Abstract
Objective
Cia3 is a locus on rat chromosome 4 that regulates severity and joint damage in collagen and pristane-induced arthritis (CIA and PIA). This study aimed to refine the Cia3 gene-containing interval towards gene identification and obtain insights into its mode of action.
Methods
Five DA.F344(Cia3) subcongenic strains were generated and studied in PIA and CIA. Levels of antibodies against type II collagen (both allo- and autoantibodies) were measured. Joints and synovial tissues were collected 32 days after the induction of PIA (chronic stage) for histology and qPCR for IL-1β and matrix metalloproteases (MMPs).
Results
Three subcongenics sharing the centromeric Cia3d interval were protected, while two subcongenics sharing the telomeric Cia3g interval, which did not overlap with Cia3d, were also protected, developing significantly less severe CIA and PIA. DA.F344(Cia3) and DA.F344(Cia3d) congenics with PIA preserved a normal joint architecture, while DA rats had pronounced synovial hyperplasia, angiogenesis, inflammatory infiltration, bone or cartilage erosions. DA.F344(Cia3d) and DA.F344(Cia3g) strains had significantly lower synovial levels of IL-1β (5-fold), MMP-1 (expressed predominantly in DA), MMP-3 (79-fold) and MMP-14 (21-fold) and reduced levels of pathogenic autoantibodies against type II collagen, compared with DA.
Conclusions
We have identified two new arthritis severity and articular damage loci within Cia3. These loci regulate pathogenic processes in two different models of RA, and the identification of these genes has the potential to generate new targets for therapies aimed at reducing disease severity and articular damage, and for prognostication in RA.
Keywords: Autoimmunity, Rheumatoid arthritis, animal model, erosion, IL-1β
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic autoimmune disease with an overall prevalence of 0.5–1% in most populations. RA has a strong genetic component (1), and several MHC and non-MHC genes have been recently associated with disease susceptibility (2, 3). In contrast to susceptibility, very little is known about the genetic regulation of disease severity and joint damage in RA (4), and the available multi-institutional family-based or case-control cohorts used in genome-wide studies were not specifically designed to address this issue. We consider that the identification of genes specifically involved in the regulation of disease severity and joint damage have a greater potential to generate useful targets for the development of new therapies aimed at preserving joint architecture and function.
We have previously identified several disease severity and articular damage quantitative trait loci (QTL) in rat models of RA (5–7). One of the identified QTLs, Cia3, was mapped to rat chromosome 4 in an intercross between the arthritis-susceptible DA and arthritis-resistant F344 rat strains studied for collagen-induced arthritis (CIA). Studies in congenic rats where the F344-derived arthritis resistance alleles at Cia3 were introduced into the arthritis-susceptible DA strain genome background, as in DA.F344(Cia3) congenic rats, determined that Cia3 also regulates arthritis severity in pristane, oil (6) and adjuvant-induced arthritis (8) (PIA, OIA and AIA, respectively). Cia3 co-localizes with QTLs involved in the regulation of arthritis in other rat intercrosses (9–11), and in other models of autoimmune diseases in rats and mice (12, 13). The Cia3 syntenic regions in the human genome also contain loci regulating different forms of autoimmune diseases (12, 13), including RA (14), suggesting that it harbors genes relevant not only to RA, but possibly to other diseases as well.
In order to localize and reduce the interval containing the arthritis-regulatory gene, and to characterize its regulatory effects in arthritis severity, joint histology and synovial tissue cytokine gene expression, and production of autoantibodies against collagen, Cia3 subcongenics were generated and studied for their susceptibility to and severity of PIA and CIA, two well-established models of autoimmune erosive arthritis. In the present study we describe the discovery that Cia3 is accounted for by at least two different genes that operate independently to regulate disease severity.
MATERIAL AND METHODS
Rats
Specific pathogen-free DA (DA/BklArb) (arthritis-susceptible) and F344 (F344/Hsd, Harlan, Indianapolis, IN) (arthritis-resistant) inbred rat strains were used in the breeding of the congenic and subcongenic strains. DA rats were originally purchased from Bantin & Kingman, Inc. (Fremont, CA), maintained at the Arthritis and Rheumatism Branch, NIAMS, NIH, and then transferred to the Feinstein Institute for Medical Research (FIMR) (former North Shore-LIJ Research Institute) (DA/BklArbNsi) and used as controls. All the experiments involving animals were reviewed and approved by the FIMR Institutional Animal Care and Use Committee.
Construction of the genotype-guided Cia3 QTL-congenic and subcongenic lines
A 75.47 Mb interval, containing the original 35 cM two logarithm of odds (LOD) support interval comprising Cia3, was introgressed from F344 into the DA rats through eight backcrosses (BC8) followed by at least five intercrosses, as previously described (6). Subcongenic lines covering the Cia3 interval (Figure 1) were generated for the present study. DA.F344(Cia3) congenics were backcrossed with DA rats to generate offspring heterozygous at the congenic interval. These heterozygous offspring were further backcrossed with DA, and the offspring screened for new recombinants within the Cia3 interval (see SSLP markers used on Figure 1). Offspring (BC10) heterozygous at identical recombinant segments, based on SSLP markers, were brother-sister mated, and their offspring (BC10F1) genotyped to ensure homozygozity at the expected intervals.
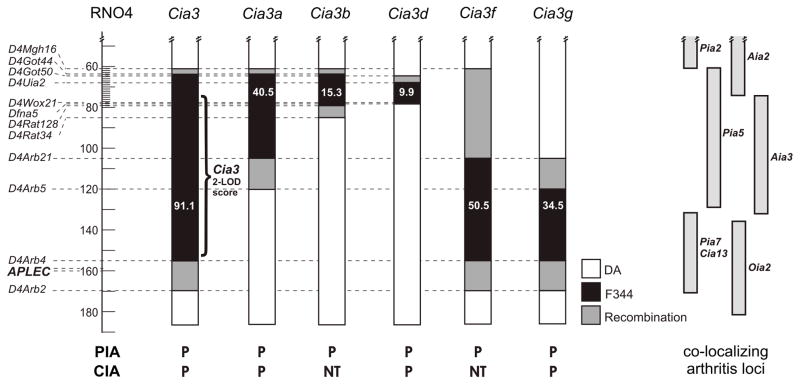
Figure 1. Markers used in the breeding of DA.F344(Cia3) congenic and subcongenics.
