Abstract
Objectives
Lower levels of serum Brain Derived Neurotrophic Factor (BDNF) is one of the best known biomarkers of depression. To identify genetic variants associated with serum BDNF, we tested the Val66Met (rs6265) functional variant and conducted a genome-wide association scan (GWAS).
Methods
In a community-based sample (N = 2054; aged 19 to 101, M = 51, SD = 15) from Sardinia, Italy, we measured serum BDNF concentration and conducted a GWAS.
Results
We estimated the heritability of serum BDNF to be 0.48 from sib-pairs. There was no association between serum BDNF and Val66Met in the SardiNIA sample and in a meta-analysis of published studies (k = 13 studies, total n = 4727, p = 0.92). Although no genome-wide significant associations were identified, some evidence of association was found in the BDNF gene (rs11030102, P = .001) and at two loci (rs7170215, P = 4.8×10−5 and rs11073742 P = 1.2×10−5) near and within NTRK3 gene, a neurotrophic tyrosine kinase receptor.
Conclusions
Our study and meta-analysis of the literature indicate that the BDNF Val66Met variant is not associated with serum BDNF, but other variants in the BDNF and NTRK3 genes might regulate the level of serum BDNF.
Keywords: brain-derived neurotrophic factor (BDNF), serum, Val66Met, NTRK3, GWAS
Introduction
Decreased brain-derived neurotrophic factor (BDNF) in serum is one of the best known biological markers of depression (Duman & Monteggia, 2006, Mossner et al., 2007), and is implicated in a number of other psychiatric and neurodegenerative disorders (Brandys et al., in press, Forlenza et al., 2010, Xiu et al., 2009, Yu et al., 2008). For example, the level of BDNF in serum is lower in patients suffering from mood disorders (Bocchio-Chiavetto et al., 2010, Diniz et al., 2010, Molendijk et al., in press, Schmidt & Duman, 2010), in subjects who experience depressive symptoms(Bus et al., 2011, Terracciano et al., in press), and in those who score high in neuroticism (Lang et al., 2004, Minelli et al., in press, Terracciano et al., in press). Given the importance of biological markers to understand the etiology and pathophysiological mechanisms of depressive disorders (Mossner et al., 2007), the aim of this study is to identify genetic variants associated with serum BDNF.
In a family-based cohort (N = 2054) we measured serum BDNF concentration and genotyped subjects using high-density arrays. We first estimated the heritability of serum BDNF from sib-pairs because a genetic association assumes that peripheral levels of BDNF are under some degree of genetic control. Next, we examined the role of the common SNP rs6265 in the BDNF gene, a Val66Met functional variant found to influence BDNF expression (Egan et al., 2003). We tested whether Met carriers have lower serum BDNF in one of the largest samples to date. In addition, given the mixed evidence for an association between the Val66Met variant and serum BDNF (Lang et al., 2009, Minelli et al., in press, Ozan et al., 2010, Yu et al., 2008), we conducted a meta-analysis to provide a quantitative summary of the current and published studies. Finally, in addition to testing the Val66Met polymorphism, we conducted a genome-wide association scan (GWAS) to identify new common variants potentially associated with serum BDNF.
Methods and Materials
Sample description
As described in detail elsewhere (Pilia et al., 2006, Sutin et al., 2010, Terracciano et al., 2010b), the SardiNIA project is an ongoing multidisciplinary study conducted in the Ogliastra province of Sardinia, Italy. This sample is derived from one of the oldest and largest genetically isolated populations (Cavalli-Sforza et al., 1994). The relatively uniform genetic background can provide robust associations by reducing the risk of false associations due to population stratification. In this study, we present results based on 2054 subjects (62% women; Mean age = 51.4, SD = 15.2) who were assayed for serum levels of BDNF and genotyped for the Val66Met variant. Genome-wide association analyses were performed on a subsample of subjects (n = 1668) with available data. The only exclusion criteria were being younger than 14 years old and being from regions other than Sardinia. Participants were not screened for psychiatric disorders. Each subject signed a consent form prior to their inclusion in the study approved by the institutional review boards in Italy and the USA. The study has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki.
