Abstract
The assembly of distinct proteins into tight junctions results in the formation of a continuous barrier that regulates the paracellular flux of water, ions and small molecules across epithelia. The claudin protein family encompasses numerous major structural components of tight junctions. These proteins specify the permeability characteristics of tight junctions and consequently, some of the physiological properties of epithelia. Furthermore, defective claudin expression has been found to correlate with some diseases, tumor progression, and defective morphogenesis. Investigating the pattern of claudin expression during embryogenesis or in certain pathological conditions is necessary to begin disclosing the role of these proteins in health and disease. This study analyzed the expression of several claudins during mouse pancreas organogenesis and in pancreatic intraepithelial neoplasias of mouse and human origin. Our results underscored a distinctive, dynamic distribution of certain claudins in both the developing pancreas and the pancreatic epithelium undergoing neoplastic transformation.
Keywords: Claudin, pancreas, pancreatic intraepithelial neoplasia, development, tight junction
Introduction
Epithelial morphogenesis comprises the various processes by which epithelia contribute to organ formation and body shape. These complex events play a central role in development, regeneration and cancer. Epithelial cell membranes are subdivided into apical and basolateral domains, and are interconnected by junctions that linked them to each other (Schock and Perrimon, 2002). Establishing and maintaining this specific architecture is necessary both to form coherent sheets of cells, and to confer barrier properties to epithelia.
The space between neighboring epithelial cells is separated into apical and basal compartments by specialized apical structures called tight junctions (TJs) (Cereijido et al., 2007). TJs consist of multiple protein complexes that surround the cell, and extend across the lipid bilayer to form an anastomosing network between adjacent cells (Gupta and Ryan, 2010). TJs form dynamic barriers regulating the paracellular flux of small molecules, and water between epithelial cells by forming a size and charge selective pore (Furuse, 2010). TJs can be modified in response to environmental, physiological or pharmacological cues, and alterations in their structure have been associated with various human pathologies including autoimmune diseases, congenital deafness, renal diseases, tumor progression and allergies (Cereijido et al., 2007).
Various proteins specifically localize to TJs, where they regulate actin structure, cell-cell adhesion and tissue permeability (Anderson and Van Itallie, 2010). Claudins are four-pass transmembrane proteins constituting the structural and functional building blocks of TJs (Angelow et al., 2008). Claudins determine the permeability of small molecules across TJs, and are broadly expressed in embryonic and adult epithelia. The expression of specific combinations of claudins in different epithelia suggests that a combinatorial ‘claudin code’ might determine the size and ion specificity of TJs (Gupta and Ryan, 2010).
Studies performed in cultured cells suggest that claudins participate in TJ formation and that individual claudins vary in their ability to form tight or leaky barriers in epithelia (Amasheh et al., 2002; Anderson and Van Itallie, 2010). Mutations in several human CLAUDIN genes have been associated with skin, kidney and ear diseases (Simon et al., 1999; Wilcox et al., 2001; De Benedetto et al., 2011). Likewise, abnormal CLAUDIN expression was reported in gastrointestinal diseases (Zeissig et al., 2007; Schulzke et al., 2009) and tumors (Aung et al., 2006; Hewitt et al., 2006; Kulka et al., 2009; Nemeth et al., 2009) in humans.
Claudin protein activity also appears necessary for proper tissue morphogenesis. For instance, in Xenopus embryos, overexpression of Xcla (the homologue of human CLAUDIN 4) was shown to affect heart looping (Brizuela et al., 2001), whereas blocking the function of both claudin 4 and claudin 6 in the mouse trophoectoderm led to blastocyst collapse (Moriwaki et al., 2007). Likewise, lumen formation in the neuroepithelium (Zhang et al., 2010) or in the gut epithelium (Bagnat et al., 2007) of zebrafish embryos was shown to require the expression of claudin 5a and claudin 15, respectively.
Cytoskeletal reorganization, cell polarization and the assembly of intercellular junctional complexes (including TJs) are processes intimately involved with the formation of tubular structures in various organs, including the pancreas (Hick et al., 2009; Kesavan et al., 2009). In mouse embryos, pancreas morphogenesis initiates at around embryonic (E) day 9.5–10.5 with the emergence of two epithelial rudiments in the foregut region of the endoderm (Guney and Gannon, 2009). Shortly thereafter, groups of pancreatic cells acquire apicobasal polarity and form microlumens; these structures then propagate, fuse and assemble into tubular complexes (Hick et al., 2009; Kesavan et al., 2009). The result of this process is an intricate ductal network that interconnects the acinar cells (producers of digestive enzymes), collects the secreted zymogens, and transports these products to the duodenum (Grapin-Botton, 2005). In addition to ducts and acinar cells, the mature pancreas also contains numerous islets, which produce hormones necessary to preserve glucose homeostasis (Slack, 1995).
Recent studies have begun to unravel how certain proteins (e.g., Cdc42) contribute to establishing the apicobasal polarity in the early pancreatic epithelium (Kesavan et al., 2009). However, the role that individual claudins play during pancreas morphogenesis remains practically unknown. This study analyzed the expression of both claudin transcripts and claudin proteins during mouse pancreas development. In addition, potential changes in claudin protein expression during the initial stages of pancreatic neoplastic transformation were also investigated. We anticipate that this information will assist future efforts aiming to uncover the role of claudin proteins in health and disease in the pancreas.
Results and Discussion
Dynamic distribution of Cldn transcripts during mouse pancreas organogenesis
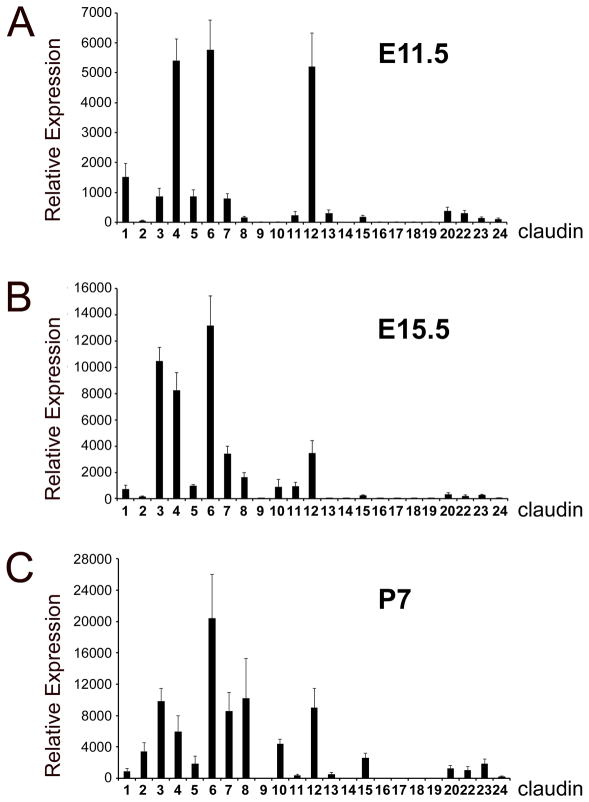
While the expression of some claudins was reported in the pancreas of adult rodents (Rahner et al., 2001; D’Souza et al., 2009), almost no information is available on the distribution of these proteins in the mammalian pancreas during development (Anderson et al., 2008; Kesavan et al., 2009). We used quantitative Real-Time PCR (qRT-PCR) to analyze the expression of transcripts of all Cldn genes in mouse pancreatic tissues dissected at three developmental stages: E11.5, E15.5 and postnatal (P) day 7. For each time point, the expression levels of individual Cldn transcripts were determined based on their relative abundance to GAPDH (Fig. 1). Five different patterns of Cldn transcript expression across the previous developmental stages were observed (Table 1). The first group (A) included Cldn1, -3, -4, -5, -6, -7, -8 and -12 was expressed at all stages analyzed in the pancreas. The second group (B) represented by Cldn2 was expressed in this tissue primarily after birth. The third group (C) included Cldn11, -15, -20, -22 and -23 had low but detectable expression throughout pancreas development. The fourth group (D) included Cldn10, -13 and -24 had detectable pancreatic expression at least at one developmental stage. The fifth group (E) represented by Cldn9, -14, -16, -17, -18 and -19 was not detected in the pancreas during development or after birth. However, within this group, Cldn18 expression was noticed in the stomach at E15.5 (Suppl. Fig. 1A), and both Cldn16 and -19 were detected in the kidney at P5 (Suppl. Fig. 1B). Immunohistochemistry analyses also showed high expression of claudin 18 proteins in the E15.5 posterior stomach (Supp. Fig. 1A), and lack of this protein in pancreatic tissues of mouse embryos or neonates (data not shown). These contrasting differences in claudin 18 expression could indicate differences in paracellular permeability between developing gastric and pancreatic epithelia. In summary, our qRT-PCR results uncovered a specific, dynamic distribution of Cldn transcripts throughout development in mouse pancreatic tissues (Table 1).