Numbers indicate interval distance in megabases (Mb) (http://www.ensembl.org/Rattus_norvegicus/). Black filling indicates homozygous F344 alleles (F/F), white filling indicates homozygous DA alleles (D/D), and grey area indicates the region where recombination took place. Right side of the figure shows co-localizing arthritis loci. PIA=pristane-induced arthritis; CIA= collagen-induced arthritis; AIA=adjuvant-induced arthritis; OIA=oil-induced arthritis. P=protected; NT=not tested.
Homozygous subcongenics were used to expand the subcongenic lines. Experiments were done with offspring from second to fifth intercrosses (BC10F2-F7, and Cia3d experiments were further confirmed with BC12F2-F5).
Genotyping
Tail tips were excised from 3–4 week-old rats, and DNA extracted with the DNeasy kit (Qiagen, Valencia, CA). PCR conditions have been previously reported, and were set up in 10μl reactions (15). GENESCAN 3.1 software (ABI) was used for fluorescent-labeled PCR products’ data extraction and allele assignment. All genotypes were manually checked by two readers and questionable readings re-checked or repeated. For marker details, see the Rat Genetic Database (http://www.niams.nih.gov/rtbc/ratgbase/index.htm) and the Rat Genome Database (http://www.rgd.mcw.edu).
Induction of PIA
Eight to twelve week-old rats received 150 μl of pristane (2,6,10,14-tetramethylpentadecane, SIGMA-Aldrich Chemical Co., Milwaukee, WI) by intradermal injection (day zero) (6, 16, 17). The dose was divided in two injection sites at the base of the tail.
Induction of CIA
Bovine type II collagen (BII; Chondrex, Redmond, WA) was dissolved overnight in 0.1N acetic acid at 4°C (2mg/ml) and emulsified with incomplete Freund’s adjuvant (IFA, Difco, Detroit, MI) to a final concentration of 1mg/ml. Eight to twelve-week old rats were injected intradermally at the base of the tail with 2mg/kg weight of BII divided into six injection sites (day zero), and a booster injection of 100μg BII/IFA administered on day seven (18). Serum was obtained on day 18 and stored at −80°C until tested.
Arthritis scoring
We used a previously described arthritis scoring system (5, 15, 18) that evaluates individual joints and measures arthritis severity according to joint size as follows: a) interphalangeal, metacarpophalangeal and metatarsophalangeal joints in each one of the four lateral digits were scored 0=no arthritis; 1=arthritis present; b) wrist, mid-forepaw, ankle and midfoot joints were scored 0=normal; 1=minimal swelling; 2=moderate swelling; 3=severe swelling; 4=severe swelling and non-weight bearing. The scores from all involved joints were added (maximum score per rat=80). The same observer obtained the arthritis scores on days 0, 14, 18, 21, 24, 28 and 31 following induction. The arthritis severity index (ASI), which is a measure of disease severity over time (area under the curve), was determined for each animal by adding the individual arthritis scores obtained over the course of the experiment. We have previously shown that the ASI correlates with histological changes and damage (7, 17).
Histology and histological scoring
At the end of the arthritis observation period (day 32), the right hind paw was fixed in 10% formaldehyde. Paws were then decalcified with a solution containing hydrochloric acid and 0.1M EDTA (Cal-Ex, Fisher Scientific, Fairlawn, NJ). Tissues were sectioned, embedded in paraffin, and slides prepared and stained with hematoxylin-eosin and safranin-O. Two slides per rat were scored without knowledge of strain identity. We used a recently described comprehensive histological scoring system (17). Briefly, tibio-talar, talus-calcaneal and midfoot joints were histologically scored for the following parameters:
Synovial inflammation. Five high-power magnification fields (HMF) were scored for the percentage of infiltrating mononuclear cells as follows: 0=absent; 1=mild (1–10%); 2=moderate (11–50%); 3=severe (51–100%). The mean of the five HMF was used for analyses.
Synovial hyperplasia. 0=absent; 1=mild (5–10 layers); 2=moderate (11–20 layers); 3=severe (>20 layers).
Extension of pannus formation based on the reader’s impression. 0=absent; 1=mild; 2=moderate; 3=severe.
Synovial fibrosis. 0=absent; 1=mild (1–10%); 2=moderate (11–50%); 3=severe (51–100%).
Synovial vascularity (angiogenesis). The number of vessels was counted in five HMF of synovial tissue, and the mean used for analyses.
Cartilage erosion. Percentage of the cartilage surface that was eroded: 0=absent; 1=mild (1–10%); 2=moderate (11–50%); 3=severe (51–100%).
Cartilage degradation. Based on safranin-O staining of proteoglycans, and described as the percentage of the cartilage that lost its staining: 0=none; 1=mild loss (1–10%); 2=moderate loss (11–50%); 3=severe loss (51–100%).
Bone erosion. 0=none; 1=minor erosion(s) observed only at HMF; 2=moderate erosion(s) observed at low magnification; 3=severe transcortical erosion(s).
Quantitative real-time PCR (qPCR)
Ankle synovial tissue obtained after the completion of the arthritis observation period (day 32) was immediately frozen in liquid nitrogen. Tissues were subsequently homogenized and total RNA isolated with the RNeasy Kit (Qiagen) and digested with DNase (Qiagen), according to the manufacturer’s protocol. 200ng of total RNA from each sample were used for cDNA synthesis (SUPERSCRIPT II Kit, Invitrogen, Carlbad, CA). qPCR, methodology, primers and probes used for IL-1β, matrix metalloprotease-1 (MMP-1), MMP-3 and MMP-14, as well as GAPDH have been previously reported (7, 17, 19). Briefly, cDNA was optimized for relative gene expression by qPCR. TaqMan (ABI) 5′exonuclease assay and Roche Universal Probe Library (Roche) were used for qPCR. GAPDH was used as endogenous control. PCR reaction mixture contained 1X mastermix (Eurogentec, San Diego, CA), 200nM of forward and reverse primers, 100nM of gene-specific probes, and 150–200ng of cDNA. All samples were run in duplicates in an ABI 7700 Sequence Detection System (ABI), and the mean used for the analyses. Ct (threshold cycle) values were obtained and analyzed with the Sequence Detection System (SDS) software version 1.9.1 (ABI). The Ct value is inversely related to the starting template copy number. Relative expression in synovial tissues was adjusted for GAPDH in each sample (ΔCt). ΔCt values were compared using the t-test. Fold-change differences in gene expression between DA and subcongenics were compared with the 2−ΔΔCt method (20).
