Abstract
The small multidrug resistance transporters represent a unique model system for studying the mechanism of secondary active transport and membrane protein evolution. However, this seemingly simple protein has been highly controversial. Recent studies have provided experimental evidence that EmrE exists as an asymmetric dimer that exchanges between identical inward- and outward-facing states. Re-examination of the published literature in light of these findings fills in many details of the microscopic steps in the transport cycle. Future work will need to examine how the symmetry observed in vitro affects EmrE function in the asymmetric environment of its native E. coli membrane.
Introduction
At first glance, small multidrug resistance (SMR) transporters appear to be an ideal system to study the microscopic steps of the transport cycle, representing a minimal system for secondary active transport. SMR transporters are also proposed to represent an intermediate point in the evolution of larger transporters with internal dual-repeat structure[1]. The small size was expected to make SMR transporters more amenable to detailed biochemical and biophysical characterization, but the best-studied of these transporters, EmrE, has not turned out be a simple model system[2-5]. Like many integral membrane proteins, EmrE is quite sensitive to its environment, and is only stably folded and functional when reconstituted into a suitable membrane or membrane-mimetic environment[2, 3, 6-9]. Significant controversy has arisen, particularly with regard to the structure and topology of EmrE. Several years ago, reviews by key labs [2, 3] summarized the structural and biochemical data on either side of the issue. This review focuses on recent progress in studies of EmrE, especially its mechanism. This includes not just the structure(s), but also the dynamics of EmrE as it moves through the various steps in the transport cycle.
Dissecting the structural and dynamic steps of the transport cycle of SMR transporters is of great interest not just for understanding the mechanism of secondary active transport, but also because SMR proteins contribute to bacterial drug resistance and biofilm formation[10]. Like other SMR transporters, EmrE is a dimer located in the bacterial inner membrane. It exports polyaromatic cations across this membrane, conferring resistance to a wide variety of compounds. Drug export is driven by the H+ gradient, with export of each polyaromatic cation coupled to import of two protons. Coupled antiport is achieved by competition for binding to E14 in TM1 (transmembrane helix 1), which binds protons (2H+ per dimer), or helps stabilize the positive charge on the polyaromatic cation substrate in the deprotonated state (see reviews [2, 3]).
The single-site alternating access model was proposed decades ago and is supported by a wealth of biochemical evidence in the case of EmrE[2, 11-14]. It has remained primarily a cartoon model due to the difficulty of working with membrane proteins under conditions suitable for detailed biophysical and structural biology studies. In the last decade however, technical advances have enabled studies of EmrE with crystallography, cryoelectron microscopy (cryoEM), EPR, NMR, and single-molecule FRET. Complete understanding of the transport mechanism requires knowledge of the structures of the individual states, their relative stability (thermodynamics) and the rates of transitions between them (kinetics). New data on EmrE structure and dynamics support an antiparallel topology and the symmetric exchange mechanism insightfully proposed several years ago by Fleishman et al[14].
Structure and Topology of EmrE
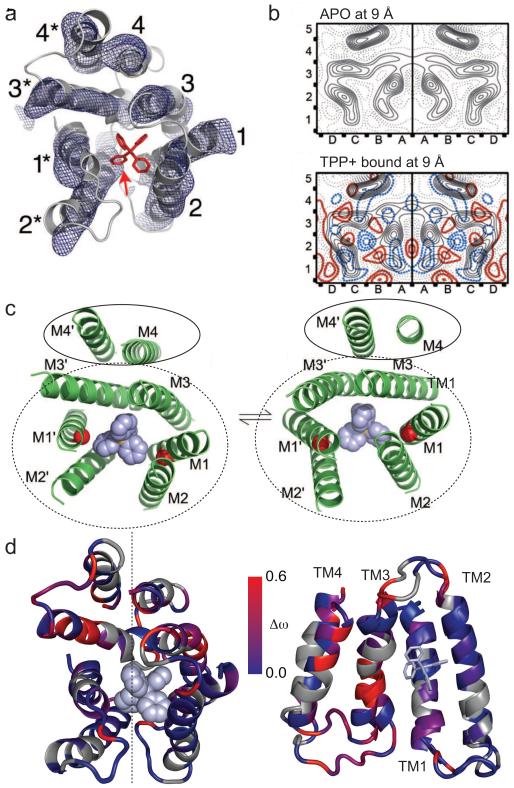
The structure of EmrE has been studied both with substrate bound and in the absence of polyaromatic cations. The first cryoEM structure of tetraphenylphosphonium (TPP+)-bound EmrE revealed an 8-helix bundle with an asymmetric arrangement in dimyristoylphosphatidylcholine (DMPC) lipid bilayers[15]. This asymmetry was also present in the crystal structure of EmrE determined at 3.8Å resolution (Fig. 1a), which unambiguously demonstrated that the asymmetry arose from an antiparallel topology within the homodimer[16]. However, concerns arose that the unusual topology could be an artifact of the crystallization conditions[17].