Figure 1. Relative abundance of Cldn transcripts in developing or postnatal mouse pancreatic tissues.
QRT-PCR analysis was performed with specific primers recognizing all Cldn family members and RNA from pancreata dissected at embryonic days (E) 11.5 (A) and E15.5 (B), and postnatal (P) 7 (C). For all experiments, expression levels were normalized to GAPDH housekeeping gene expression. Individual bars represent mean expression across three individual tissues (P7) or three individual tissue pools (E11.5 and E15.5). Error bars represent standard error from the three individual experiments.
Table 1.
Summary of claudin transcript expression in mouse developing pancreata.
| CLAUDIN TRANSCRIPT | E11.5 | E15.5 | P5 | EXPRESSION PATTERN |
|---|---|---|---|---|
| Cldn1 | ++ | + | + | A |
| Cldn2 | Very low | Very low | ++ | B |
| Cldn3 | ++ | ++++ | +++ | A |
| Cldn4 | ++++ | ++++ | ++ | A |
| Cldn5 | ++ | + | + | A |
| Cldn6 | ++++ | ++++ | ++++ | A |
| Cldn7 | ++ | ++ | +++ | A |
| Cldn8 | + | + | +++ | A |
| Cldn9 | n.d. | n.d. | n.d. | E |
| Cldn10 | n.d. | + | ++ | D |
| Cldn11 | + | + | Very low | C |
| Cldn12 | ++++ | ++ | +++ | A |
| Cldn13 | + | n.d. | Very low | D |
| Cldn14 | n.d. | n.d. | n.d. | E |
| Cldn15 | + | Very low | + | C |
| Cldn16 | n.d. | n.d. | n.d.1 | E1 |
| Cldn17 | n.d. | n.d. | n.d. | E |
| Cldn18 | n.d. | n.d.2 | n.d. | E2 |
| Cldn19 | n.d. | n.d. | n.d.1 | E1 |
| Cldn20 | + | Very low | + | C |
| Cldn22 | + | Very low | + | C |
| Cldn23 | + | Very low | + | C |
| Cldn24 | + | n.d. | Very low | D |
A: Detected at all developmental stages.
B: Predominantly expressed after birth.
C: Low but detectable expression at all stages (compared to other claudins).
D: Detectable expression at least at one developmental stage.
E: Undetected at all stages analyzed
Detected in the kidney.
Detected in the stomach.
n.d.: not detected.
Cell-type restricted expression of claudin proteins in the developing pancreas
The E10.5 pancreas is a multilayered, non-polarized epithelium mainly consisting of multipotent progenitors that express the homeodomain transcription factor Pdx1, and give rise to endocrine, acinar and ductal cells (Guney and Gannon, 2009). These precursors normally initiate differentiation between E13.5–E15.5 of embryogenesis, and this process continues throughout the rest of gestation (Pan and Wright, 2011). Full maturation of the mouse pancreas is completed around weaning (Pan and Wright, 2011)
We used commercial anti-claudin antibodies to investigate the spatial and temporal expression pattern of claudin proteins in mouse pancreata dissected at E10.5, E12.5, E15.5, E18.5 and P5. Of several antibodies tested, only those recognizing claudin 1, -2, -3, -5, -7 or -18 were suitable for immunostaining (Supp. Table 1).
Claudin 1
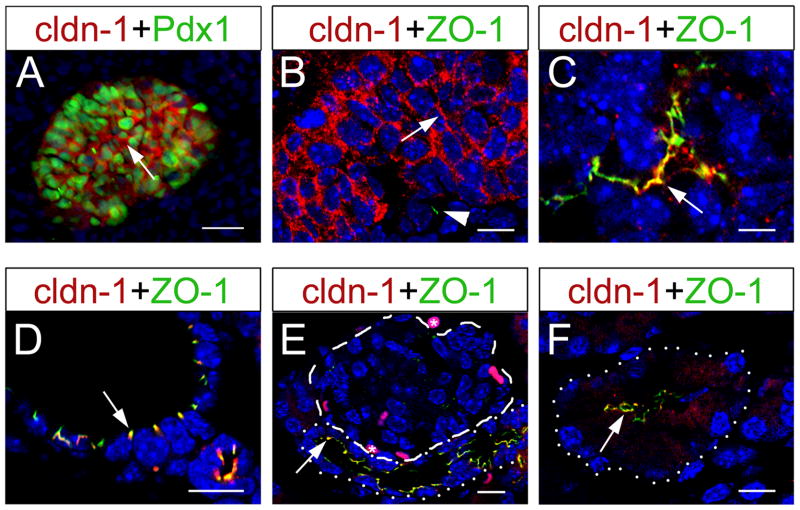
Concurring with our qRT-PCR data (Fig. 1 and Table 1), the results of immunofluorescence showed expression of claudin-1 proteins in nearly every Pdx1+ cell of E10.5 pancreata (Fig. 2A). Claudin 1 extensively localized throughout the membrane of the E10.5 unpolarized pancreatic epithelium, which was also nearly devoid of ZO-1 expression (Fig. 2B). By E12.5, apicobasal polarity is established in some areas of the pancreas (Hick et al., 2009; Kesavan et al., 2009). At this stage, claudin 1 proteins and ZO-1 proteins co-localized towards the lumen of the pancreatic epithelium (Fig. 2C), in discrete areas probably representing incipient TJs. Claudin 1 expression rapidly decayed after E12.5 and was no longer visible in the embryonic pancreas after E15.5 (data not shown). At P5, claudin 1 was again expressed in the pancreas, co-localizing extensively with ZO-1 in TJs of both ductal cells (Fig. 2D, E) and acinar cells (Fig. 2F). In contrast, islet cells were devoid of claudin 1 expression (Fig. 2E).
Figure 2. Claudin 1 expression is restricted to early embryonic and postnatal stages in the mouse pancreas.