Measurement of anti-collagen antibodies
Serum samples collected on day 18 after the induction of CIA were assayed for IgG antibodies against bovine and against rat type II collagen using commercially available ELISA Kits (Chondrex), according to the manufacturer’s instructions, and results shown as IgG U/ml.
Statistical analyses
Males and females were initially studied separately for their arthritis severity, and in the absence of gender-specific effects were then combined for analysis. Non-normally distributed data were compared with ANOVA on ranks with a pairwise multiple comparison procedure (Dunn’s method) for multiple groups, or with the Mann-Whitney test for two group comparisons. The t-test was used to compare normally distributed data (qPCRs). A p-value of 0.05 or less was regarded as significant. All statistical analyses were done with SigmaStat 3.0 (SPSS, Chicago, IL).
RESULTS
DA.F344(Cia3) congenic rats are protected and develop a significantly milder form of PIA and CIA
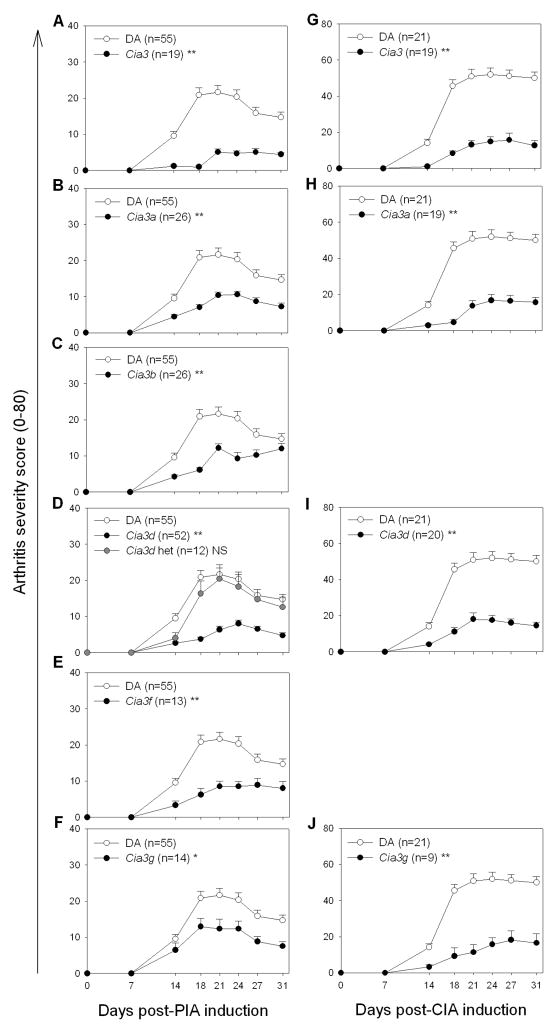
Both genders were similarly protected, and therefore male and female data were combined for analysis. The introgression of the F344-derived Cia3 interval into the DA background, as in the DA.F344(Cia3) congenics (figure 1), was associated with a highly significant reduction of 81% in median PIA ASI, compared with DA rats (figure 2A and 3A) [median ASI: DA=95, DA.F344(Cia3)=18, p 0.001; table 1]. DA.F344(Cia3) congenics were also similarly protected in CIA (figure 2G), and had an 84.9% lower median ASI compared with DA rats [Median ASI: DA=265, DA.F344(Cia3)=40, p 0.001; table 1].
Figure 2. Arthritis severity clinical scores in DA and congenic/subcongenic rats during a 31-day observation period.
Males and females had similar disease severity scores in each strain, and therefore were combined for analyses. Left column: Pristane-induced arthritis (PIA): (A) DA and DA.F344(Cia3); (B) DA and DA.F344(Cia3a); (C) DA and DA.F344(Cia3b); (D) DA and DA.F344(Cia3d), including rats heterozygous at the congenic interval; (E) DA and DA.F344(Cia3f); (F) DA and DA.F344(Cia3g); Right column: Collagen-induced arthritis (CIA): (G) DA and DA.F344(Cia3); (H) DA and DA.F344(Cia3a); (I) DA and DA.F344(Cia3d); (J) DA and DA.F344(Cia3g). *p<0.03; **p≤0.003, Mann-Whitney test; Data shown as mean ± SEM.
Figure 3. Clinical and histological characteristics of DA and DA.F344(Cia3) congenic and subcongenic strains.
(A) DA rats developed severe arthritis with pronounced ankle and midfoot swelling and erythema (arrow), while (B) DA.F344(Cia3b), (C) DA.F344(Cia3d) and (D) DA.F344(Cia3g) developed very mild arthritis. (E–F) DA rats had extensive synovial hyperplasia, pannus formation, cartilage and bone invasion and erosion, contrasting with normal joint architecture in (G) DA.F344(Cia3) congenics, and (H) DA.F344(Cia3d) subcongenics. (Ankles were collected on day 32 post-pristane injection; hematoxylin-eosin staining; 100X magnification)
Table 1.
Arthritis Severity Scores for DA and Cia3 congenic and subcongenic rats.
| Model | Strain | n | ASI 1 |
P-value 2 | ASI reduction | ||
|---|---|---|---|---|---|---|---|
| median | 25% | 75% | |||||
| PIA | DA | 55 | 95.0 | 43.0 | 144.5 | ||
| Cia3 | 19 | 18.0 | 16.0 | 26.0 | ≤ 0.001 | 81.1% | |
| Cia3a | 26 | 46.0 | 32.3 | 57.8 | ≤ 0.001 | 51.6% | |
| Cia3b | 26 | 49.5 | 36.3 | 66.5 | 0.003 | 47.9% | |
| Cia3d | 46 | 38.5 | 23.5 | 54.8 | ≤ 0.001 | 59.5% | |
| Cia3f | 13 | 33.0 | 31.0 | 43.0 | 0.002 | 65.3% | |
| Cia3g | 14 | 54.0 | 35.5 | 70.3 | 0.031 | 43.2% | |
| CIA | DA | 21 | 265.0 | 210.0 | 342.0 | --- | |
| Cia3 | 19 | 40.0 | 31.0 | 87.5 | ≤ 0.001 | 84.9% | |
| Cia3a | 19 | 56.0 | 20.5 | 107.0 | ≤ 0.001 | 78.9% | |
| Cia3d | 20 | 64.5 | 40.8 | 112.8 | ≤ 0.001 | 75.7% | |
| Cia3g | 9 | 47.0 | 28.0 | 139.0 | ≤ 0.001 | 82.3% | |
Arthritis Severity Index (sum of arthritis scores obtained over time).