Measurement of serum BDNF concentration
As described previously (Terracciano et al., in press) blood samples were drawn from subjects in the morning after an overnight fast. A total of 5.5ml of blood were collected in anticoagulant-free tubes. The blood samples were processed within 2 hours and serum was stored at −80°C until they were assayed, between 3 and 13 months (7 months average). BDNF concentrations in serum were assayed with the commercially available BDNF Emax® ImmunoAssay System (Promega, Madison, WI), following the manufacturer’s protocol. The microplate reader SpectraMax Plus384 (Molecular device) was used to record the absorbance at 450 nm. All BDNF assays were performed in the same laboratory by the same biologists (ML and MGP). This BDNF enzyme-linked immunosorbent assay (ELISA) has high specificity, with less than 3% cross-reactivity with other related neurotrophic factors (NGF, NT-3, and NT-4). This assay has a minimum sensitivity of 15.6 pg/ml of BDNF. To measure concentrations within the range of the standard curve, the BDNF concentration was examined on serum samples diluted 1:70 using the manufacturer recommended buffer. The concentration of each sample was determined in reference to standard curves from 7.8 to 500 pg/ml BDNF (R2 ≥ 0.94), which were examined in duplicate in each plate (Coefficient of Variation < 10%). As further control, we used the spike-and-recovery method to evaluate potential interference of our biological samples with the assay. For a subset of samples we measured the concentrations of the un-spiked aliquot and compared it with three aliquots spiked by adding 31.25, 62.5, and 125 pg/ml of exogenous BDNF. The recovery rate of the spiked BDNF in the measured samples exceeded 95%. The BDNF concentration values were normally distributed, ranging from 1 to 27 ng/ml, M = 14.07, SD = 3.04, Median = 14.06.
Genotyping
DNA was extracted from blood and genotyping was done with a combination of Affymetrix 10K, 500K, and 1M arrays and the Illumina Cardio-Metabochip array, as described in detail elsewhere (Sanna et al., 2008, Terracciano et al., 2010b, Terracciano et al., 2010c, Terracciano et al., 2010d). BRLMM was used for genotype calling, and quality controls included sample call rate > 95%, and SNP exclusions criteria were Hardy- Weinberg equilibrium ≤ 10−6, SNP call rate ≤ 90%, and minor allele frequency < 5%. The rs6265 was assessed on both the Affymetrix and Illumina arrays, which showed perfect genotyping match except for two individuals who were excluded from the analyses.
Statistical analyses
We calculated descriptive statistics as means ± SD or percentages, as appropriate. Univariate and multivariate analyses of variance were used to evaluate differences between Val66Met genotypes on serum BDNF concentration. As covariates we used age, sex, cigarette smoking, antidepressant use, and obesity (BMI ≥ 30). To increase genomic coverage, we imputed autosomal SNPs based on the CEU sample from the Haplotype Mapping Project (http://www.hapmap.org). SNPs with poor imputation quality (r2 ≤ .3 were excluded. MACH v1.015 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) was used to impute genotypes. The GWAS analyses were performed using the fastAssoc option in MERLIN (Chen & Abecasis, 2007). The association test implemented in MERLIN estimates the additive effect of genotyped and imputed SNPs in the context of a variance component model to adjust for relatedness among individuals.
Meta-analysis
To identify studies for inclusion in the meta-analysis, we searched the PubMed database (www.ncbi.nlm.nih.gov/pubmed) through 1 June 2011 using the following combinations of terms “BDNF,” “brain-derived neurotrophic factor,” “serum,” “Val66Met,” and “rs6265”. The reference lists of the studies identified and those of other relevant articles were examined to find additional studies. For studies that presented only limited statistics in their article, we sent an e-mail to the corresponding author to request the missing data. We obtained the necessary information from all studies we identified except for two (Tramontina et al., 2007; Zhou et al., 2011). We performed a fixed-effects as well as random-effects model meta-analysis, and heterogeneity was evaluated using the Q statistics. Publication bias was assessed using the Egger test. Stratified analyses to assess the potential moderating effect of sample ancestry were conducted for European and Asian samples. Similarly, analyses were conducted separately for studies with community-based cohorts and clinical samples. Data were analyzed using the “Comprehensive Meta Analysis” (Version 2) software package. The standard p < 0.05 (two-tails) was used as a threshold for statistical significance.