Figure 1. Antiparallel structure of EmrE.
a) The asymmetric cryoEM structure (mesh surface, red arrow indicates density corresponding to TPP+) and antiparallel crystal structure (gray ribbon, TPP+ in red) of TPP+-bound EmrE overlay quite well. Transmembrane helices are numbered 1-4 and 1*-4* in the two monomers. Reproduced from [16], copyright (2007) National Academy of Sciences, U.S.A. b) 2D cryoEM map of apo (top) and TPP+-bound EmrE (bottom) show similar asymmetric arrangements of the TM helices. The red/blue contours correspond to positive/negative difference density between the two maps (apo minus TPP+). The maps highlight the close packing of two asymmetric dimers in the 2D crystals (right/left boxes), which places parallel monomers from separate dimers in close proximity. Reproduced from[30] with permission. c) Top view of the conformational exchange model proposed by Fleishman et al.[14], with TM helices numbered M1-M4 and M1′-M4′ in each monomer. The three substrate-binding helices reorient, TM3 kinks, and the substrate-binding domain (dotted oval) moves slightly relative to TM4 to interconvert the structures of the two monomers in the asymmetric dimer. This also swaps which end of the substrate-binding domain is open to the aqueous compartment. Reproduced from [14] with permission. d) Differences in chemical shift between monomer A and monomer B (Δω) are plotted on the structure using the scale shown. A top view is shown on the left side. On the right, monomer A is rotated 180° about an axis in the plane of the membrane (dotted line) and the substrate-binding domains of each monomer are aligned. This illustrates that the largest chemical shift differences are exactly where backbone structure or helix-helix contact differ two monomers. Reproduced from [25] with permission.
Antiparallel topology within the homodimer was unexpected because membrane proteins generally insert in a particular orientation following the “positive inside rule”. However, EmrE does not have a significant charge bias. Indeed, recent work has demonstrated that small alterations in charge distribution, even at C-terminal positions, can alter the distribution of monomer orientations within the membrane[1, 18]. Several accessibility studies revealed both termini-in and termini-out orientations for the monomers[19, 20], with an equal probability of either orientation in E. coli[20], the native environment. These experiments demonstrated that an antiparallel topology was possible, but did not directly assess the topology within the dimer.
Extensive EPR studies of TPP+-bound EmrE reconstituted into liposomes provide additional structural restraints with site-specific resolution[8]. Spin label mobility, O2 and NiEDDA accessibility were used to map transmembrane helices and define lipid- and water-accessible regions. This is slightly complicated by the fact that EmrE labeled at a single site has two spin-labels per dimer, and in the case of an asymmetric dimer the parameters will reflect an average of the two environments. Nevertheless, this data plus the distance between spin labels is generally consistent with the single antiparallel TPP+-bound crystal structure. A few positions report two distance distributions, but these are located where addition of the spin label may affect packing of the protein, and lower substrate affinity or in vivo drug resistance activity is reported for EmrE with spin labels in these positions. More significantly, several regions have discrepancies indicative of slight differences in helix tilt (TM2, C-terminal region of TM3) or position relative to the membrane surface (loop between TM3 and TM4). These variations may arise from errors in the details of the crystal structure due to its limited resolution or from structural adaptation of EmrE to different membrane/detergent environments. The latter possibility is suggested by variation in EmrE’s substrate affinity with lipid type[9, 21]. Importantly, the EPR data on EmrE in liposomes is consistent with the general topology and structure determined by cryoEM and crystallography.
