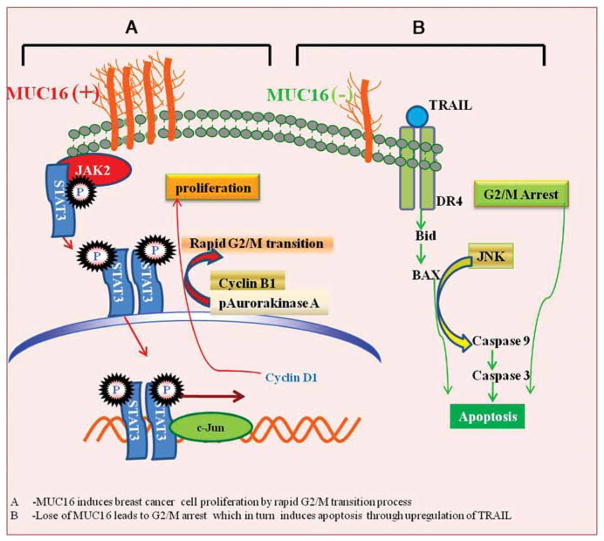
In conclusion, our results indicate that MUC16, although not expressed in the non-neoplastic ducts, is overexpressed in breast cancer tissues and its expression correlates positively with the stage of the disease. Data from our functional studies demonstrated that MUC16 promotes breast cancer cell proliferation and tumor development. Further, MUC16 interacts with JAK2 and mediates STAT3 and c-Jun phosphorylation, which in turn promotes breast cancer cell proliferation via Cyclin D1 upregulation (Figure 7). MUC16 also regulates breast cancer proliferation by promoting rapid G2/M transition processes. In addition, MUC16 inhibits apoptosis in breast cancer cells by inhibiting TRAIL-mediated extrinsic apoptotic signaling (Figure 7). Overall, our results suggest that MUC16 has a dual role in breast cancer cell proliferation by interacting with JAK2 and inhibiting the apoptotic process through downregulation of TRAIL.
Figure 7.
A schematic diagram of MUC16 involved in the process of breast cancer proliferation. Expression of MUC16 (indicated by +) induces breast cancer cell proliferation via its interaction with the non-receptor tyrosine kinase JAK2 and this interaction mediates phosphorylation of transcription factor STAT3, which may transactivate c-Jun for Cyclin D1 expression. MUC16 also regulates G2/M transition process through Cyclin B1 and phosphorylation of Aurora kinase A. Decreased MUC16 expression (indicated by −) results in an accumulation of breast cancer cells at the G2/M phase of the cell cycle, which in turn leads to apoptosis of breast cancer cells through JNK signaling.
Materials and methods
Cell culture and transfection
MDA MB 231 and HBL100 cells were procured from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and antibiotics. The cultures were incubated in a humidified atmosphere at 37 °C with 5% CO2. Endogenously expressed MUC16 was stably knocked down using a MUC16 shRNA construct (pSUPER-Retro-MUC16-sh—provided by Dr Ilene K Gipson from Harvard Medical School) in MDA MB 231 and HBL 100 breast cancer cells by stable transfection method. Using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), the MUC16 shRNA and scramble vector were transfected into phoenix cells, a packaging cell line that produces high-titer retrovirus in culture. MDA MB 231 and HBL 100 cells were then seeded in 24-well plates at 2 × 104 cells per well and grown to 40% confluence in serum-free growth medium. Tissue culture medium from transfected phoenix cells was filtered 48 h after transfection, and the viral supernatant was used to infect the cultures of subconfluent MDA MB 231 and HBL 100 cells after the addition of 4 mg/ml polybrene. Isolated clones were obtained using antibiotic selection (puromycin 3 μg/ml) and were further expanded to confluent levels to obtain stably transfected cells.