(A) Expression of the homeodomain protein Pdx1 (green) labels multipotent progenitors of the E10.5 pancreatic primordium. At this stage, all Pdx1+ cells co-express claudin 1 (red; arrow). (B) Claudin 1 (red; arrow) localizes to the lateral membrane of the E10.5 pancreatic epithelium. At this stage, expression of the tight junction marker ZO-1 (green; arrowhead) is nearly absent in the pancreas. (C) By E12.5, when apicobasal polarity has been established, claudin 1 co-localizes with ZO-1 (arrow) in primitive pancreatic ducts. (D,E) At P5, claudin 1 co-localizes with ZO-1 at TJ complexes in ducts (arrows; dotted line in E), but not in islets (dashed line in E). (F) Also at this stage, claudin 1 co-localizes with ZO-1 in acinar cells (arrow; dotted lined). (E) Auto-fluorescent red blood cells are marked with asterisks. (F) Non-specific claudin 1 cytoplasmic staining is observed in acinar cells at P5. Images in B–F were taken with a confocal microscope, and cell nuclei were stained with DAPI (blue). Scale bars: 25 μm (A), 10 μm (B,D–F), 5 μm (C).
We conclude that claudin 1 expression precedes ZO-1 expression and the formation of TJs in progenitors of the developing pancreas. These results suggest that claudin 1 function could be necessary to establish TJs, form microlumens, or both in the early pancreatic epithelium. Our finding that claudin-1 becomes re-expressed after birth and localizes specifically to TJs of ductal and acinar cells in the pancreas, also indicates that this protein could contribute to establishing paracellular permeability in those cells. Similar to our study, CLAUDIN 1 expression was also reported in both, acinar and ductal cells in the adult human pancreas (Tsukahara et al., 2005; Borka et al., 2007).
Claudin 2
QRT-PCR results showed barely detectable expression of Cldn2 transcripts in E11.5 or E15.5 mouse pancreata (Fig. 1 and Table 1). Likewise, immunostaining results did not show claudin 2 protein expression in pancreatic epithelia dissected between E10.5 and E15.5 (data not shown). In contrast, claudin 2 was abundantly expressed in the kidney epithelium at E15.5 (data not shown).
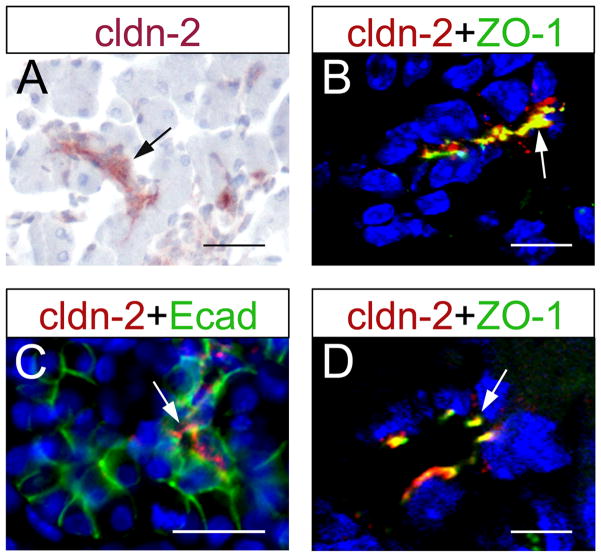
Different from early embryonic stages, we detected claudin 2 protein expression in the developing ducts of E18.5 pancreata (Fig. 3A, B), and in the lumen of the ductal pancreatic epithelium at P5 (Fig. 3C, D). Confocal microscopy results also showed co-expression of claudin 2 and ZO-1 proteins in TJs of pancreatic ductal epithelial cells at both, E18.5 (Fig. 3B) and P5 (Fig. 3D). The relative late expression of claudin 2 rules out a potential role of this protein in microlumen formation. However, it remains plausible that claudin 2 contributes to some aspects of pancreatic duct physiology because this protein is expressed in the pancreatic ducts of both, adult rats (Rahner et al., 2001) and adult humans (Aung et al., 2006; Borka et al., 2007).
Figure 3. Claudin 2 expression is restricted to ductal epithelial cells during late stages of pancreas organogenesis and in the postnatal organ.
(A,C) Claudin 2 expression is observed only in ductal epithelial cells in pancreatic tissues dissected at E18.5 (arrow in A) or P5 (arrow in C; E-cadherin [Ecad] staining demarcates the pancreatic epithelium). At both E18.5 (B) and P5 (D), claudin 2 co-localized with ZO-1 at tight junctions (arrows). Images in B and D were taken with a confocal microscope. Cell nuclei were stained with hematoxylin (A) and DAPI (blue, B–D). Scale bars: 25 μm (A,C), 10 μm (B), 5 μm (D).
Claudin 3
Cldn3 transcripts were detected in pancreata dissected at E11.5, E15.5 and P7 (Fig. 1 and Table 1) using qRT-PCR. Similar to claudin 1 (Fig. 2A), Pdx1+ progenitors of E10.5 pancreata expressed claudin 3 proteins all through the epithelial membrane (Fig. 4A,B). Like claudin 1, claudin 3 redistributed and co-localized with ZO-1 (presumably in nascent TJs), in the lumen of E12.5 pancreata (Fig. 4C). Different from claudin 1, claudin 3 proteins remained strongly expressed towards the lumen of tubular structures in the pancreas at E15.5 (Fig. 4D). These results are consistent with the Kesavan et al. (Kesavan et al., 2009) report, showing claudin 3 protein expression in TJs during remodeling of the pancreatic tubular network at E15.5. Confocal microscopy results corroborated extensive co-localization of claudin 3 and ZO-1 in TJ’s of pancreatic epithelia at both E15.5 (Fig. 4D) and E18.5 (Fig. 4E).
Figure 4. Claudin 3 is broadly expressed in developing pancreatic epithelia but becomes restricted to ductal and acinar cells after birth.
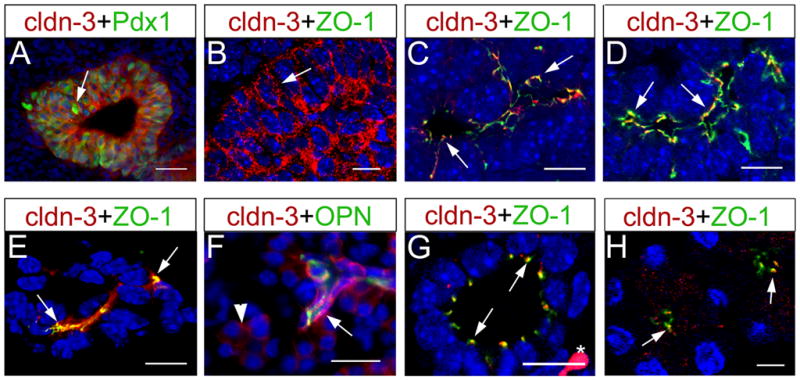
(A) Claudin 3 is co-expressed with Pdx1 (arrow) in multipotent progenitors of E10.5 pancreata. (B) At this stage, claudin 3 (red; arrow) localizes to the lateral membrane of pancreatic cells. (C) At E12.5, following polarization of the pancreatic primordium, claudin 3 (red; arrows) co-localizes with ZO-1 (green) at tight junctions in primitive ducts. (D,E) Claudin 3 and ZO-1 coexpression is noticed towards the lumen of the emerging pancreatic ductal network at E15.5 (arrows in D), and in ductal epithelial cells at E18.5 (arrows in E). (F) Claudin 3 is expressed at high levels in ductal cells (Opn+ cells; arrows), and at lower levels in acinar cells (arrowhead), in P5 pancreata. (G,H) Claudin 3 co-localizes with ZO-1 in both ductal (arrows in G) and acinar (arrows in H) cells in P5 pancreata. Asterisk in G indicates an autofluorescent red blood cell. B–H are confocal images (cell nuclei were stained with DAPI [blue]). Scale bars: 25 μm (A), 20 μm (F), 10 μm (B–E,G), 5 μm (H).