A number of experiments have seemed to support parallel topology. Genetic fusions designed to force either parallel or antiparallel topology are both functional[20, 22], although it is difficult to rule out formation of a dimer of dimers as seen in cryoEM (Fig. 1b). Antiparallel dimerization of a parallel fusion could lead to a more stable oligomer, preventing swapping in of inactive monomers under conditions optimized for wildtype EmrE.
Further support for parallel topology has been drawn from cross-linking of single cysteine mutants (K22C, H110C) with short dimaleimide cross-linkers, although the cross-linking efficiencies have varied[20, 23, 24]. Cross-linking due to transient proximity rather than a stable structure is always a possibility, and in the case of a membrane protein, the ratio of protein to lipid/detergent is key. Until recently, no attempts had been made to cross-link antiparallel EmrE[25]. Using a single cysteine mutant (S107C) and a heterobifunctional cross-linker, cross-linking to the single native lysine (K22) will only occur between antiparallel monomers. Amazingly, this cross-linking proceeds with 100% efficiency in less than 30 minutes at room temperature with an EmrE:dodecylmaltoside (DDM) ratio of 1:200 (1 dimer: 400 DDM, lipid gives same results[25]).
The most unambiguous determination of topology requires direct measurement of the relative orientation of the two monomers within the dimer. Single-molecule FRET measurements of TPP+-bound EmrE labeled at a single position per monomer accomplished this task[25]. For each labeling site, a single FRET efficiency was observed that was consistent with donor and acceptor on opposite sides of the membrane, indicating an antiparallel topology within the dimer.
What about substrate-free EmrE? Characterization of “apo” EmrE is complicated by the pKa of the active site glutamate. This pKa has been reported between 7.3-8.5[26-28], and care must be taken to assess whether “apo” structures represent the proton-bound or truly apo state. The crystal structure of substrate-free EmrE is almost certainly an artifact of the crystallization conditions[16]. The cryoEM maps reveal changes in TM helix tilt, but otherwise the same general arrangement within the helical bundle in the presence or absence of substrate[29, 30]. The EPR data are also in agreement with a generally similar structure with only changes in helix tilt, packing density, and loop position in the “apo” state compared to the TPP+-bound state[8]. Thus, at least at low resolution, the apo structure is similar to the TPP+ bound state (Fig. 1b).
Symmetry and Conformational Exchange During Transport
CryoEM studies first suggested the flexible nature of EmrE. EmrE bound to different substrates has a common asymmetric 8-helix bundle in each case with differences in the helix tilt depending on which substrate was bound [30]. Upon examining the initial cryoEM data and antiparallel crystal structure, Fleishman et al. noted a pseudo 2-fold axis in the plane of the membrane (Fig. 1c) and proposed that the two monomers in the asymmetric dimer could simply interchange conformations (AB dimer to BA dimer)[14] (Fig. 2). This achieves inward-to outward-facing conformational exchange with identical inward- and outward-facing states. Importantly, this model reconciles the asymmetric antiparallel structural data with the seemingly symmetric behavior of active site residues in biochemical assays[28, 31], since only one active site conformation is present.
Figure 2. Single-site alternating access model for antiport by EmrE.
The crystal structure of EmrE (PDB 3B5D) is used to represent the TPP+-bound states of EmrE. As proposed by Fleishman[14] and confirmed by NMR[25], the structures of the inward- and outward-facing conformations are identical. Based on cryoEM[29] and EPR[8] data, the same structure is used to model the protonated form, although there will be some changes in helix tilt and loop positions. It is assumed that the protonated state also has identical inward- and outward-facing conformations, although this has not been experimentally confirmed. Rate constants for substrate binding and release are konTPP+ = 4.9 × 106 M−1 s−1, koff TPP+ ≈ 10 s−1 (estimated from graph), TPP+-induced koffH+ ≈ kon TPP+ (stopped flow, DDM micelles, pH 7.0, 25 °C)[28], and the rate constant for conformational exchange in the TPP+-bound state, k, has been reported as 5 s−1 (NMR, 45 °C, isotropic bicelles) [25] and 1. 5 s−1 (stopped flow, DDM micelles, 25 °C)[28]. k’ has not been measured.