Mann-Whitney test (comparisons versus DA)
The protective effect was detected at day 14 (figures 2A and 2G), and persisted throughout the observation period, suggesting that the arthritis gene located within Cia3 regulates both early and chronic stages during the course of disease pathogenesis.
Cia3 contains two arthritis severity genes: Identification of Cia3d
Rats subcongenics for the centromeric segments of Cia3, DA.F344(Cia3a), DA.F344(Cia3b) and DA.F344(Cia3d) (figure 1) were protected in PIA and had significantly lower arthritis scores (figure 2B, 2C, 2D and figure 3B and 3C), with a respective 51.6%, 49.7%, and 59.5% reduction in median ASI compared with DA [median ASI: DA=95, DA.F344(Cia3a)=46, p 0.001; DA.F344(Cia3b)=49.5, p=0.003; DA.F344(Cia3d)=38.5, p 0.001; all comparisons with DA; table 1]. These three subcongenics had a similar reduction in arthritis scores, compared with DA, and shared a 9.9 Mb region containing F344 alleles between D4Uia2 and D4Wox22 (figure 1).
Two centromeric subcongenics DA.F344(Cia3a) (figure 2H) and DA.F344(Cia3d) (figure 2I) were studied in CIA, and both were protected with significantly 78.9% and 75.7% lower median ASI, respectively, compared with DA [Median ASI: DA=265, DA.F344(Cia3a)=56, p≤0.001; DA.F344(Cia3d)=64, p≤0.001; table 1].
These observations suggest that the interval shared by Cia3a, Cia3b and Cia3d, which is the 9.9 Mb Cia3d locus itself, contains an arthritis gene that regulates both PIA and CIA.
Cia3d operates in a dominant manner to increase arthritis severity
In order to determine the mode of inheritance of Cia3d, homozygous DA.F344(Cia3d) rats were backcrossed with DA, and the offspring (BC11) heterozygous at Cia3d, studied in PIA experiments. Cia3d heterozygous rats were not protected and had ASIs that were similar to DA (figure 2D), suggesting that the DA-derived arthritis severity alleles at this locus operate in a dominant manner.
Identification of Cia3g
The magnitude of the PIA protection detected in DA.F344(Cia3) congenics (81%) was more pronounced that in DA.F344(Cia3a), DA.F344(Cia3b) or DA.F344(Cia3d) subcongenics (49–59%). This observation suggested the existence of yet another arthritis gene located in the telomeric portion of Cia3. To address that possibility, DA.F344(Cia3f) and DA.F344(Cia3g) homozygous subcongenic strains (figure 1) were generated from DA.F344(Cia3), and studied in PIA. Both subcongenics were significantly protected in PIA, with 65.3% and 43.2% reduction in median ASI compared with DA (figure 2E and 2F, and figure 3D) [median ASI: DA=95, DA.F344(Cia3f)=33, p=0.002; DA.F344(Cia3g)=54, P=0.031; all comparisons versus DA; table 1].
DA.F344(Cia3g) was also significantly protected in CIA and had an 82.3% lower median ASI, compared with DA [Median ASI: DA=265, DA.F344(Cia3g)=47, p 0.001] (figure 2J).
DA.F344(Cia3g) is contained within DA.F344(Cia3f), and therefore its 34.5Mb interval between D4Arb5 and D4Arb4, and its adjacent recombination regions, contain the arthritis gene.
Taken together, these results show that Cia3 contains at least two distinct arthritis genes, and both genes regulate disease severity in two different models of autoimmune erosive arthritis, CIA and PIA. These observations suggest that these two genes regulate processes common and central to the pathogenesis of arthritis.
Histological studies in PIA synovial tissues
Males and females within each strain had similar histological findings and therefore, data from both genders were combined for analyses. DA rats with PIA had a highly abnormal joint histological architecture (table 1; figure 3E and F), with pronounced synovial hyperplasia and pannus formation, increased number of synovial vessels (angiogenesis), synovial infiltration with mononuclear cells, and extensive cartilage and bone erosive changes, and cartilage loss of proteoglycans.
Both DA.F344(Cia3) (figure 3G) and DA.F344(Cia3d) (figure 3H) preserved a nearly normal joint histological architecture with significantly lower histological scores compared with DA (table 2). These histological findings further support the reduced clinical arthritis severity observed in the congenics, and suggest that the disease genes contained within Cia3 regulate the pathogenesis of synovial hyperplasia, pannus formation and joint destruction in arthritis.
Table 2.
Histology scoring of DA, DA.F344(Cia3) and DA.F344(Cia3d) congenics with pristane-Induced arthritis.
| DA (n=10) | Cia3 (n=8) 1 | Cia3d (n=13) 2 | |
|---|---|---|---|
| Inflamm. infiltrate (0–3) | 1.94 ± 0.17 | 0.40 ± 0.11 | 0.32 ± 0.08 |
| Synovial hyperplasia (0–3) | 3.00 ± 0.00 | 1.25 ± 0.37 | 0.92 ± 0.31 |
| Pannus (0–3) | 2.80 ± 0.13 | 0.63 ± 0.26 | 0.46 ± 0.14 |
| Fibrosis (0–3) | 2.30 ± 0.15 | 0.75 ± 0.41 | 0.15 ± 0.10 |
| Vessels/HMF 3 | 12.40 ± 1.27 | 6.20 ± 0.90 | 7.31 ± 0.55 |
| Cartilage erosions (0–3) | 2.90 ± 0.10 | 0.38 ± 0.26 | 1.08 ± 0.21 |
| Bone erosions (0–3) | 3.00 ± 0.00 | 0.75 ± 0.25 | 0.92 ± 0.34 |
Means±SEM; DA vs. Cia3 p ≤ 0.0016, (t-test).
DA vs. Cia3d p ≤ 0.0006 (t-test).