Immunoblot analysis
For immunoblotting, MDA MB 231 and HBL 100 cells were lysed in RIPA buffer (50mM Tris–HCl, 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS) containing protease inhibitors (1mM phenyl–methyl sulphonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin). Samples were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinyldifluoride membranes (Millipore Coporation, Bedford, MA, USA) for immunodetection. After quick washing in PBST (phosphate-buffered saline (PBS) and 0.1% Tween 20), the membranes were blocked in 5% non-fat dry milk in PBS for at least 1 h and then incubated with primary antibodies MUC16 (mouse, 1:4000, DAKO, Carpinteria, CA, USA), JAK2 (Rabbit, 1:1000), pSTAT3 (Tyr705) (mouse), c-Jun (rabbit, 1:2000), Cyclin D1 (rabbit, 1:1500), Cyclin E (mouse, 1:1500), Cyclin A (rabbit, 1:1500), Cyclin B1 (rabbit, 1:2000), phospho-Aurora kinase A (rabbit, 1:2000), Bcl-2 (mouse, 1:1500), TRAIL (rabbit, 1:2000) JNK (rabbit, 1:2000), pJNK (rabbit, 1:2000), caspase 3 (mouse, 1:2000), caspase 9 (rabbit, 1:2000) and anti-β-actin (mouse) (diluted in 2% bovine serum albumin in PBS) overnight at 4 °C. Then the membranes were washed (3 × 10min) in PBST at room temperature and probed with the appropriate secondary antibodies at 1:2000 dilutions for 1 h at room temperature and washed 3 × 10min with PBST. The signal was detected with the ECL chemiluminescence kit (Amersham Bioscience, Amersham Place Little Chalfont, Buckinghamshire, UK).
Immunoprecipitation analysis
Equal amounts of protein (500 μg) were incubated overnight with anti-MUC16, anti-Jak2, MUC1 (mouse monoclonal) and MUC4 (mouse monoclonal) antibodies in a 500 μl total volume. Protein A+ G-Sepharose beads (Sigma-Aldrich Corp., St Louis, MO, USA) were added to the lysate-antibody mix and incubated on a rotating platform for 3 h at 4 °C and then washed four times with lysis buffer. The immunoprecipitates or total cell lysates were electrophoretically resolved on SDS–polyacrylamide gel electrophoresis (8%). Resolved proteins were transferred onto the polyvinyldifluoride membrane. After quick washing in PBST, the membranes were blocked in 5% non-fat dry milk in PBS for at least 1 h and then incubated with primary antibodies (anti-MUC16 and Jak2). The immunoblots were washed five times (5 × 10 min), incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies, washed five times (5 × 10 min), reacted with enhanced chemiluminescence ECL reagent (Amersham Biosciences) and exposed to X-ray film to detect the signal.
Confocal immunofluorescence microscopy
MDA MB 231 cells with MUC16 knockdown and the control vector were grown on sterilized cover slips for 30 h. Cells were washed with Hanks buffer containing 0.1M HEPES, fixed in ice-cold methanol at −20 °C for 2 min and blocked with 10% goat serum (Jackson Immunoresearch Labs, Inc., West Grove, PA, USA) for at least 30 min. After the blocking step and a quick wash in PBS, cells were incubated with antibodies for MUC16, JAK2, Cyclin E, Cyclin A, p21, Cyclin B1 and phospho-Aurora kinase A for 60 min at room temperature. Then, cells were washed (4 × 5 min each washing) with PBS and incubated with fluorescein isothiocyanate-conjugated anti-mouse and Texas red-conjugated anti-rabbit secondary anti-bodies (Jackson Immunoresearch labs, Inc.) for 30 min at room temperature in the dark. 4′, 6-diamidino-2-phenylindole was used for nuclear staining. Cells were washed again (5 × 5 min) and mounted on glass slides in anti-fade Vecta-shield mounting medium (Vector Laboratories, Burlingame, CA, USA). Laser confocal microscopy was performed using an LSM 510 microscope (Carl Zeiss GmbH, Jena, Germany).
Growth kinetics assay
Growth kinetics assays were performed as previously described (Singh et al., 2004).
In vivo tumor growth
Tumorigenicity assays were performed as previously described (Singh et al., 2004).
Synchronization and cell cycle analysis
MDA MB 231 and HBL 100 cells were grown in 100mm plates, and thymidine (Sigma) was added to the culture medium at a final concentration of 2mM for 12 h. Following two washes with serum-free medium, the cells were released from the thymidine block by culturing in fresh medium containing 24mM 2 deoxycytidine. After 9 h of incubation, the second thymidine block was initiated and completed after 14 h. The cells were released from the block by washing in warm phosphate buffered saline and replacing with complete culture medium. At different time points, the cells were fixed in 70% ethanol. After fixation, the cells were left on ice (~45 min) and then centrifuged. The pellets were resuspended in Telford’s reagent (90mM EDTA, 2.5mU of RNase A/ml, 50mg of propidium iodide/ml and 0.1% Triton X-100 in PBS). After incubating in an ice bath for ~2 h, the total DNA content was analyzed using the fluorescence-activated cell sorting method.
Apoptosis assay
A total of 2 × 106 cells were seeded in 60mm petri dishes and allowed to grow for 48 h. The cells were then trypsinized and washed with PBS twice. Apoptosis was measured using the annexin V-fluorescein isothiocyanate apoptosis detection kit (Roche Diagnostics, Indianapolis, IN, USA). Apoptosis was detected by staining the cells with annexin V and propidium iodide solution followed by flow cytometry.