The EPR studies of the Mchaourab lab[8] provided the first insight into EmrE flexibility with site-specific resolution. In the context of a dynamic asymmetric homodimer, such as EmrE, the EPR parameters reflect an average state. However, careful analysis of O2 and NiEDDA accessibility and spin-label mobility, allowed the authors to determine that the N-terminal region of TM3 is tightly packed while the C-terminal portion is kinked and more flexible leading into a highly dynamic loop between TM3 and TM4 that samples both lipid and aqueous environments. They also found that TPP+ binding orders TM3 and rearranges the TM3-4 loop, suggesting that TM4 must also be dynamic. These findings are consistent with the cryoEM inspired model (Fig. 2), where kinking of TM3 is important for opening and closing the transport pathway during exchange between the inward and outward-facing states and TM4 moves relative to the substrate-binding domain. Furthermore, mutations of TM4 in Hsmr, an EmrE homolog, found some mutations at the center of TM4 did not alter dimerization strength but reduced transport activity[32]. This suggests a functional role, perhaps as a pivot point, in accord with the EPR dynamics in this region.
Solid-state NMR experiments on EmrE in liposomes revealed asymmetry of the active site residue, E14[33, 34], but heterogeneity, likely due to the dynamic nature of EmrE, precluded more detailed analysis of the structure. More recently, solution NMR[25] has been used to measure the dynamics of EmrE in the TPP+-bound state solubilized in bicelles. Two sets of peaks were observed in the NMR spectra with equal populations, representing the two distinct monomer structures within the asymmetric dimer. Chemical shift difference mapping (Fig. 1 d) revealed the largest differences between the monomers exactly at the positions where the backbone conformation (TM3 kink) or TM helix packing (TM4 relative to substrate-binding domain, central TM4 pivot point) differs between the two monomers in the asymmetric dimer structure.
The single molecule FRET data are also consistent with the proposed conformational exchange model[25]. This model predicts a single conformation of EmrE in solution, thus a single FRET efficiency for EmrE labeled with donor and acceptor at a single position. Furthermore, there should be no change in FRET efficiency over time even though the timescale of conformational change is ideal for measuring dynamics by single molecule FRET, as discussed below. This is exactly what was observed at multiple labeling positions.
Kinetic Analysis of Steps in the Transport Cycle
To measure substrate on- and off-rates by following the tryptophan fluorescence of EmrE the Schuldiner lab performed stopped flow fluorescence studies[28]. Tryptophan quenching occurs upon substrate binding, and this effect was tracked to tryptophan 63 in the active site using tryptophan knockouts[28, 35]. With identical inward- and outward-facing conformations, only a single species is present in solution, consistent with the single on- and off-rates measured.
These experiments detected an additional slow component in experiments probing substrate binding at high pH[28]. Creation of a single-tryptophan mutant retaining only tryptophan 63 enhanced the signal change, permitting quantitative analysis of this second slower component at high pH. The rate constant for this component was 1.5 s−1 at 25 °C for EmrE solubilized in DDM micelles, and it was independent of pH and substrate concentration. Thus, the authors suggested that it may reflect a conformational exchange process occurring after substrate is bound to the protein.