High Magnification Field - 400X
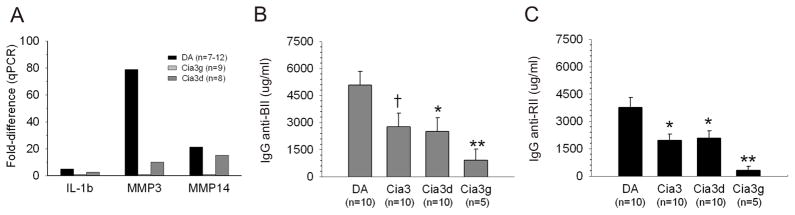
Synovial tissue levels of IL-1βmRNA were reduced in DA.F344(Cia3d) and DA.F344(Cia3g) congenics compared with DA rats
qPCR analysis revealed that synovial levels of IL-1βmRNA were nearly five-fold higher in DA (figure 4A), compared with DA.F344(Cia3d) congenics [ΔCt: DA=7.6, DA.F344(Cia3d)=9.54; p=0.001, Mann-Whitney test, figure 4A] (The ΔCt is inversely correlated with the number of copies of mRNA in the tissue, and each Ct cycle difference is equivalent to a nearly two-fold difference in mRNA levels). The expression of IL-1β was also lower in DA.F344(Cia3g) compared with DA, [ΔCt: DA.F344(Cia3g)=8.3], with a nearly 2-fold difference, but did not reach statistical significance (p=0.56; t-test), suggesting that the two loci regulate synovitis through difference mechanisms (figure 4A).
Figure 4. Synovial tissue levels of IL-1β, MMP-3 and MMP14 mRNA, allo and autoantibodies against type II collagen in DA and Cia3 strains.
(A) qPCR analyses of synovial tissues from rats with PIA showing DA.F344(Cia3d) (n=8), and DA.F344(Cia3g) (n=9) rats with significantly lower levels of IL-1β, MMP-3 and MMP-14 mRNA compared with DA (n=12). Fold-differences were determined with the 2−ΔΔCt method, and are shown compared with DA.F344(Cia3d) as reference. ΔCt results were used for comparisons with DA (all with p≤0.001, except for Cia3g IL-1β which had p=0.56; t-test). (B) Levels (measured at day 18 post-induction; μg IgG/mL) of antibodies against bovine type II collagen (anti-BII) and (C) autoantibodies against rat type II collagen (BII) were significantly lower in DA.F344(Cia3), DA.F344(Cia3d and in DA.F344(Cia3g) strains. (ANOVA on ranks with a pairwise multiple comparison procedure [Dunn’s method]; *p≤ 0.05; ** p≤ 0.01; † p=0.054); results shown as mean±S.E.M..
DA.F344(Cia3d) and DA.F344(Cia3g) have significantly lower synovial tissue levels of MMP-1, MMP-3 and MMP-14 mRNA compared with DA rats
MMP-1 was expressed in 86% (6/7) of DA synovial tissues, but only in 12.5% (1/8) of DA.F344(Cia3d) (P=0.01, Fisher’s exact test) and in 33% (3/9) DA.F344(Cia3g) congenics (P=0.06, Fisher’s exact test). Levels of MMP-3 and MMP-14 mRNA were 78-fold and 21-fold higher in DA, compared with DA.F344(Cia3d) [MMP-3 ΔCt: DA=27.65, DA.F344(Cia3d)=33.95; p= 0.00004; MMP-14 ΔCt: DA=28.26, DA.F344(Cia3d)=32.67; p= 0.004, t-test, figure 4A], and 8-fold and 1.4-fold when DA was compared with DA.F344(Cia3g) congenics [MMP-3 ΔCt: DA=27.65, DA.F344(Cia3g)=30.66; p= 0.00006; MMP-14 ΔCt: DA=27.65, DA.F344(Cia3d)=28.76; p= 0.007, t-test, figure 4A].
Reduced levels of allo antibodies against bovine and autoantibodies against rat type II collagen in Cia3 congenic and subcongenic strains tested for CIA
High levels of IgG antibodies against bovine type II collagen (BII) were detected in all strains immunized with BII/IFA, confirming appropriate immunization (figure 4B). There was concordance between median levels of anti-BII and mean levels of autoantibodies anti-rat type II collagen (RII) in each strain (figure 4B and 4C), and levels of anti-BII (p=0.054) and anti-RII (p≤0.05) were lower in DA.F344(Cia3) congenics compared with DA.
The arthritis-protected subcongenic strains DA.F344(Cia3d) and DA.F344(Cia3g) also had significantly reduced levels of anti-BII and anti-RII (p≤0.05 and p≤0.01, respectively) compared with DA (figures 4B and 4C). DA.F344(Cia3g) had a more pronounced reduction in levels of antibodies anti-BII and anti-RII than DA.F344(Cia3d), again suggesting differences in gene-specific mode of action.
Cia3g is not explained by the variants in antigen-presenting lectin-like receptor gene complex (APLEC) previously associated with arthritis and the Oia2 locus
We have sequenced the coding regions of the APLEC genes previously associated with arthritis severity in a DAxPVG intercross and congenics (Dcar1, Dcir2, Dcir1 and Mincle) (21). DA and F344, the parental strains used to discover Cia3g, had the same alleles at all of the amino-acid changing SNPs originally described between DA and PVG, suggesting that another new gene(s) account for this locus’ effect in arthritis severity.
DISCUSSION
RA is associated with increased risk for disability and deformities, reduced survival and reduced income (22). Disease severity and articular damage are associated with increased risk for disability, joint deformities and reduced life expectancy in patients with RA (23–25). Therefore, genes implicated in the regulation of disease severity and articular damage are expected to generate important new targets for therapies aimed at preserving the joint architecture and function. Yet, little is known about severity or articular damage genes in arthritis.
In the present study, we report the generation of Cia3 subcongenic strains and the identification and localization of two new arthritis severity and articular damage regulatory loci, Cia3d, the most centromeric 9.9 Mb interval of Cia3, and a second locus in the most telomeric segment contained within the 59.5 Mb Cia3g interval. Cia3d and Cia3g regulate disease severity of two different models of autoimmune erosive arthritis, CIA and PIA. The observation that the effect of these two loci is not limited to a single model suggests that they regulate processes that are central to arthritis pathogenesis, thus making them potentially even more relevant to human disease.