Expression of claudin 3 was previously reported in acinar and ductal cells of the adult rat pancreas (Rahner et al., 2001), and in acinar, ductal and islet cells of the adult human pancreas (Borka et al., 2007; Comper et al., 2009). We detected claudin 3 protein expression in TJs of the duct epithelium (Fig. 4F,G) and acinar cells (Fig. 4F,H) but not in islet cells (data not shown) in mouse pancreatic tissues dissected at P5. These results indicate that in the postnatal mouse pancreas, claudin 3 expression is restricted to duct epithelial and acinar cells, although it remains plausible that islet cells start expressing claudin 3 in this organ of mice after P5.
Claudin 5
Claudin 5 is the major component of TJs in endothelial cells, and functions to generate a proper blood-brain barrier (Nitta et al., 2003). QRT-PCR analysis revealed low expression of Cldn5 transcripts in E11.5, E15.5 and P7 pancreata (Fig. 1 and Table 1). However, immunofluorescence results identified claudin-5 protein expression exclusively in endothelial cells, but not in epithelial cells of pancreata dissected between E13.5-E15.5 (data not shown). In contrast, claudin 5 protein expression was detected in both ductal cells (Fig. 5A) and acinar cells (Fig. 5B) in pancreata dissected at E18.5 and P5. In these tissues, claudin 5 proteins localized almost exclusively to TJs, which were identified with antibodies recognizing ZO-1 (Fig. 5A, B). Our data are consistent with previous reports showing claudin 5 expression in rat acini (Rahner et al., 2001) and human ductal and acinar cells (Comper et al., 2009). Endothelial cells associated with the postnatal pancreas also expressed high claudin 5 levels (data not shown). We conclude that claudin 5 and claudin 2 have similar temporal expression patterns in the pancreas, because these two proteins start to be noticed in this organ only close to birth. However, their distribution in pancreatic tissues is somewhat different since claudin 2 is exclusively expressed in duct epithelial cells, whereas claudin 5 is expressed in both duct and acinar cells.
Figure 5. Claudin 5 expression is restricted to tight junctions of the exocrine cells.
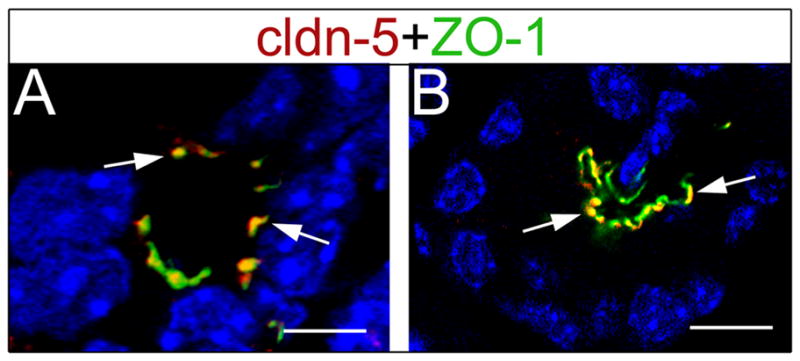
(A) Claudin 5 co-localizes with ZO-1 at tight junctions in ductal cells (arrows) of E18.5 pancreata. (B) Claudin 5 also co-localizes with ZO-1 in acinar cells (arrows) of P5 pancreata. Images were taken with a confocal microscope, and cell nuclei were stained with DAPI (blue). Scale bars: 5 μm (A), 10 μm (B).
Claudin 7
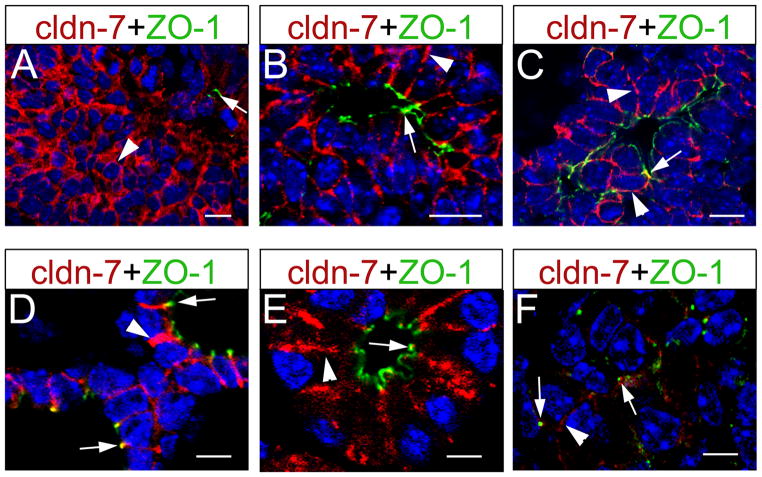
The expression of claudin 7 in mammalian embryonic pancreatic tissues has not been reported. Our qRT-PCR data showed Cldn7 transcript expression in pancreata dissected at E11.5, E15.5 and P7 (Fig. 1 and Table 1). Using confocal microscopy, we uncovered claudin 7 protein expression throughout the membrane of the E10.5 pancreatic epithelium (Fig. 6A) and in both, the lateral membranes and TJs (i.e., in ZO-1+ structures) of E12.5 (Fig. 6B) and E15.5 (Fig. 6C) pancreata. Claudin 7 proteins were also broadly distributed through most of the pancreatic epithelium at E18.5 (data not shown).
Figure 6. Claudin 7 localizes to both tight junctions and the lateral epithelial membrane in developing pancreata.
(A) Claudin 7 (red; arrowhead) is broadly expressed in the lateral membrane of E10.5 pancreatic epithelia (limited ZO-1 expression [green and arrow] indicates absence of tight junctions in this tissue). (B) Claudin 7 proteins localize to tight junctions (ZO1+) in primitive ductal structures (arrow), and to lateral membranes (arrowhead) of most epithelial cells, in E12.5 pancreata. (C) Claudin 7 also localizes to tight junctions of the developing ducts (arrow), and to lateral epithelial membranes (arrowheads), in E15.5 pancreata. (D,E) At P5, most claudin 7 proteins appear distributed in the lateral membranes of ductal (arrowhead in D) and acinar (arrowhead in E) cells, along with limited co-localization with ZO-1 in ductal and acinar tight junctions (arrows in D and E, respectively). (F) Similar distribution of claudin 7 proteins (albeit at lower levels) is observed in islet cells of P5 pancreata (arrows indicate tight junctions; arrowhead indicates the lateral membrane). All images were taken with a confocal microscope, and cell nuclei were stained with DAPI (blue). Scale bars: 10 μm (A–C), 5 μm (D–F).
High claudin 7 expression persisted in the lateral membrane of both ductal cells (Fig. 6D) and acinar cells (Fig. 6E) in P5 pancreata. In these tissues, claudin 7 also co-localized extensively with ZO-1 in TJs of duct epithelial cells, whereas there was limited association of claudin 7 and ZO-1 in acinar TJs (Fig. 6D, E). Low but detectable claudin 7 expression was also noticed in the lateral membrane and possibly the TJs of islet cells (Fig. 6F) in P5 pancreata.
Our finding that a substantial portion of claudin 7 proteins distributes along the lateral membrane of pancreatic epithelial cells is consistent with previous reports describing similar expression of this protein in mammary and bronchial epithelium of mammals (Coyne et al., 2003; Blackman et al., 2005). Arguably, in addition to its potential contribution to TJ function, claudin 7 could have additional roles in the lateral membrane because this protein was shown to associate with cell adhesion proteins in cultured rat pancreatic adenocarcinoma and colorectal tumor lines (Kuhn et al., 2007).
In summary, we uncovered a dynamic expression pattern of some members of the claudin protein family during mouse pancreas organogenesis (Table 2). One group of claudins (1, 3 and 7) was broadly expressed throughout the unpolarized early pancreatic epithelium, but became restricted to TJs (together with ZO-1) when microlumens began to form in those tissues. Loss-of-function experiments should address whether expression of claudin 1, -3 and -7 in progenitors of the pancreas is necessary for TJ formation in this organ.