Results
Heritability of serum BDNF
To estimate the heritability of BDNF levels in our sample, we examined the correlation among 428 sib-pairs, one pair from each nuclear family with available data. The sib correlation was 0.24, which, divided by the coefficient of relationship (0.5), gives an estimated heritability of 0.48 for serum BDNF.
Association analyses of serum BDNF and the Val66Met genotypes in the SardiNIA sample
After quality controls, 2054 subjects assayed for serum BDNF had valid Val66Met (rs6265) genotyping. There were 844 individuals with a GG (Val/Val) genotype, 955 with AG (Val/Met), and 255 with the AA (Met/Met), in agreement with the Hardy- Weinberg equilibrium (χ2 = 0.36, P > 0.05). The frequency of the G (Val) allele was 64%, which is lower than the frequencies found in other European populations (~81%).
The concentration of serum BDNF was 14.13 ng/ml (SD = 3.0) for the Val/Val genotype, 14.00 ng/ml (SD = 3.1) for the Val/Met, and 14.11 ng/ml (SD = 3.1) for the Met/Met; there was no significant difference between the groups (P = 0.67). We further compared the Met carriers (Val/Met + Met/Met) with the homozygous Val/Val group, and found no significant association (P = 0.46). There was no association even after controlling for relevant covariates, including age, sex, cigarette smoking, antidepressant use, and obesity (P = 0.71).
Meta-analysis
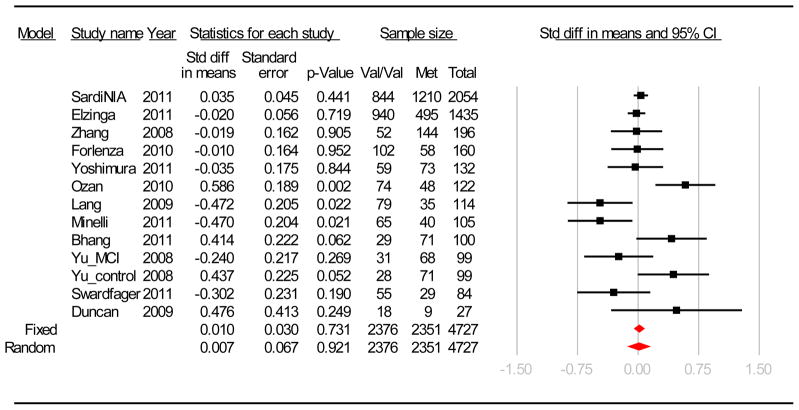
In addition to the data from the SardiNIA sample, we identified 11 articles for inclusion in the meta-analysis (Bhang et al., 2011, Duncan et al., 2009, Elzinga et al., 2011, Forlenza et al., 2010, Lang et al., 2009, Minelli et al., in press, Ozan et al., 2010, Swardfager et al., 2011, Yoshimura et al., 2011, Yu et al., 2008, Zhang et al., 2008), for a total of k = 13 independent samples, ranging in size from 27 to 2054, and for a total of 4727 subjects (2351 Met carriers and 2376 Val/Val carriers). The mean and SD of serum BDNF for each sample included in the meta-analysis are reported in supplementary Table 1. There was significant heterogeneity across studies (Q = 32.35, df = 12, P = 0.001; I2 = 62.96). Both the fixed and random effect estimates were not significant (see Figure 1). When the studies were examined separately based on ethnicity (Asian: k = 5, Z = 0.874, P = 0.38; European: k = 8, Z = 0.043, P = 0.97) or sample type (cohort studies: k = 5, Z = 0.244, P = 0.81; clinical studies: k = 8, Z = 0.242, P = 0.81) the meta-analyses revealed no significant association. This lack of association was not due to any single study and even without the SardiNIA study, there was no significant association (P = 0.81). No evidence of publication bias was indicated by the Egger’s test (B0 = 0.02, 95%CI = −1.67 to 1.71, P = 0.98) and year of publication was not associated with effect size (P = 0.89).