In addition to evaluating clinical arthritis severity during a 31-day observation period, a comprehensive histological analysis determined that the presence of F344 alleles at Cia3 or only at Cia3d was associated with preservation of a normal joint architecture, with no synovial hyperplasia or pannus formation, no significant synovial inflammatory infiltration or angiogenesis, and no cartilage or bone erosive damage. Synovial angiogenesis is a critical event in the pathogenesis of autoimmune arthritis and in the development of synovial hyperplasia (26, 27). Similarly, synovial hyperplasia and pannus formation are typically associated with cartilage and bone erosions in arthritis (17, 28), and are partially dependent on the inflammatory mononuclear cellular infiltrate and the cytokines these infiltrating cells produce (29–32). Moreover, several of the cytokines produced by the synovial tissues and infiltrating mononuclear cells have angiogenic properties (33). Therefore, while we cannot determine at this point which specific cellular and molecular events the Cia3d and Cia3g genes regulate, our observations demonstrate that these genes control fundamental events required for the development of synovial inflammation, arthritis and articular damage.
Both DA.F344(Cia3d) and DA.F344(Cia3g) subcongenics had reduced synovial levels of IL-1β mRNA compared with DA, and synovial levels of IL-1β mRNA correlate with their respective protein levels in rats (34). The reduced levels of IL-1β, even at a later time-point during disease course (day 32), demonstrate that the regulatory effects of these two loci persist beyond the early stages of disease and into the chronic stages. IL-1β is a key pro-inflammatory cytokine involved in the pathogenesis of rodent arthritis (17, 35–37) and RA (38–40).
IL-1β is known to induce the expression of IL-1β itself, IL-6, and MMPs implicated in articular damage such as those expressed in significantly reduced levels in Cia3d and Cia3g congenics (41, 42). Furthermore, IL-1β induces both synovial hyperplasia and angiogenesis, features associated with erosive arthritis (41, 42). Therefore, the reduction in levels of IL-1β could explain the significantly reduced expression of MMP-1, MMP-3 and MMP-14 observed in Cia3d and Cia3g congenics. These MMPs have been implicated in cartilage and bone damage in RA and rodent models of RA, and therefore, the reduced expression of IL-1β and MMPs explains at least part of the histological joint protection observed in the subcongenics. Interestingly, while both subcongenics had reduced synovial levels of IL-1β mRNA, the reduction was more pronounced in DA.F344(Cia3d) than in DA.F344(Cia3g).
While levels of IL-1β were more significantly reduced in DA.F344(Cia3d), levels of antibodies anti-BII (immunogen) and pathogenic autoantibodies anti-RII were more significantly reduced in DA.F344(Cia3g) than in DA.F344(Cia3d). These observations suggest that Cia3g could be involved in B cell responses, or processes of the immune response required for B cell responses such as antigen presentation or T cell cognate interactions. B cells and antibody-producing plasma cells are present in arthritic synovial tissues (16, 43), and are required for disease development (44). Autoantibodies such as anti-CCP, rheumatoid factors and anti-type II collagen antibodies are commonly detected in RA patients and correlate with disease severity and joint damage (45, 46). Additionally, therapies targeting B cells have been highly effective in RA (47).
Therefore, our results show that Cia3d had a more significant effect on the expression of IL-1β, while Cia3g had a more significant effect on levels of autoantibodies. These observations suggest that the two genes have preferential effects on different pathogenic processes.
The Cia3 syntenic region in the human genome contains loci regulating different forms of autoimmune diseases (12–14). Cia3d is syntenic to a region on human chromosome 7p not known to contain RA genes. Cia3g is syntenic to a RA susceptibility region on chromosome 7q (14), and co-localizes with five different arthritis severity loci previously identified in rats, including another CIA locus, Cia13 (48), the oil-induced arthritis locus Oia2 (21), Aia3 (8), Pia5 and with Pia7 (49). Allelic variants in coding regions of four different APLEC genes were associated with the Oia2 locus (21). However, DA and F344 had no non-synonimous coding region differences in the chromosome 4 APLEC genes implicated in Oia2. It is still possible that a non-coding polymorphism in one of the APLEC genes exists, and therefore we cannot completely exclude that one of the APLEC genes accounts for the Cia3g locus. However, since the original polymorphisms were in coding regions (21), as in the case of most disease-causing polymorphisms, and we used the same susceptible strain, we considered that it is more likely that Cia3g is a new arthritis severity gene and not the same as Oia2/APLEC.
In conclusion, the present study determined that Cia3 contains two different and independent arthritis severity and articular damage regulatory genes. The Cia3d interval contains genes that regulate arthritis severity, synovial hyperplasia, pannus formation, synovial inflammation and angiogenesis and joint damage, processes central to disease pathogenesis underscoring the potential relevance of the gene identification. We are in the process of reducing the critical regions containing Cia3d and Cia3g to ≤1Mb for positional cloning of the specific genes. The identification of these genes will generate novel new targets for the development of new therapies aimed at reducing joint damage and disease severity, as well as potentially new tools for prognostication.
Acknowledgments
Funded by the National Institutes of Health grants number R01-AR46213 (NIAMS) and R01-AI54348 (NIAID) to Dr. P. Gulko.