Inhibition of JNK1/2 in breast cancer cell MDA MB 231
MUC16-knockdown MDA MB 231 cells (2 × 106) were seeded, and after 12 h treated with JNK inhibitor. The JNK1/2 inhibitor SP600125 (20, 40 and 80 nM) was used to treat MUC16-knockdown MDA MB 231 cells for 12 h. After the inhibition of JNK1/2, protein was extracted from the cells for further western blot analysis.
Supplementary Material
Acknowledgments
We are supported by grants from the National Institutes of Health (CA78590, CA111294, CA133774 and CA131944) and Department of Defense (BC101014). We thank Dr Jessica Mercer, Editorial Grants Associate at UNMC, for carefully editing this manuscript. We also acknowledge Erik Moore and Kavita Mallya for their technical support. We also thank Janice A Tayor and James R Talaska of the confocal laser scanning microscope core facility at UNMC for their support.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Bafna S, Singh AP, Moniaux N, Eudy JD, Meza JL, Batra SK. MUC4, a multifunctional transmembrane glycoprotein, induces oncogenic transformation of NIH3T3 mouse fibroblast cells. Cancer Res. 2008;68:9231–9238. doi: 10.1158/0008-5472.CAN-08-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC, Jr, Xu FJ, Yu YH, Barnhill S, Zhang Z, Mills GB. CA 125: the past and the future. Int J Biol Markers. 1998;13:179–187. doi: 10.1177/172460089801300402. [DOI] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock TD, Spurr-Michaud SJ, Tisdale AS, Heimer SR, Gilmore MS, Ramesh V, et al. Functions of MUC16 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2007;48:4509–4518. doi: 10.1167/iovs.07-0430. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. The functional contrariety of JNK. Mol Carcinog. 2007;46:591–598. doi: 10.1002/mc.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, et al. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Singh AP, Chakraborty S, Chauhan SC, Bafna S, Meza JL, et al. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells. Cancer Res. 2008;68:2065–2070. doi: 10.1158/0008-5472.CAN-07-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Cheng A, Chen YQ, Hymel A, Hanson EP, Kimmel L, et al. The amino terminus of JAK3 is necessary and sufficient for binding to the common gamma chain and confers the ability to transmit interleukin 2-mediated signals. Proc Natl Acad Sci USA. 1997;94:6910–6915. doi: 10.1073/pnas.94.13.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, Prasad U, Shankar S, Hamstra DA, Shanaiah M, Chenevert TL, et al. Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci USA. 2000;97:1754–1759. doi: 10.1073/pnas.030545097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV. Roles and regulation of stat family transcription factors in human breast cancer. Am J Pathol. 2004;165:1449–1460. doi: 10.1016/S0002-9440(10)63403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrick JL, Konishi I, Geary SM, Parmley TH, Quirk JG, Jr, O’Brien TJ. CA125 phosphorylation is associated with its secretion from the WISH human amnion cell line. Tumour Biol. 1997;18:278–289. doi: 10.1159/000218041. [DOI] [PubMed] [Google Scholar]
- Firmbach-Kraft I, Byers M, Shows T, la-Favera R, Krolewski JJ. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- Fujimoto K, Hosotani R, Doi R, Wada M, Lee JU, Koshiba T, et al. Induction of cell-cycle arrest and apoptosis by a novel retinobenzoic-acid derivative, TAC-101, in human pancreatic-cancer cells. Int J Cancer. 1999;81:637–644. doi: 10.1002/(sici)1097-0215(19990517)81:4<637::aid-ijc21>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, et al. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000;60:1426–1433. [PubMed] [Google Scholar]
- Ginsberg M, Czeko E, Muller P, Ren Z, Chen X, Darnell JE., Jr Amino acid residues required for physical and cooperative transcriptional interaction of STAT3 and AP-1 proteins c-Jun and c-Fos. Mol Cell Biol. 2007;27:6300–6308. doi: 10.1128/MCB.00613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- He L, Yang H, Ma Y, Pledger WJ, Cress WD, Cheng JQ. Identification of Aurora-A as a direct target of E2F3 during G2/M cell cycle progression. J Biol Chem. 2008;283:31012–31020. doi: 10.1074/jbc.M803547200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Constantinescu SN, Lodish HF. The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol Cell. 2001;8:1327–1338. doi: 10.1016/s1097-2765(01)00401-4. [DOI] [PubMed] [Google Scholar]