Figure 1.
Forest plot for Val66Met (rs6265) and serum BDNF association studies included in the meta-analysis.
The squares represent the standardized mean difference on serum BDNF between the groups with the Met variant and those with the Val/Val genotypes. The error bars represent the 95% confidence interval.
Additional studies have examined the association between the Val66Met variant and BDNF in samples of plasma, whole blood, and amniotic fluid. The level of BDNF in plasma and serum are not closely correlated (r = 0.21; n = 482)(Terracciano et al., in press), and less is known for the other fluids, so these studies were not included in the meta-analyses. However, the inclusion of these studies would not change the above results, given that the Val66Met was not associated with levels of BDNF in the two studies that examined plasma (Jiang et al., 2009, Terracciano et al., 2010a), nor in the two studies that examined whole blood (Trajkovska et al., 2007, Vinberg et al., 2009). The only interesting exception is a relatively small study that found lower BDNF in amniotic fluids of Met carriers (Cattaneo et al., 2010).
Genome-wide association results for serum BDNF concentrations
GWAS analyses were performed on 1668 subjects with both serum BDNF and genome-wide scan data. We tested 2,325,980 autosomal imputed and genotyped SNPs and found no association that exceeded the commonly accepted threshold for genome-wide significance (P < 5×10−8). The inflation factor λ was 1.027, indicating that the distribution of p-values was not inflated by population stratification. The SNPs with the strongest signals (P < 5×10−5) are presented in Supplementary Table 1. The genotyped SNP rs17008416 showed the strongest signal of association (P = 6.8×10−7; G allele: frequency 89%, Beta = −0.95, SE = 0.19). This SNP maps on chromosome 4, about 500Kb from the CDS1 and WDFY3 genes.
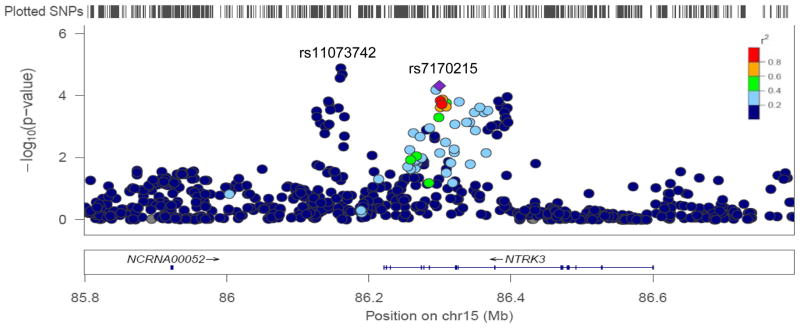
Among the strongest effects, we found an association with the genotyped SNP rs7170215 (P = 4.8×10−5), which maps within the neurotrophic tyrosine kinase receptor, type 3 (NTRK3) gene. The A allele (frequency 56%) was associated with lower serum BDNF concentrations (Beta = −0.51, SE = 0.13), an effect that explained approximately 1% of the variance in serum BDNF. As shown in Figure 2, an additional association signal was present at the 5′ of NTRK3, with the strongest effect observed for the imputed SNP rs11073742 (P = 1.2×10−5; T allele: frequency 65%, Beta = −0.56, SE = 0.13). The two SNPs rs7170215 and rs11073742 within and near NTRK3 are independent loci, with no linkage disequilibrium (R2 = 0.00). Other relatively strong effects were found for SNPs within the IFRD1, TRHR, GRM8, CDH4, and CDH23 genes (see supplementary Table 1).
Figure 2.
Regional plot of the NTRK3 loci
Shown is the SNP association with serum BDNF concentration for the NTRK3 loci (with −log10 P values on the y axis and the SNP genomic position on the x axis). The index SNP rs7170215 is denoted with a purple diamond. SNPs are colored to reflect LD with the index SNP (pairwise r2 values from HapMap CEU). Gene and microRNA annotations are from the UCSC genome browser. LocusZoom (http://csg.sph.umich.edu/locuszoom/) was used for this plot.