Table 2.
Cell-type specific expression pattern of claudin 1, -2, -3, -5, and -7 in mouse developing pancreata.
| E10.5 | E12.5 | E15.5 | E18.5 | P5 | |
|---|---|---|---|---|---|
| claudin 1 | + 1 | + 2 | − | − | + 3 |
| claudin 2 | − | − | − | + 4 | + 4 |
| claudin 3 | + 1 | + 2 | + 2 | + 3 | + 3 |
| claudin 5 | − | − | − | + 3 | + 3 |
| claudin 7 | + 1 | + 5 | + 5 | + 6 | + 7 |
Expression pattern:
Lateral membrane of Pdx1+ progenitors
TJs of luminal epithelial cells
TJs of ductal and acinar cells
TJs of ductal cells
Lateral membrane (epithelium) and TJs (lumen)
Lateral membrane and TJs of ductal and acinar cells
Lateral membrane and TJs of ductal, acinar and islet cells
We also found that as organogenesis proceeds, some claudins become allocated to specific cell types in the pancreas. For instance, in postnatal pancreata claudin 2 was restricted to duct epithelial cells, whereas claudin 1, -3 and -5 were expressed in both ductal and acinar cells. On the other hand, islet cells of the postnatal pancreas expressed claudin-7 but not claudin 1, -2, -3 or -5. Interestingly, claudin 7 not only localized to TJs but also was abundantly expressed throughout the periphery of pancreatic epithelial cells. Overall, the specific pattern of claudin expression underscored in our study suggests that these proteins could be important components of morphogenetic processes during pancreas development.
Restricted expression of claudin proteins in pancreatic duct epithelium undergoing neoplastic transformation
Pancreatic Intraepithelial Neoplasias (PanINs, or mPanINs in mouse) are microscopic papillary or flat, noninvasive intraepithelial neoplasms arising in the smaller pancreatic ducts (Hruban et al., 2001). These epithelial entities –representing discrete stages in early pancreatic tumor progression– undergo a series of histological changes that classify them into three grades: PanIN-1A/B, PanIN-2 or PanIN-3 (Hruban et al., 2001). PanINs progress from low to high-grade lesions as the epithelial cells lose polarity and achieve nuclear atypia. Increasing genomic instability due to oncogenic mutations and loss of tumor suppressor function further enhances the potential of these neoplasms to evolve into pancreatic tumors (Hezel et al., 2006).
Variable expression of some CLAUDIN proteins was reported in epithelial cells of PanINs or pancreatic adenocarcinoma (PDA) samples of human origin (Nichols et al., 2004; Hewitt et al., 2006; Borka et al., 2007). These results were consistent with the notion that cancer progression is accompanied by a disruption in tight junctions, and suggested that both the levels and the claudin expression pattern could have diagnostic value (Morin, 2005).
Our finding that claudin 1, -2, -3, -5 and -7 had distinctive expression patterns in developing pancreatic epithelia prompted us to investigate the expression of these proteins in similar tissues undergoing neoplastic transformation. Two mouse models were used for this purpose: Pdx1Cre;LSL-KrasG12D mice and Ptf1aCre;LSL-KrasG12D mice, which express an oncogenic form of Kras (KrasG12D) throughout the pancreas and consequently, develop lesions recapitulating the entire spectrum from PanIN to invasive cancer (Hingorani et al., 2003). Our study also analyzed the expression of claudin-18 in pancreata of Pdx1Cre;LSL-KrasG12D and Ptf1aCre;LSL-KrasG12D mice, because this protein was recently identified as a novel marker of PanINs and infiltrating PDA in both, humans and mice (Karanjawala et al., 2008; Sanada et al., 2010; Ray et al., 2011).
Claudin expression in PanIN-1A, PanIN-1B and PanIN-2 lesions
H&E staining identified numerous PanIN-1A lesions in the pancreas of both, Pdx1Cre;LSL-KrasG12D and Ptf1aCre;LSL-KrasG12D mice that were about 1 year old. These neoplasms had flat, columnar epithelium, basally located nuclei and abundant cytoplasm (Fig. 7A). H&E staining results also identified mPanIN-1B lesions –with characteristic papillary or pseudostratified architecture (Fig. 7H)– and a few mPanIN-2 lesions –with both, papillary cytoarchitecture and features indicative of nuclear atypia, including loss of polarity and nuclear crowding (Fig. 7O)– in Pdx1Cre;LSL-KrasG12D and Ptf1aCre;LSL-KrasG12D pancreata.
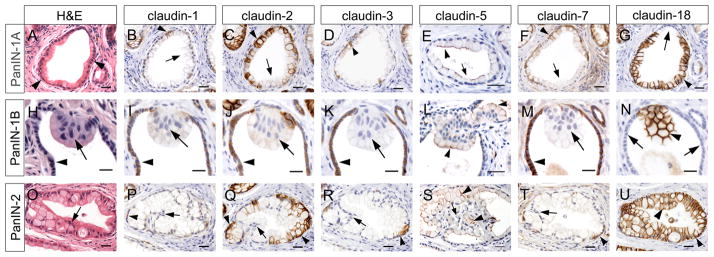
Figure 7. Variable expression of claudin proteins in pancreatic epithelia undergoing neoplastic transformation.
(A) H&E staining reveals tall cells accumulating cytoplasm (arrowheads) in PanIN-1A lesions. (H) Papillary architecture (arrow) of epithelial cells identifies a PanIN-1B lesion within a relatively normal duct (arrowhead). (O) Loss of nuclear polarity (arrow) indicative of more advanced transformation is observed in a PanIN-2 lesion. (B,D) Only very few cells express claudin 1 (arrowhead in B) or claudin 3 (arrowhead in D) in PanIN-1A lesions. (C,E,F) Claudin 2 (C), -5 (E), and -7 (F) proteins are detected in some PanIN-1A lesions (arrowheads), but not all neoplastic epithelial cells (arrows). (G) Claudin 18 expression (arrowheads) is broadly detected in epithelia of PanIN-1A lesions (arrowheads), but not in epithelial cells retaining normal ductal morphology (arrow). (I,J,K,M) Epithelial cells of a PanIN-1B lesion are devoid of claudin 1 (I), -2 (J), -3 (K), and -7 (M) protein expression (arrows). Adjacent normal ductal epithelial cells are positive for these claudin proteins (arrowheads). (L,N) Expression of claudin 5 (L) and claudin 18 (N) is noticed (arrowhead) in epithelia of PanIN-1B neoplasms (notice lack of claudin-18 expression in the normal duct epithelium, arrows in N). (P,R,T) Very few cells retaining basolateral nuclear polarity express claudin 1 (P), -3 (R), and -7 (T) in PanIN-2 lesions (arrowheads). In contrast, epithelial regions showing nuclear atypia are devoid of these proteins (arrows). (Q) Claudin 2 is expressed in regions of polarized nuclei (arrowheads), but absent where nuclear atypia is observed (arrow) in PanIN-2 epithelia. (S) Few cells retain claudin 5 expression (arrowheads) in PanIN-2 neoplasms (arrow indicate lack of claudin 5 immunoreactivity). (U) Claudin-18 expression (arrowheads) is broadly detected in epithelia of PanIN-2 lesions. (A–D,F,G), (H–K,M,N) and (O–R,T,U) respectively represent adjacent sections from the same pancreatic specimen. (E,L,S) are sections from the same pancreatic specimen. Cell nuclei were stained with hematoxylin. Scale bars: 10 μm (H–K, M,N), and 25 μm (A–G,L, O–U).