The details of the conformational exchange process were elucidated in recent solution NMR studies of EmrE solubilized in isotropic bicelles and saturated with TPP+ [25]. ZZ-exchange experiments unambiguously demonstrated exchange between two sets of peaks corresponding to the two monomer conformations in the asymmetric dimer. A rate constant of 5 s−1 was determined for the exchange process at 45 °C, and residues throughout the protein moved at the same rate, indicating a global conformational change. Accessibility mapping with a water-soluble paramagnetic probe demonstrated asymmetric water accessibility between the two monomer conformations, consistent with the TPP+-bound crystal structure closed to one side of the membrane and open to the other. Strikingly, these results are exactly what would be predicted for the model of AB to BA exchange in antiparallel asymmetric EmrE: two sets of peaks with equal population corresponding to the two distinct monomer conformations in the asymmetric dimer with exchange between those states.
Evidence for a transient occluded state where water is excluded from the binding site comes from transport assays performed on EmrE as well as its homolog, TBsmr[34, 36]. Upon initiation of a pH gradient, a relatively fast substrate binding process was observed by following the changes in the ethidium fluorescence, and an additional linear fluorescence increase was attributed to the transport process. Careful controls excluded ethidium fluorescence changes due to different environments inside and outside the liposome, upon binding to lipids, or upon changes in concentration. The fluorescence increase and high fluorescence anisotropy indicate a rigid, water-excluded environment as the source of the transport signal. Thus, this work provides experimental evidence that SMR transporters must transiently form a water-occluded intermediate state.
Conclusion
Movement of substrate from one side of the membrane to the other is a key step in the transport cycle. According to the single-site alternating access model, this is accomplished by conformational exchange of the transporter from an inward-facing to an outward-facing state while substrate is bound in the pore. This has now been directly observed with atomic resolution for TPP+-bound EmrE in isotropic bicelles using solution NMR spectroscopy[25]. Interestingly, the same conformational exchange process appears to be detected in stopped-flow fluorescence studies aimed at characterizing substrate binding[28]. Although the fluorescence experiments likely reflect formation of a water-occluded intermediate, as required for active transport, the NMR data are well fit by a two-state exchange model indicating that this intermediate state only exists transiently.
The experimental results obtained by many labs with independently purified samples of EmrE reconstituted into different membrane mimetic environments are amazingly consistent in support of the structure and mechanism shown in figures 1 and 2. The symmetry of EmrE has important functional consequences for the energetics of the transport process, motivating in vivo studies. Attempts to characterize functional effects of EmrE and its mutants in cells have had variable results, complicated by differences in protein expression level[37], efficiency of proper folding and localization, and compensatory function of other MDR efflux proteins with overlapping substrate specificities[20, 38]. Future studies will need to develop methods to control for these variables in order to determine if the dynamic symmetry observed in vitro is maintained in the asymmetric environment of the native E. coli membrane.
Highlights.
EmrE is oriented both ways in its native E. coli membrane
Antiparallel dimer topology confirmed by EPR, smFRET, and cross-linking
NMR dynamics provides experimental evidence for AB-BA exchange model
Can now fill in structures and rate constants for many steps in the transport cycle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rapp M, Seppälä S, Granseth E, von Heijne G. Emulating membrane protein evolution by rational design. Science. 2007;315:1282–1284. doi: 10.1126/science.1135406. [DOI] [PubMed] [Google Scholar]
- •2.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. Comprehensive review of the biochemistry of EmrE by the lab that discovered EmrE and has performed many of the biochemical studies. However, conflicting data are interpreted to support function of both parallel and antiparallel EmrE dimers. This conflicts with all the structural studies by other labs that show a single species in solution, not a mixture of parallel and antiparallel topologies.