The GWAS analyses included 29 SNPs that map in the BDNF gene. As reported above, rs6265 was not associated with serum BDNF. However, two SNPs in perfect LD (R2 = 1) showed a nominally significant association: the genotyped rs11030102 (P = 0.0015, C allele: frequency 91%, Beta = 0.66, SE = 0.21) and the imputed rs11030107 (P = 0.0016, A allele: frequency 91%, Beta = 0.66, SE = 0.21). A third imputed SNP, rs10835211, which was in high LD with the rs11030102 and rs11030107 (R2 = 0.96), was also associated with serum BDNF (P = 0.0027, G allele: frequency 91%, Beta = 0.62, SE = 0.21).
The GWAS analyses included 365 SNPs that map in the neurotrophic tyrosine kinase receptor type 2 (NTRK2), which has the highest affinity for BDNF. None of the SNPs would pass a stringent gene-based Bonferroni correction (0.05/365 = 0.00013). The strongest effect within NTRK2 was found for rs4419891 (P = 0.0011). Interestingly, the NTRK2 intronic SNP rs1565445, which has been associated with lithium response in bipolar patients (Bremer et al., 2007), was genotyped in our sample and showed some evidence of association with serum BDNF (P = 0.0040, A allele: frequency 71%, Beta = −0.39, SE = 0.14).
Discussion
The aim of this study was to identify genetic factors that influence the concentration of BDNF in serum, which we estimated to be about 50% heritable in our cohort. Serum levels of BDNF were unrelated to the Val66Met in the current sample and in the meta-analysis of published studies. Although the GWAS did not reveal any common variant that passed the genome-wide significance threshold, the results suggest previously unidentified SNPs in the BDNF and NTRK3 genes that should be considered as candidates in future studies.
Despite much interest and a large number of studies on the topic, the meta-analysis of the current and published studies rejects the hypothesis that the Val66Met variant has a direct effect on serum BDNF. This study adds to other large studies and meta-analyses that have found no association between the Val66Met and the neuroticism personality trait (de Moor et al., in press, Terracciano et al., 2010c), mood disorders (Schumacher et al., 2005), ADHD (Sanchez-Mora et al., 2010), schizophrenia (Schizophrenia Research Forum, 2011), and Alzheimer’s Disease (Alzheimer Research Forum, 2011). Compared to these complex traits and diseases, serum BDNF is a biological marker or endophenotype (Gottesman & Gould, 2003) and thus might have stronger genetic associations compared to more distant phenotypes, such as mood disorders or personality traits. The results of this study, however, provide no evidence that the genetics of circulating BDNF in serum is any simpler. No variant explained large variance in serum BDNF. If there is a genetic basis for serum BDNF, as suggested by the heritability estimate, it is likely to be several common variants, each with a small effect size. Larger studies that use a GWAS approach will be required to identify such common variants.
To our knowledge, we present the results of the first GWAS of serum BDNF. Among the most interesting associations, we found SNPs (e.g., rs7170215 and rs11073742) that map within and near NTRK3, a tyrosine kinase receptor that mediates the neurotrophic/TRK signal transduction pathway. NTRK3 encodes for the TrkC receptor, which binds Neurotrophin-3 but not BDNF (Lamballe et al., 1991). If replicated, this finding suggests that NTRK3 receptor might play a role in regulative feedback on the expression and storage of BDNF. NTRK3 has been proposed as a candidate gene for a number of psychiatric disorders, including major depressive disorder (Feng et al., 2008, Verma et al., 2008), bipolar disorder (Athanasiu et al., 2011), and anxiety disorder (Muinos-Gimeno et al., 2009). We also identified a nominally significant association with rs11030102 (P = 0.0015; and two other SNPs in high LD, rs11030107 and rs10835211), which maps in the BDNF gene, and as such is a candidate variant that might regulate the level of BDNF in serum.