Abbreviations
- ASI
arthritis severity index
- CIA
collagen-induced arthritis
- PIA
pristane-induced arthritis
- QTL
quantitative trait locus or loci
- RA
rheumatoid arthritis
References
- 1.Gregersen PK, Plenge RM, Gulko PS. Genetics of rheumatoid arthritis. In: Firestein G, Panayi G, Wollheim FA, editors. Rheumatoid arthritis. 2. New York: Oxford University Press; 2006. pp. 3–14. [Google Scholar]
- 2.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marinou I, Maxwell JR, Wilson AG. Genetic influences modulating the radiological severity of rheumatoid arthritis. Ann Rheum Dis. 2010;69(3):476–82. doi: 10.1136/ard.2009.117721. [DOI] [PubMed] [Google Scholar]
- 5.Gulko PS, Kawahito Y, Remmers EF, Reese VR, Wang J, Dracheva SV, et al. Identification of a new non-major histocompatibility complex genetic locus on chromosome 2 that controls disease severity in collagen- induced arthritis in rats. Arthrititis Rheum. 1998;41(12):2122–31. doi: 10.1002/1529-0131(199812)41:12<2122::AID-ART7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Remmers EF, Joe B, Griffiths MM, Dobbins DE, Dracheva SV, Hashiramoto A, et al. Modulation of multiple experimental arthritis models by collagen-induced arthritis quantitative trait loci isolated in congenic rat lines: different effects of non-major histocompatibility complex quantitative trait loci in males and females. Arthritis Rheum. 2002;46(8):2225–34. doi: 10.1002/art.10439. [DOI] [PubMed] [Google Scholar]
- 7.Brenner M, Meng HC, Yarlett NC, Joe B, Griffiths MM, Remmers EF, et al. The Non-MHC Quantitative Trait Locus Cia5 Contains Three Major Arthritis Genes That Differentially Regulate Disease Severity, Pannus Formation, and Joint Damage in Collagen- and Pristane-Induced Arthritis. J Immunol. 2005;174(12):7894–903. doi: 10.4049/jimmunol.174.12.7894. [DOI] [PubMed] [Google Scholar]
- 8.Joe B, Cannon GW, Griffiths MM, Dobbins DE, Gulko PS, Wilder RL, et al. Evaluation of quantitative trait loci regulating severity of mycobacterial adjuvant-induced arthritis in monocongenic and polycongenic rats: identification of a new regulatory locus on rat chromosome 10 and evidence of overlap with rheumatoid arthritis susceptibility loci. Arthritis Rheum. 2002;46(4):1075–85. doi: 10.1002/art.10164. [DOI] [PubMed] [Google Scholar]
- 9.Vingsbo-Lundberg C, Nordquist N, Olofsson P, Sundvall M, Saxne T, Pettersson U, et al. Genetic control of arthritis onset, severity and chronicity in a model for rheumatoid arthritis in rats. Nat Genet. 1998;20(4):401–4. doi: 10.1038/3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorentzen JC, Glaser A, Jacobsson L, Galli J, Fakhrai-rad H, Klareskog L, et al. Identification of rat susceptibility loci for adjuvant-oil-induced arthritis. Proc Natl Acad Sci U S A. 1998;95(11):6383–7. doi: 10.1073/pnas.95.11.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuya T, Salstrom JL, McCall-Vining S, Cannon GW, Joe B, Remmers EF, et al. Genetic dissection of a rat model for rheumatoid arthritis: significant gender influences on autosomal modifier loci. Hum Mol Genet. 2000;9(15):2241–50. doi: 10.1093/oxfordjournals.hmg.a018915. [DOI] [PubMed] [Google Scholar]
- 12.Wilder R, Remmers E, Kawahito Y, Gulko P, Cannon G, Griffiths M. Genetic factors regulating experimental arthritis in mice and rats. In: Theophilopoulos A, editor. Current Directions in Autoimmunity. Basel: Karger; 1999. pp. 121–65. [DOI] [PubMed] [Google Scholar]
- 13.Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, et al. Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci U S A. 1998;95(17):9979–84. doi: 10.1073/pnas.95.17.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos CI, Chen WV, Lee A, Li W, Kern M, Lundsten R, et al. High-density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes Immun. 2006;7(4):277–86. doi: 10.1038/sj.gene.6364295. [DOI] [PubMed] [Google Scholar]
- 15.Meng H, Griffiths M, Remmers E, Kawahito Y, Li W, Neisa R, et al. Identification of two novel female-specific non-MHC loci regulating collagen-induced arthritis severity and chronicity, and evidence of epistasis. Arthritis Rheum. 2004;50(8):2695–705. doi: 10.1002/art.20366. [DOI] [PubMed] [Google Scholar]
- 16.Vingsbo C, Sahlstrand P, Brun J, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: A new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol. 1996;149:1675–83. [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner M, Meng H, Yarlett N, Griffiths M, Remmers E, Wilder R, et al. The non-MHC quantitative trait locus Cia10 contains a major arthritis gene and regulates disease severity, pannus formation and joint damage. Arthritis Rheum. 2005;52(1):322–32. doi: 10.1002/art.20782. [DOI] [PubMed] [Google Scholar]
- 18.Remmers EF, Longman RE, Du Y, O’Hare A, Cannon GW, Griffiths MM, et al. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nat Genet. 1996;14(1):82–5. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- 19.Laragione T, Brenner M, Li W, Gulko PS. Cia5d regulates a new fibroblast-like synoviocyte invasion-associated gene expression signature. Arthritis Res Ther. 2008;10(4):R92. doi: 10.1186/ar2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Lorentzen JC, Flornes L, Eklow C, Backdahl L, Ribbhammar U, Guo JP, et al. Association of arthritis with a gene complex encoding C-type lectin-like receptors. Arthritis Rheum. 2007;56(8):2620–32. doi: 10.1002/art.22813. [DOI] [PubMed] [Google Scholar]
- 22.Gulko PS, Winchester RJ. Rheumatoid arthritis. In: Austen KF, Frank MM, Atkinson JP, Cantor H, editors. Samter’s Immunologic Diseases. 6. Baltimore: Lippincott, Williams & Wilkins; 2001. pp. 427–63. [Google Scholar]
- 23.van Zeben D, Breedveld FC. Prognostic factors in rheumatoid arthritis. J Rheumatol Suppl. 1996;44:31–3. [PubMed] [Google Scholar]
- 24.Gossec L, Dougados M, Goupille P, Cantagrel A, Sibilia J, Meyer O, et al. Prognostic factors for remission in early rheumatoid arthritis: a multiparameter prospective study. Ann Rheum Dis. 2004;63(6):675–80. doi: 10.1136/ard.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfe F, Mitchell DM, Sibley JT, Fries JF, Bloch DA, Williams CA, et al. The mortality of rheumatoid arthritis. Arthritis Rheum. 1994;37(4):481–94. doi: 10.1002/art.1780370408. [DOI] [PubMed] [Google Scholar]