- 3.Korkhov VM, Tate CG. An emerging consensus for the structure of EmrE. Acta Crystallogr D Biol Crystallogr. 2009;65:186–192. doi: 10.1107/S0907444908036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner S, Granot D, Mordoch SS, Ninio S, Rotem D, Soskin M, Tate CG, Yerushalmi H. Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. News Physiol Sci. 2001;16:130–134. doi: 10.1152/physiologyonline.2001.16.3.130. [DOI] [PubMed] [Google Scholar]
- 5.Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Miller D, Charalambous K, Rotem D, Schuldiner S, Curnow P, Booth PJ. In vitro unfolding and refolding of the small multidrug transporter EmrE. J Mol Biol. 2009;393:815–832. doi: 10.1016/j.jmb.2009.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Bay DC, Budiman RA, Nieh MP, Turner RJ. Multimeric forms of the small multidrug resistance protein EmrE in anionic detergent. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamem.2009.12.017. [DOI] [PubMed] [Google Scholar]
- ••8.Amadi ST, Koteiche HA, Mishra S, McHaourab HS. Structure, dynamics and substrate-induced conformational changes of the multidrug transporter EmrE in liposomes. J Biol Chem. 2010;285:26710–26718. doi: 10.1074/jbc.M110.132621. EPR study of both substrate-free and TPP+-bound EmrE in liposomes. Substrate binding and in vivo activity controls shown for complete cysteine scan. Mobility of spin label and accessibility to both aqueous and lipid phase of spin label at each residue throughout the protein highlight general similarity between susbtrate-free and TPP+-bound state, with increased dynamics in the “apo” state. Data support an antiparallel topology.
- 9.Charalambous K, Miller D, Curnow P, Booth PJ. Lipid bilayer composition influences small multidrug transporters. BMC Biochem. 2008;9:31. doi: 10.1186/1471-2091-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumura K, Furukawa S, Ogihara H, Morinaga Y. Roles of multidrug efflux pumps on the biofilm formation of escherichia coli K-12. Biocontrol Sci. 2011;16:69–72. doi: 10.4265/bio.16.69. [DOI] [PubMed] [Google Scholar]
- 11.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 12.Yerushalmi H, Schuldiner S. A model for coupling of H(+) and substrate fluxes based on “time-sharing” of a common binding site. Biochemistry. 2000;39:14711–14719. doi: 10.1021/bi001892i. [DOI] [PubMed] [Google Scholar]
- •13.Rotem D, Schuldiner S. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J Biol Chem. 2004;279:48787–48793. doi: 10.1074/jbc.M408187200. [DOI] [PubMed] [Google Scholar]
- 14.Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J Mol Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 15.Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 2003;22:6175–6181. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YJ, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuldiner S. When biochemistry meets structural biology: the cautionary tale of EmrE. Trends Biochem Sci. 2007;32:252–258. doi: 10.1016/j.tibs.2007.04.002. [DOI] [PubMed] [Google Scholar]
- •18.Seppala S, Slusky J, Lloris-Garcera P, Rapp M, Von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. How EmrE is inserted in the membrane equally in both orientations is an important question since it functions as an antiparallel homodimer. A series of charge bias mutants demonstrates that EmrE is finely balanced. The data reveal that charge bias acts globally, and even C-terminal residues added to the protein at the end of synthesis can alter the topology of the entire protein.
- 19.Nara T, Kouyama T, Kurata Y, Kikukawa T, Miyauchi S, Kamo N. Anti-parallel membrane topology of a homo-dimeric multidrug transporter, EmrE. J Biochem. 2007;142:621–625. doi: 10.1093/jb/mvm169. [DOI] [PubMed] [Google Scholar]
- •20.Nasie I, Steiner-Mordoch S, Gold A, Schuldiner S. Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters. J Biol Chem. 2010;285:15234–15244. doi: 10.1074/jbc.M110.108746. Analysis of in vivo topology of EmrE that provides quantitative data showing equal populations in each orientation. Functional analysis of knockout and mutant EmrE highlights the complexity of MDR knockout effects, with different depending on expression level, cell line, and which other MDR transporters are present. Results are interpreted in terms of highly promiscuous dimerization among different SMR transporters, although no evidence is shown for direct interaction between the monomers.