In future studies, it could be fruitful to examine the role of rarer genetic variants and the effect of physiological and environmental factors on circulating levels of BDNF, extending research on the effect of antidepressant drugs and treatments (Brunoni et al., 2008, Molendijk et al., in press), the role of common stressor such as harmful life events (Elzinga et al., 2011), and those of health-promoting behaviors, such as physical activity (Mattson et al., 2004). BDNF expression could also be down-regulated by post-transcriptional processes or epigenetic mechanisms, such as DNA methylation or histone acetylation (Roth et al., 2009). By not accounting for the above factors, this study was limited to the analyses of main effects of common genetic variants on serum BDNF. Another potential limitation is the lack of psychiatric screening of the SardiNIA sample. However, at least for the Val66Met, the meta-analysis found no difference between cohort-based and clinical samples.
In summary, the data from our sample and a meta-analysis indicate that there is no association between the much-studied Val66Met variant and serum BDNF, while the GWAS provided interesting new candidate variants. These results extend our knowledge on the genetics of serum BDNF, which appear more complex than previously anticipated.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Conflict of interest: The authors declare that they have no financial interest or other conflict of interest to disclose.
References
- Alzheimer Research Forum. 2011 http://www.alzforum.org/res/com/gen/alzgene/meta.asp?geneID=109&polyID=334.
- Athanasiu L, Mattingsdal M, Melle I, Inderhaug E, Lien T, Agartz I, et al. Intron 12 in NTRK3 is associated with bipolar disorder. Psychiatry Res. 2011;185:358–362. doi: 10.1016/j.psychres.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Bhang S, Ahn JH, Choi SW. Brain-derived neurotrophic factor and serotonin transporter gene-linked promoter region genes alter serum levels of brain-derived neurotrophic factor in humans. J Affect Disord. 2011;128:299–304. doi: 10.1016/j.jad.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L, Bagnardi V, Zanardini R, Molteni R, Nielsen GM, Placentino A, et al. Serum and plasma BDNF levels in major depression: A replication study and meta-analyses. World J Biol Psychiatry. 2010;11:763–773. doi: 10.3109/15622971003611319. [DOI] [PubMed] [Google Scholar]
- Brandys MK, Kas MJ, van Elburg AA, Campbell IC, Adan RA. A meta-analysis of circulating BDNF concentrations in anorexia nervosa. World J Biol Psychiatry. doi: 10.3109/15622975.15622011.15562244. (in press) [DOI] [PubMed] [Google Scholar]
- Bremer T, Diamond C, McKinney R, Shehktman T, Barrett TB, Herold C, et al. The pharmacogenetics of lithium response depends upon clinical co-morbidity. Mol Diagn Ther. 2007;11:161–170. doi: 10.1007/BF03256238. [DOI] [PubMed] [Google Scholar]
- Brunoni AR, Lopes M, Fregni F. A systematic review and meta-analysis of clinical studies on major depression and BDNF levels: implications for the role of neuroplasticity in depression. Int J Neuropsychopharmacol. 2008;11:1169–1180. doi: 10.1017/S1461145708009309. [DOI] [PubMed] [Google Scholar]
- Bus BA, Tendolkar I, Franke B, de Graaf J, Heijer MD, Buitelaar JK, et al. Serum brain-derived neurotrophic factor: Determinants and relationship with depressive symptoms in a community population of middle-aged and elderly people. World J Biol Psychiatry. 2011 doi: 10.3109/15622975.15622010.15545187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A, Bocchio-Chiavetto L, Zanardini R, Marchina E, Bellotti D, Milanesi E, et al. BDNF Val66Met polymorphism and protein levels in amniotic fluid. BMC Neurosci. 2010;11:16. doi: 10.1186/1471-2202-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Toshiko T, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. doi: 10.1038/mp.2010.1128. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz BS, Teixeira AL, Talib LL, Mendonca VA, Gattaz WF, Forlenza OV. Serum brain-derived neurotrophic factor level is reduced in antidepressant-free patients with late-life depression. World J Biol Psychiatry. 2010;11:550–555. doi: 10.3109/15622970903544620. [DOI] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Duncan LE, Hutchison KE, Carey G, Craighead WE. Variation in brain-derived neurotrophic factor (BDNF) gene is associated with symptoms of depression. J Affect Disord. 