- 26.Storgard CM, Stupack DG, Jonczyk A, Goodman SL, Fox RI, Cheresh DA. Decreased angiogenesis and arthritic disease in rabbits treated with an alphavbeta3 antagonist. J Clin Invest. 1999;103(1):47–54. doi: 10.1172/JCI3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sone H, Kawakami Y, Sakauchi M, Nakamura Y, Takahashi A, Shimano H, et al. Neutralization of vascular endothelial growth factor prevents collagen-induced arthritis and ameliorates established disease in mice. Biochem Biophys Res Commun. 2001;281(2):562–8. doi: 10.1006/bbrc.2001.4395. [DOI] [PubMed] [Google Scholar]
- 28.Kleinau S, Erlandsson H, Holmdahl R, Klareskog L. Adjuvant oils induce arthritis in the DA rat. I. Characterization of the disease and evidence for an immunological involvement. J Autoimmun. 1991;4(6):871–80. doi: 10.1016/0896-8411(91)90050-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miagkov AV, Kovalenko DV, Brown CE, Didsbury JR, Cogswell JP, Stimpson SA, et al. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95(23):13859–64. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fava RA, Olsen NJ, Postlethwaite AE, Broadley KN, Davidson JM, Nanney LB, et al. Transforming growth factor beta 1 (TGF-beta 1) induced neutrophil recruitment to synovial tissues: implications for TGF-beta-driven synovial inflammation and hyperplasia. J Exp Med. 1991;173(5):1121–32. doi: 10.1084/jem.173.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langdon C, Kerr C, Hassen M, Hara T, Arsenault AL, Richards CD. Murine oncostatin M stimulates mouse synovial fibroblasts in vitro and induces inflammation and destruction in mouse joints in vivo. Am J Pathol. 2000;157(4):1187–96. doi: 10.1016/S0002-9440(10)64634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung BP, McInnes IB, Esfandiari E, Wei XQ, Liew FY. Combined effects of IL-12 and IL-18 on the induction of collagen- induced arthritis. J Immunol. 2000;164(12):6495–502. doi: 10.4049/jimmunol.164.12.6495. [DOI] [PubMed] [Google Scholar]
- 33.Szekanecz Z, Szegedi G, Koch AE. Angiogenesis in rheumatoid arthritis: pathogenic and clinical significance. J Investig Med. 1998;46(2):27–41. [PubMed] [Google Scholar]
- 34.Rioja I, Bush KA, Buckton JB, Dickson MC, Life PF. Joint cytokine quantification in two rodent arthritis models: kinetics of expression, correlation of mRNA and protein levels and response to prednisolone treatment. Clin Exp Immunol. 2004;137(1):65–73. doi: 10.1111/j.1365-2249.2004.02499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joosten LA, Helsen MM, Saxne T, van De Loo FA, Heinegard D, van Den Berg WB. IL-1 alpha beta blockade prevents cartilage and bone destruction in murine type II collagen-induced arthritis, whereas TNF-alpha blockade only ameliorates joint inflammation. J Immunol. 1999;163(9):5049–55. [PubMed] [Google Scholar]
- 36.van de Loo AA, Arntz OJ, Bakker AC, van Lent PL, Jacobs MJ, van den Berg WB. Role of interleukin 1 in antigen-induced exacerbations of murine arthritis. Am J Pathol. 1995;146(1):239–49. [PMC free article] [PubMed] [Google Scholar]
- 37.Makarov SS, Olsen JC, Johnston WN, Anderle SK, Brown RR, Baldwin AS, Jr, et al. Suppression of experimental arthritis by gene transfer of interleukin 1 receptor antagonist cDNA. Proc Natl Acad Sci U S A. 1996;93(1):402–6. doi: 10.1073/pnas.93.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Genant HK, Watt I, Cobby M, Bresnihan B, Aitchison R, et al. A multicenter, double-blind, dose-ranging, randomized, placebo-controlled study of recombinant human interleukin-1 receptor antagonist in patients with rheumatoid arthritis: radiologic progression and correlation of Genant and Larsen scores. Arthritis Rheum. 2000;43(5):1001–9. doi: 10.1002/1529-0131(200005)43:5<1001::AID-ANR7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Chomarat P, Vannier E, Dechanet J, Rissoan MC, Banchereau J, Dinarello CA, et al. Balance of IL-1 receptor antagonist/IL-1 beta in rheumatoid synovium and its regulation by IL-4 and IL-10. J Immunol. 1995;154(3):1432–9. [PubMed] [Google Scholar]
- 40.Neidhart M, Gay RE, Gay S. Anti-interleukin-1 and anti-CD44 interventions producing significant inhibition of cartilage destruction in an in vitro model of cartilage invasion by rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2000;43(8):1719–28. doi: 10.1002/1529-0131(200008)43:8<1719::AID-ANR7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 41.Ghivizzani SC, Kang R, Georgescu HI, Lechman ER, Jaffurs D, Engle JM, et al. Constitutive intra-articular expression of human IL-1 beta following gene transfer to rabbit synovium produces all major pathologies of human rheumatoid arthritis. J Immunol. 1997;159(7):3604–12. [PubMed] [Google Scholar]
- 42.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117(14):3720–32. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dechanet J, Merville P, Durand I, Banchereau J, Miossec P. The ability of synoviocytes to support terminal differentiation of activated B cells may explain plasma cell accumulation in rheumatoid synovium. J Clin Invest. 1995;95(2):456–63. doi: 10.1172/JCI117685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svensson L, Jirholt J, Holmdahl R, Jansson L. B cell-deficient mice do not develop type II collagen-induced arthritis (CIA) Clin Exp Immunol. 1998;111(3):521–6. doi: 10.1046/j.1365-2249.1998.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook AD, Rowley MJ, Mackay IR, Gough A, Emery P. Antibodies to type II collagen in early rheumatoid arthritis. Correlation with disease progression. Arthritis Rheum. 1996;39(10):1720–7. doi: 10.1002/art.1780391015. [DOI] [PubMed] [Google Scholar]
- 46.Vencovsky J, Machacek S, Sedova L, Kafkova J, Gatterova J, Pesakova V, et al. Autoantibodies can be prognostic markers of an erosive disease in early rheumatoid arthritis. Ann Rheum Dis. 2003;62(5):427–30. doi: 10.1136/ard.62.5.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards JC, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350(25):2572–81. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 48.Griffiths MM, Wang J, Joe B, Dracheva S, Kawahito Y, Shepard JS, et al. Identification of four new quantitative trait loci regulating arthritis severity and one new quantitative trait locus regulating autoantibody production in rats with collagen-induced arthritis. Arthritis Rheum. 2000;43(6):1278–89. doi: 10.1002/1529-0131(200006)43:6<1278::AID-ANR10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 49.Rintisch C, Kelkka T, Norin U, Lorentzen JC, Olofsson P, Holmdahl R. Finemapping of the arthritis QTL Pia7 reveals co-localization with Oia2 and the APLEC locus. Genes Immun. 2010;11(3):239–45. doi: 10.1038/gene.2010.2. [DOI] [PubMed] [Google Scholar]