Immunohistochemistry results uncovered broad expression of claudin 2 (Fig. 7C) and claudin 18 proteins (Fig. 7G) in mPanIN-1A neoplasms, although claudin 18 proteins had more prominent distribution throughout these lesions compared to claudin 2 proteins. Interestingly, claudin 18 was not detected in portions of the PanIN-1A epithelium with normal ductal morphology (arrow in Fig 7G). Similar to these results, PanIN-1A lesions of human origin abundantly expressed both CLAUDIN 2 (Suppl. Fig. 2A,C) and CLAUDIN 18 (Suppl. Fig. 2B,D) proteins. Our results showing specific activation of claudin-18 expression in epithelial cells of early PanINs validate that this protein is a unique, reliable marker of early neoplastic transformation in pancreatic tissues of both, mice and humans (Karanjawala et al., 2008; Sanada et al., 2010; Ray et al., 2011).
The mPanIN-1A epithelium also displayed claudin 5 (Fig. 7E) and claudin 7 (Fig. 7F) immunoreactivity, although these proteins were not distributed as extensively as were claudin 2 and claudin 18. In contrast, only a few epithelial cells in mPanIN-1A neoplasms expressed claudin 1 (Fig. 7B) and claudin 3 (Fig. 7D). These data indicate that a distinctive pattern of claudin protein expression is established during early neoplastic transformation in the pancreas, which is different from that of normal pancreatic epithelia.
Similar to mPanIN-1A, the papillary epithelium that identifies mPanIN-1B lesions (Figs. 7H) was positive for claudin 5 (Fig. 7L) and claudin 18 (Fig. 7N) proteins. On the other hand, PanIN-1B neoplasms were completely devoid of claudin 1 (Fig. 7I), -2 (Fig. 7J), -3 (Fig. 7K) and 7 (Fig. 7M) immunoreactivity (arrows in Fig. 7I,J,K,M), while expression of these proteins was clearly noticeable in the normal ductal epithelium surrounding those lesions (arrowheads in Fig. 7I,J,K,M). Therefore, downregulation of claudin 1, -2, -3, and -7 appears to accompany the PanIN-1A to PanIN-1B transition.
We uncovered extensive claudin 18 protein expression throughout the entire mPanIN-2 epithelium (Fig. 7U). Claudin 5 proteins were also detected in mPanIN-2 neoplasms, albeit only in areas where nuclear atypia was not very prominent (Fig. 7S, arrowhead). Some areas in mPanIN-2 lesions also displayed claudin 2 immunoreactivity (Fig. 7Q, arrowhead), although expression of this protein was restricted to epithelial cells that retained cell polarity. In contrast, all mPanIN-2 lesions completely lacked or were nearly devoid of claudin 1 (Fig. 7P), -3 (Fig. 7R) and -7 proteins (Fig. 7T). These collective data suggest that changing the repertoire of claudin proteins could be important for neoplastic progression in the pancreas.
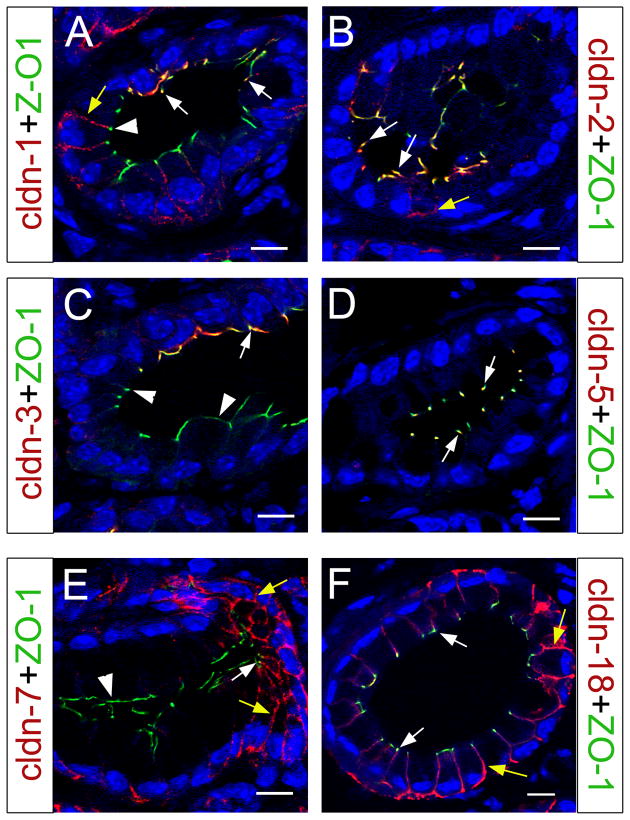
We used confocal microscopy to identify the subcellular localization of claudin 1, -2, -3, -5, -7 and -18 in mPanIN-1A neoplasms. Different from postnatal pancreata, where claudin-1 and claudin 2 proteins exclusively localized to TJs (Table 2 and Figs. 2 and 3), these proteins were found associated with both TJs (arrows in Fig. 8A,B) and the lateral epithelial membrane (yellow arrows in Fig. 8A,B) in mPanIN-1A lesions. Interestingly, in mPanIN-1A areas with notorious cytoplasm accumulation claudin 1 proteins were detected in the lateral membrane but not in TJs (arrowhead in Fig. 8A). Confocal microscopy results also revealed limited distribution of claudin 3 proteins in TJs (arrow in Fig. 8C) of the mPanIN-1A epithelium. These results are different from postnatal pancreata because in these tissues claudin 3 broadly co-localized to TJs of ductal and acinar cells (Fig. 4G,H). On the other hand, similar to postnatal pancreata (Fig. 5) claudin 5 was extensively distributed in TJs of the mPanIN-1A epithelium (Fig. 8D).
Figure 8. Subcellular localization of claudin proteins in PanIN-1A lesions.
(A) PanIN-1A lesion showing claudin 1 co-localization with ZO-1 in TJs of cells with normal morphology (arrows). In more transformed cells with abundant cytoplasm claudin 1 does not co-express with ZO-1 (arrowhead) but appears distributed to the lateral membrane (yellow arrow). (B) Claudin 2 mainly localizes to TJs (white arrows) in PanIN-1A epithelia, although a few cells also exhibit lateral distribution of this protein (yellow arrow). (C) Claudin 3 proteins specifically localize to TJs (white arrow) of normal appearing PanIN-1A epithelial cells (arrow; arrowheads indicate portions of the transformed epithelium devoid of claudin 3 expression). (D) Claudin 5 is detected in all TJs in the PanIN-1A epithelium (white arrows). (E) Claudin 7 proteins localize to both TJs (white arrow) and the lateral membrane (yellow arrows) in portions of the PanIN-1A epithelium, whereas is absent in TJs of cells with abundant cytoplasm (arrowhead). (F) Claudin 18 proteins extensively localize throughout the PanIN-1A epithelium, both in TJs (white arrows) and in the lateral membrane (yellow arrows). All images were taken with a confocal microscope, and cell nuclei were stained with DAPI (blue). Scale bar: 10 μm.
This study uncovered expression of claudin-7 in both TJs and the lateral membrane of pancreatic cells at postnatal stages (Fig. 6). We noticed similar subcellular localization of claudin 7 in portions of the PanIN-1 epithelium with relatively normal appearance (yellow and white arrows in Fig. 8E). In contrast, in PanIN-1A areas with abundant cytoplasm claudin 7 was not detected in TJs (arrowhead in Fig. 8E), and was only weakly expressed in the lateral cellular membrane (Fig. 8E). Finally, when we looked at claudin 18 subcellular localization in mPanIN-1A epithelia we found that compared to all the previous claudins, this protein had the most extensive subcellular distribution in both TJs (white arrows in Fig. 8F) and the lateral membrane (yellow arrows in Fig. 8F).