2009;115:215–219. doi: 10.1016/j.jad.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Elzinga BM, Molendijk ML, Oude Voshaar RC, Bus BA, Prickaerts J, Spinhoven P, et al. The impact of childhood abuse and recent stress on serum brain-derived neurotrophic factor and the moderating role of BDNF Val66Met. Psychopharmacology (Berl) 2011;214:319–328. doi: 10.1007/s00213-010-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Vetro A, Kiss E, Kapornai K, Daroczi G, Mayer L, et al. Association of the neurotrophic tyrosine kinase receptor 3 (NTRK3) gene and childhood-onset mood disorders. Am J Psychiatry. 2008;165:610–616. doi: 10.1176/appi.ajp.2007.07050805. [DOI] [PubMed] [Google Scholar]
- Forlenza OV, Diniz BS, Teixeira AL, Ojopi EB, Talib LL, Mendonca VA, et al. Effect of brain-derived neurotrophic factor Val66Met polymorphism and serum levels on the progression of mild cognitive impairment. World J Biol Psychiatry. 2010;11:774–780. doi: 10.3109/15622971003797241. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang R, Liu Y, Zhang Y, Chen ZY. BDNF Val66Met polymorphism is associated with unstable angina. Clin Chim Acta. 2009;400:3–7. doi: 10.1016/j.cca.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Lamballe F, Klein R, Barbacid M. trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell. 1991;66:967–979. doi: 10.1016/0092-8674(91)90442-2. [DOI] [PubMed] [Google Scholar]
- Lang UE, Hellweg R, Gallinat J. BDNF serum concentrations in healthy volunteers are associated with depression-related personality traits. Neuropsychopharmacology. 2004;29:795–798. doi: 10.1038/sj.npp.1300382. [DOI] [PubMed] [Google Scholar]
- Lang UE, Hellweg R, Sander T, Gallinat J. The Met allele of the BDNF Val66Met polymorphism is associated with increased BDNF serum concentrations [letter] Mol Psychiatry. 2009;14:120–122. doi: 10.1038/mp.2008.80. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Maudsley S, Martin B. BDNF and 5-HT: a dynamic duo in age-related neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 2004;27:589–594. doi: 10.1016/j.tins.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Minelli A, Zanardini R, Bonvicini C, Sartori R, Pedrini L, Gennarelli M, et al. BDNF serum levels, but not BDNF Val66Met genotype, are correlated with personality traits in healthy subjects. Eur Arch Psychiatry Clin Neurosci. doi: 10.1007/s00406-00011-00189-00403. (in press) [DOI] [PubMed] [Google Scholar]
- Molendijk ML, Bus BA, Spinhoven P, Penninx BW, Kenis G, Prickaerts J, et al. Serum levels of brain-derived neurotrophic factor in major depressive disorder: state-trait issues, clinical features and pharmacological treatment. Mol Psychiatry. doi: 10.1038/mp.2010.1098. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner R, Mikova O, Koutsilieri E, Saoud M, Ehlis AC, Muller N, et al. Consensus paper of the WFSBP Task Force on Biological Markers: biological markers in depression. World J Biol Psychiatry. 2007;8:141–174. doi: 10.1080/15622970701263303. [DOI] [PubMed] [Google Scholar]
- Muinos-Gimeno M, Guidi M, Kagerbauer B, Martin-Santos R, Navines R, Alonso P, et al. Allele variants in functional MicroRNA target sites of the neurotrophin-3 receptor gene (NTRK3) as susceptibility factors for anxiety disorders. Hum Mutat. 2009;30:1062–1071. doi: 10.1002/humu.21005. [DOI] [PubMed] [Google Scholar]
- Ozan E, Okur H, Eker C, Eker OD, Gonul AS, Akarsu N. The effect of depression, BDNF gene val66met polymorphism and gender on serum BDNF levels. Brain Res Bull. 2010;81:61–65. doi: 10.1016/j.brainresbull.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, et al. Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PloS Genetics. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early- life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mora C, Ribases M, Ramos-Quiroga JA, Casas M, Bosch R, Boreatti-Hummer A, et al. Meta-analysis of brain-derived neurotrophic factor p.Val66Met in adult ADHD in four European populations. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:512–523. doi: 10.1002/ajmg.b.31008. [DOI] [PubMed] [Google Scholar]
- Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Research Forum. 2011 http://www.szgene.org/meta.asp?geneID=2&polyID=28.