- 21.Curnow P, Lorch M, Charalambous K, Booth PJ. The reconstitution and activity of the small multidrug transporter EmrE is modulated by non-bilayer lipid composition. J Mol Biol. 2004;343:213–222. doi: 10.1016/j.jmb.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Steiner-Mordoch S, Soskine M, Solomon D, Rotem D, Gold A, Yechieli M, Adam Y, Schuldiner S. Parallel topology of genetically fused EmrE homodimers. EMBO J. 2008;27:17–26. doi: 10.1038/sj.emboj.7601951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soskine M, Steiner-Mordoch S, Schuldiner S. Crosslinking of membrane-embedded cysteines reveals contact points in the EmrE oligomer. Proc Natl Acad Sci USA. 2002;99:12043–12048. doi: 10.1073/pnas.192392899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soskine M, Mark S, Tayer N, Mizrachi R, Schuldiner S. On parallel and antiparallel topology of a homodimeric multidrug transporter. J Biol Chem. 2006;281:36205–36212. doi: 10.1074/jbc.M607186200. [DOI] [PubMed] [Google Scholar]
- ••25.Morrison EA, Dekoster GT, Dutta S, Clarkson M, Vafabakhsh R, Bahl A, Kern D, Ha T, Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. doi: 10.1038/nature10703. in Press. Solution NMR and single molecule FRET experiments on TPP+-bound EmrE solubilized in isotropic bicelles quantitatively measure the rate of conversion between inward- and outward-facing states, and show that those two states have identical asymmetric antiparallel structures. First direct experimental evidence for the AB to BA dimer exchange model.
- 26.Muth TR, Schuldiner S. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 2000;19:234–240. doi: 10.1093/emboj/19.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soskine M, Adam Y, Schuldiner S. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J Biol Chem. 2004;279:9951–9955. doi: 10.1074/jbc.M312853200. [DOI] [PubMed] [Google Scholar]
- 28.Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S. The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc Natl Acad Sci USA. 2007;104:17989–17994. doi: 10.1073/pnas.0704425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tate CG, Ubarretxena-Belandia I, Baldwin JM. Conformational changes in the multidrug transporter EmrE associated with substrate binding. J Mol Biol. 2003;332:229–242. doi: 10.1016/s0022-2836(03)00895-7. [DOI] [PubMed] [Google Scholar]
- •30.Korkhov VM, Tate CG. Electron crystallography reveals plasticity within the drug binding site of the small multidrug transporter EmrE. J Mol Biol. 2008;377:1094–1103. doi: 10.1016/j.jmb.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinglass AB, Soskine M, Vazquez-Ibar JL, Whitelegge JP, Faull KF, Kaback HR, Schuldiner S. Exploring the role of a unique carboxyl residue in EmrE by mass spectrometry. J Biol Chem. 2005;280:7487–7492. doi: 10.1074/jbc.M413555200. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen BE, Rath A, Deber CM. The assembly motif of a bacterial small multidrug resistance protein. J Biol Chem. 2009;284:9870–9875. doi: 10.1074/jbc.M900182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal V, Fink U, Schuldiner S, Reif B. MAS solid-state NMR studies on the multidrug transporter EmrE. Biochim Biophys Acta. 2007;1768:3036–3043. doi: 10.1016/j.bbamem.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Lehner I, Basting D, Meyer B, Haase W, Manolikas T, Kaiser C, Karas M, Glaubitz C. The key residue for substrate transport (Glu14) in the EmrE dimer is asymmetric. J Biol Chem. 2008;283:3281–3288. doi: 10.1074/jbc.M707899200. [DOI] [PubMed] [Google Scholar]
- 35.Elbaz Y, Tayer N, Steinfels E, Steiner-Mordoch S, Schuldiner S. Substrate-induced tryptophan fluorescence changes in EmrE, the smallest ion-coupled multidrug transporter. Biochemistry. 2005;44:7369–7377. doi: 10.1021/bi050356t. [DOI] [PubMed] [Google Scholar]
- 36.Basting D, Lorch M, Lehner I, Glaubitz C. Transport cycle intermediate in small multidrug resistance protein is revealed by substrate fluorescence. FASEB J. 2008;22:365–373. doi: 10.1096/fj.07-9162com. [DOI] [PubMed] [Google Scholar]
- 37.McHaourab HS, Mishra S, Koteiche HA, Amadi SH. Role of sequence bias in the topology of the multidrug transporter EmrE. Biochemistry. 2008;47:7980–7982. doi: 10.1021/bi800628d. [DOI] [PubMed] [Google Scholar]
- 38.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]