- Huang Y, Walstrom A, Zhang L, Zhao Y, Cui M, Ye L, et al. Type I interferons and interferon regulatory factors regulate TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected macrophages. PLoS One. 2009;4:e5397. doi: 10.1371/journal.pone.0005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Hardy S, Morgan DO. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- Korgun ET, Celik-Ozenci C, Acar N, Cayli S, Desoye G, Demir R. Location of cell cycle regulators cyclin B1, cyclin A, PCNA, Ki67 and cell cycle inhibitors p21, p27 and p57 in human first trimester placenta and deciduas. Histochem Cell Biol. 2006;125:615–624. doi: 10.1007/s00418-006-0160-y. [DOI] [PubMed] [Google Scholar]
- Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- Lee S, Helfman DM. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J Biol Chem. 2004;279:1885–1891. doi: 10.1074/jbc.M306968200. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li G, Gustafson-Brown C, Hanks SK, Nason K, Arbeit JM, Pogliano K, et al. c-Jun is essential for organization of the epidermal leading edge. Dev Cell. 2003;4:865–877. doi: 10.1016/s1534-5807(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Lin HH, Chen JH, Huang CC, Wang CJ. Apoptotic effect of 3,4-dihydroxybenzoic acid on human gastric carcinoma cells involving JNK/p38 MAPK signaling activation. Int J Cancer. 2007;120:2306–2316. doi: 10.1002/ijc.22571. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- Moritani S, Ichihara S, Hasegawa M, Endo T, Oiwa M, Yoshikawa K, et al. Serous papillary adenocarcinoma of the female genital organs and invasive micropapillary carcinoma of the breast. Are WT1, CA125, and GCDFP-15 useful in differential diagnosis? Hum Pathol. 2008;39:666–671. doi: 10.1016/j.humpath.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Munoz-Pinedo C, Guio-Carrion A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc Natl Acad Sci USA. 2006;103:11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro EG, Jain M, Oliva E, Kamal N, Lele SM, Lynch MP, et al. Upregulation of MUC4 in cervical squamous cell carcinoma: pathologic significance. Int J Gynecol Pathol. 2009;28:127–133. doi: 10.1097/PGP.0b013e318184f3e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TJ, Beard JB, Underwood LJ, Shigemasa K. The CA 125 gene: a newly discovered extension of the glycosylated N-terminal domain doubles the size of this extracellular superstructure. Tumour Biol. 2002;23:154–169. doi: 10.1159/000064032. [DOI] [PubMed] [Google Scholar]
- Orren DK, Petersen LN, Bohr VA. Persistent DNA damage inhibits S-phase and G2 progression, and results in apoptosis. Mol Biol Cell. 1997;8:1129–1142. doi: 10.1091/mbc.8.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- Perez BH, Gipson IK. Focus on molecules: human mucin MUC16. Exp Eye Res. 2008;87:400–401. doi: 10.1016/j.exer.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy MP, Lakshmanan I, Jain M, Das S, Chakraborty S, Dey P, et al. MUC4 mucin-induced epithelial to mesenchymal transition: a novel mechanism for metastasis of human ovarian cancer cells. Oncogene. 2010;29:5741–5754. doi: 10.1038/onc.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Davis SR, Pumphrey JG, Bao J, Nau MM, Meltzer PS, et al. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res Treat. 2009;113:217–230. doi: 10.1007/s10549-008-9924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Boyce RW, bd El-Rehim D, Kurien T, Green AR, Paish EC, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol. 2005;18:1295–1304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, et al. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol. 2002;158:321–329. doi: 10.1083/jcb.200202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Pollard JW. Progesterone inhibits estrogen-induced cyclin D1 and cdk4 nuclear translocation, cyclin E- and cyclin A-cdk2 kinase activation, and cell proliferation in uterine epithelial cells in mice. Mol Cell Biol. 1999;19:2251–2264. doi: 10.1128/mcb.19.3.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- Watson CJ. Stat transcription factors in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:115–127. doi: 10.1023/a:1009524817155. [DOI] [PubMed] [Google Scholar]
- Wu YM, Nowack DD, Omenn GS, Haab BB. Mucin glycosylation is altered by pro-inflammatory signaling in pancreatic- cancer cells. J Proteome Res. 2009;8:1876–1886. doi: 10.1021/pr8008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin BW, Dnistrian A, Lloyd KO. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int J Cancer. 2002;98:737–740. doi: 10.1002/ijc.10250. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- Zhao L, Ma Y, Seemann J, Huang LJ. A regulating role of the JAK2 FERM domain in hyperactivation of JAK2(V617F) Biochem J. 2010;426:91–98. doi: 10.1042/BJ20090615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.