In summary, we found that both the abundance and the subcellular distribution of specific claudin proteins are different between normal and transformed pancreatic epithelia. These results raise the possibility that changes in paracellular permeability accompany the formation of PanIN neoplasms.
Conclusion
This study uncovered a remarkable plasticity in the expression and distribution of claudin proteins in pancreatic epithelia undergoing morphogenesis or neoplastic transformation. In the mature mammalian kidney, claudin proteins have different expression patterns along the epithelium of each nephronic segment, and experimental evidence indicates that this differential expression provides the renal epithelium with unique paracellular permeability properties (Angelow et al., 2008). Our developmental results thus suggest that allocating different claudin proteins to epithelial TJs could be important to confer specific permeability properties to acinar, ductal or islet cells. Loss-of-function or gain-of-function studies in mice will be needed to underscore the role of individual claudins or combinations of these proteins in TJ assembly and function in pancreatic epithelia. Likewise, our results also support investigating the potential role of claudin function during pancreatic neoplastic transformation.
Experimental Procedures
Animals
RNA isolation was performed on C57BL/6 wild-type mice. Immunostaining was performed on NMRI/C57BL6 wild-type mice. Ptf1a-Cre mice (Kawaguchi et al., 2002), Pdx1Cre mice (Gu et al., 2002), and LSL-Kras mice (Hingorani et al., 2003) were maintained on a C57BL6 background. All experiments were performed with approval from the St. Jude Children’s Research Hospital Animal Care and Use Committee and the Vanderbilt Institutional Animal Care and Use Committee.
Processing of embryos and pancreatic tissues
Immunostaining of frozen and paraffin sections was performed as previously described (Westmoreland et al., 2009), except antigen retrieval of paraffin slides was performed using the Antigen 2100 Retriever and R-buffer A (Electron Microscopy Sciences). De-identified human tissues were obtained from the Vanderbilt Human Tissue Acquisition and Pathology Resource, sectioned at a depth of 5 μm, deparaffinized, retrieved in 100 mM Tris, pH 10 in a Cuisinart pressure cooker with high pressure for 15 minutes and high heat for a total of 2 h. After cooling, slides were washed in PBS, blocked in 5% donkey serum, and incubated with the indicated antibodies.
Immunohistochemical analysis
Supplemental Table 1 indicates the primary antibodies used in this study. The secondary antibodies (diluted 1:250) were: Cy3-conjugated donkey anti-rabbit IgG (Jackson); Cy3-conjugated donkey anti-goat IgG (Jackson); Alexa 488-conjugated donkey anti-rat IgG (Molecular Probes); Alexa 488-conjugated goat anti-guinea pig IgG (Molecular Probes); Alexa 488-conjugated donkey anti-goat IgG (Molecular Probes); Alexa 488-conjugated donkey anti-sheep IgG (Molecular Probes). Biotinylated donkey anti-rabbit IgG antibodies were detected by using the VECTASTAIN Elite ABC kit (Vector Laboratories). Prolong Gold with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) was used for nuclear staining. Images were obtained with a Zeiss Axioskop 2 microscope, or with a confocal/Multiphoton laser-scanning Zeiss LSM 510 META microscope.
Quantitative RT-PCR
For embryonic stages, three individual pools of 10 pancreata each, dissected at E11.5 or E15.5, and a pool of 10 stomachs dissected at E15.5, were used. Three individual postnatal day 7pancreata, and one P5 kidney were used. Total cellular RNA was extracted from pancreatic tissues by TRizol reagent and PureLink mini kit (Invitrogen). cDNA synthesis was performed using 1 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen) and random hexamers according to the manufacturer’s instructions. For quantification of Cldn mRNA levels (quantitative RT-PCR), we performed Sybr Green PCR using an ABI PRISM 7900HT sequence detection system (Perkin-Elmer). Cldn expression levels were normalized against the expression of Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), using the ΔΔ ct method for comparing relative expression levels.
Supplementary Material
(A) QRT-PCR results showing the relative abundance of Cldn transcripts in the E15.5 mouse stomach. Immunohistochemistry results corroborate abundant expression of claudin 18 in the posterior stomach epithelium at E15.5. (B) QRT-PCR analysis was performed with specific primers recognizing all Cldn family members and RNA from P5 kidney. One pool of E15.5 stomachs and one P5 kidney were used for qRT-PCR. (A) Cell nuclei were stained with methyl green. Scale bar: 100 μm.
(A,C) CLAUDIN 2 protein expression (arrowheads) is restricted to some areas of the PanIN-1A epithelium (arrowheads) from human pancreatic specimens (arrows show that the papillary infoldings are devoid of CLAUDIN 2 proteins). (B,D) CLAUDIN 18 proteins are expressed in all epithelial cells (arrowheads in B,D), including the papillary infoldings (arrow in B), in human PanIN-1A lesions. (A–D) Cell nuclei were counter-stained with hematoxylin. Scale bar: 50 μm.
Acknowledgments
We thank C. Wright for the generous gift of the anti-Pdx1 antibody, Y. Ouyang and the Cell and Tissue imaging facility (St. Jude) for help with the confocal/multiphoton microscopy, and the Hartwell Center for Bioinformatics and Biotechnology (St. Jude) for RNA quality analysis. The National Institute of Diabetes and Digestive and Kidney Diseases (grant DK060542, B.S.-P.), The National Cancer Institute (grants CA123061, ALM; P50CA095103, ALM and MKW; and P30DK058404 MKW), and the American Lebanese Syrian Associated Charities (ALSAC) supported these studies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FINANCIAL SUPPORT
National Institute of Diabetes and Digestive and Kidney Diseases, NIH (grant DK060542 [BS-P]); the American Lebanese Syrian Associated Charities (BS-P); and the National Cancer Institute, NIH (grants CA123061 [ALM], P50CA095103 [ALM and MKW], and P30DK058404 [MKW]).