- Schmidt HD, Duman RS. Peripheral BDNF produces antidepressant-like effects in cellular and behavioral models. Neuropsychopharmacology. 2010;35:2378–2391. doi: 10.1038/npp.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, et al. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58:307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Sutin AR, Scuteri A, Lakatta EG, Tarasov KV, Ferrucci L, Costa PT, Jr, et al. Trait antagonism and the progression of arterial thickening: women with antagonistic traits have similar carotid arterial thickness as men. Hypertension. 2010;56:617–622. doi: 10.1161/HYPERTENSIONAHA.110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swardfager W, Herrmann N, Marzolini S, Saleem M, Shammi P, Oh PI, et al. Brain derived neurotrophic factor, cardiopulmonary fitness and cognition in patients with coronary artery disease. Brain Behav Immun. 2011;25:1264–1271. doi: 10.1016/j.bbi.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Lobina M, Piras MG, Mulas A, Cannas A, Meirelles O, et al. Lower serum BDNF associated with higher neuroticism and depressive symptoms. Psychosomatic Medicine. doi: 10.1097/PSY.0b013e3182306a4f. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Martin B, Ansari D, Tanaka T, Ferrucci L, Maudsley S, et al. Plasma BDNF concentration, Val66Met genetic variant and depression-related personality traits. Genes Brain Behav. 2010a;9:512–518. doi: 10.1111/j.1601-183X.2010.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2010b;15:647–656. doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Deiana B, Balaci L, Sanna S, et al. BDNF Val66Met is associated with introversion and interacts with 5-HTTLPR to influence neuroticism. Neuropsychopharmacology. 2010c;35:1083–1089. doi: 10.1038/npp.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010d;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovska V, Marcussen AB, Vinberg M, Hartvig P, Aznar S, Knudsen GM. Measurements of brain-derived neurotrophic factor: methodological aspects and demographical data. Brain Res Bull. 2007;73:143–149. doi: 10.1016/j.brainresbull.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Verma R, Holmans P, Knowles JA, Grover D, Evgrafov OV, Crowe RR, et al. Linkage disequilibrium mapping of a chromosome 15q25-26 major depression linkage region and sequencing of NTRK3. Biol Psychiatry. 2008;63:1185–1189. doi: 10.1016/j.biopsych.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinberg M, Trajkovska V, Bennike B, Knorr U, Knudsen GM, Kessing LV. The BDNF Val66Met polymorphism: relation to familiar risk of affective disorder, BDNF levels and salivary cortisol. Psychoneuroendocrinology. 2009;34:1380–1389. doi: 10.1016/j.psyneuen.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Xiu MH, Hui L, Dang YF, Hou TD, Zhang CX, Zheng YL, et al. Decreased serum BDNF levels in chronic institutionalized schizophrenia on long-term treatment with typical and atypical antipsychotics. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:1508–1512. doi: 10.1016/j.pnpbp.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Yoshimura R, Kishi T, Suzuki A, Umene-Nakano W, Ikenouchi-Sugita A, Hori H, et al. The brain-derived neurotrophic factor (BDNF) polymorphism Val66Met is associated with neither serum BDNF level nor response to selective serotonin reuptake inhibitors in depressed Japanese patients. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1022–1025. doi: 10.1016/j.pnpbp.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhang Z, Shi Y, Bai F, Xie C, Qian Y, et al. Association study of the decreased serum BDNF concentrations in amnestic mild cognitive impairment and the Val66Met polymorphism in Chinese Han. J Clin Psychiatry. 2008;69:1104–1111. doi: 10.4088/jcp.v69n0710. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Zhou DF, Wu GY, Cao LY, Tan YL, Haile CN, et al. BDNF levels and genotype are associated with antipsychotic-induced weight gain in patients with chronic schizophrenia. Neuropsychopharmacology. 2008;33:2200–2205. doi: 10.1038/sj.npp.1301619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.