References
- Amasheh S, Meiri N, Gitter AH, Schoneberg T, Mankertz J, Schulzke JD, Fromm M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J Cell Sci. 2002;115:4969–4976. doi: 10.1242/jcs.00165. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Van Itallie CM. Physiology and Function of Tight Junctions. In: Nelson WJ, Fuchs E, editors. Cell-Cell Junctions. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2010. pp. 123–138. [Google Scholar]
- Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, Guseh JS, Rajagopal J, Melton DA. Genetic targeting of the endoderm with claudin-6CreER. Dev Dyn. 2008;237:504–512. doi: 10.1002/dvdy.21437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung PP, Mitani Y, Sanada Y, Nakayama H, Matsusaki K, Yasui W. Differential expression of claudin-2 in normal human tissues and gastrointestinal carcinomas. Virchows Arch. 2006;448:428–434. doi: 10.1007/s00428-005-0120-2. [DOI] [PubMed] [Google Scholar]
- Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- Blackman B, Russell T, Nordeen SK, Medina D, Neville MC. Claudin 7 expression and localization in the normal murine mammary gland and murine mammary tumors. Breast Cancer Res. 2005;7:R248–255. doi: 10.1186/bcr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borka K, Kaliszky P, Szabo E, Lotz G, Kupcsulik P, Schaff Z, Kiss A. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 2007;450:549–557. doi: 10.1007/s00428-007-0406-7. [DOI] [PubMed] [Google Scholar]
- Brizuela BJ, Wessely O, De Robertis EM. Overexpression of the Xenopus tight-junction protein claudin causes randomization of the left-right body axis. Dev Biol. 2001;230:217–229. doi: 10.1006/dbio.2000.0116. [DOI] [PubMed] [Google Scholar]
- Cereijido M, Contreras RG, Flores-Benitez D, Flores-Maldonado C, Larre I, Ruiz A, Shoshani L. New diseases derived or associated with the tight junction. Arch Med Res. 2007;38:465–478. doi: 10.1016/j.arcmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Comper F, Antonello D, Beghelli S, Gobbo S, Montagna L, Pederzoli P, Chilosi M, Scarpa A. Expression pattern of claudins 5 and 7 distinguishes solid-pseudopapillary from pancreatoblastoma, acinar cell and endocrine tumors of the pancreas. Am J Surg Pathol. 2009;33:768–774. doi: 10.1097/PAS.0b013e3181957bc4. [DOI] [PubMed] [Google Scholar]
- Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1166–1178. doi: 10.1152/ajplung.00182.2003. [DOI] [PubMed] [Google Scholar]
- D’Souza T, Sherman-Baust CA, Poosala S, Mullin JM, Morin PJ. Age-related changes of claudin expression in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med Sci. 2009;64:1146–1153. doi: 10.1093/gerona/glp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, Berger AE, Zhang K, Vidyasagar S, Yoshida T, Boguniewicz M, Hata T, Schneider LC, Hanifin JM, Gallo RL, Novak N, Weidinger S, Beaty TH, Leung DY, Barnes KC, Beck LA. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127:773–786. e771–777. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M. Molecular Basis of the Core Structure of Tight Junctions. In: Nelson WJ, Fuchs E, editors. Cell-Cell Junctions. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2010. pp. 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapin-Botton A. Ductal cells of the pancreas. Int J Biochem Cell Biol. 2005;37:504–510. doi: 10.1016/j.biocel.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Guney MA, Gannon M. Pancreas cell fate. Birth Defects Res C Embryo Today. 2009;87:232–248. doi: 10.1002/bdrc.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta IR, Ryan AK. Claudins: unlocking the code to tight junction function during embryogenesis and in disease. Clin Genet. 2010;77:314–325. doi: 10.1111/j.1399-0004.2010.01397.x. [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hick AC, van Eyll JM, Cordi S, Forez C, Passante L, Kohara H, Nagasawa T, Vanderhaeghen P, Courtoy PJ, Rousseau GG, Lemaigre FP, Pierreux CE. Mechanism of primitive duct formation in the pancreas and submandibular glands: a role for SDF-1. BMC Dev Biol. 2009;9:66. doi: 10.1186/1471-213X-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS, Luttges J, Offerhaus GJ. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Illei PB, Ashfaq R, Infante JR, Murphy K, Pandey A, Schulick R, Winter J, Sharma R, Maitra A, Goggins M, Hruban RH. New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol. 2008;32:188–196. doi: 10.1097/PAS.0b013e31815701f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kesavan G, Sand FW, Greiner TU, Johansson JK, Kobberup S, Wu X, Brakebusch C, Semb H. Cdc42-mediated tubulogenesis controls cell specification. Cell. 2009;139:791–801. doi: 10.1016/j.cell.2009.08.049. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zoller M. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- Kulka J, Szasz AM, Nemeth Z, Madaras L, Schaff Z, Molnar IA, Tokes AM. Expression of tight junction protein claudin-4 in basal-like breast carcinomas. Pathol Oncol Res. 2009;15:59–64. doi: 10.1007/s12253-008-9089-x. [DOI] [PubMed] [Google Scholar]
- Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol. 2007;312:509–522. doi: 10.1016/j.ydbio.2007.09.049. [DOI] [PubMed] [Google Scholar]
- Nemeth Z, Szasz AM, Tatrai P, Nemeth J, Gyorffy H, Somoracz A, Szijarto A, Kupcsulik P, Kiss A, Schaff Z. Claudin-1, -2, -3, -4, -7, -8, and -10 protein expression in biliary tract cancers. J Histochem Cytochem. 2009;57:113–121. doi: 10.1369/jhc.2008.952291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol. 2004;121:226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Rahner C, Mitic LL, Anderson JM. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology. 2001;120:411–422. doi: 10.1053/gast.2001.21736. [DOI] [PubMed] [Google Scholar]
- Ray KC, Bell KM, Yan J, Gu G, Chung CH, Washington MK, Means AL. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One. 2011;6:e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada Y, Hirose Y, Osada S, Tanaka Y, Takahashi T, Yamaguchi K, Yoshida K. Immunohistochemical study of claudin 18 involvement in intestinal differentiation during the progression of intraductal papillary mucinous neoplasm. Anticancer Res. 2010;30:2995–3003. [PubMed] [Google Scholar]
- Schock F, Perrimon N. Molecular mechanisms of epithelial morphogenesis. Annu Rev Cell Dev Biol. 2002;18:463–493. doi: 10.1146/annurev.cellbio.18.022602.131838. [DOI] [PubMed] [Google Scholar]
- Schulzke JD, Ploeger S, Amasheh M, Fromm A, Zeissig S, Troeger H, Richter J, Bojarski C, Schumann M, Fromm M. Epithelial tight junctions in intestinal inflammation. Ann N Y Acad Sci. 2009;1165:294–300. doi: 10.1111/j.1749-6632.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- Tsukahara M, Nagai H, Kamiakito T, Kawata H, Takayashiki N, Saito K, Tanaka A. Distinct expression patterns of claudin-1 and claudin-4 in intraductal papillary-mucinous tumors of the pancreas. Pathol Int. 2005;55:63–69. doi: 10.1111/j.1440-1827.2005.01793.x. [DOI] [PubMed] [Google Scholar]
- Westmoreland JJ, Wang Q, Bouzaffour M, Baker SJ, Sosa-Pineda B. Pdk1 activity controls proliferation, survival, and growth of developing pancreatic cells. Dev Biol. 2009;334:285–298. doi: 10.1016/j.ydbio.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox ER, Burton QL, Naz S, Riazuddin S, Smith TN, Ploplis B, Belyantseva I, Ben-Yosef T, Liburd NA, Morell RJ, Kachar B, Wu DK, Griffith AJ, Friedman TB. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–172. doi: 10.1016/s0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U, Kroesen AJ, Zeitz M, Fromm M, Schulzke JD. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Piontek J, Wolburg H, Piehl C, Liss M, Otten C, Christ A, Willnow TE, Blasig IE, Abdelilah-Seyfried S. Establishment of a neuroepithelial barrier by Claudin5a is essential for zebrafish brain ventricular lumen expansion. Proc Natl Acad Sci U S A. 2010;107:1425–1430. doi: 10.1073/pnas.0911996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) QRT-PCR results showing the relative abundance of Cldn transcripts in the E15.5 mouse stomach. Immunohistochemistry results corroborate abundant expression of claudin 18 in the posterior stomach epithelium at E15.5. (B) QRT-PCR analysis was performed with specific primers recognizing all Cldn family members and RNA from P5 kidney. One pool of E15.5 stomachs and one P5 kidney were used for qRT-PCR. (A) Cell nuclei were stained with methyl green. Scale bar: 100 μm.
(A,C) CLAUDIN 2 protein expression (arrowheads) is restricted to some areas of the PanIN-1A epithelium (arrowheads) from human pancreatic specimens (arrows show that the papillary infoldings are devoid of CLAUDIN 2 proteins). (B,D) CLAUDIN 18 proteins are expressed in all epithelial cells (arrowheads in B,D), including the papillary infoldings (arrow in B), in human PanIN-1A lesions. (A–D) Cell nuclei were counter-stained with hematoxylin. Scale bar: 50 μm.