Abstract
The murine 2′-5′ oligoadenylate synthetase 1a (Oas1a) and Oas1b genes are type 1 IFN responsive genes. Oas1a is an active synthetase with broad antiviral activity mediated through RNase L. Oas1b is inactive but can inhibit Oas1a synthetase activity and mediate a flavivirus-specific antiviral activity through an unknown RNase L-independent mechanism. Analysis of promoter elements regulating gene transcription confirmed that an IFN-stimulated response element (ISRE) is required for IFN beta-activation but neither the overlapping IRF binding site present in both promoters nor the adjacent Oas1b NF-kappa B site is required. Mutation of the overlapping STAT site negatively affected IFN beta-induction of Oas1a but not of Oas1b. Also, IFN beta induction of Oas1a was STAT1- and STAT2-dependent, while induction of Oas1b was STAT1-independent but STAT2-dependent. The two promoters differ at a single nucleotide in the STAT site. The data indicate that these two duplicated genes can be differentially regulated by IFN beta.
Keywords: interferon stimulated gene, promoter analysis, Oas1a, Oas1b
Introduction
The recognition of viral dsRNA by cellular sensors in infected cells leads to activation of the NF-kappa B, IRF3 and ATF2/c-Jun transcription factors that bind cooperatively to the interferon (IFN) beta promoter and activate its transcription (Merika and Thanos, 2001). The IFN beta secreted by infected cells binds to IFN alpha/beta receptors on the surfaces of both infected and uninfected cells resulting in activation of JAK1 and Tyk2 kinases that phosphorylate STAT1 and STAT2 transcription factors. Phosphorylated STAT1 and STAT2 and IFN regulatory factor 9 (IRF-9) form the IFN stimulated gene factor 3 complex (ISGF3) that translocates to the nucleus where it binds to IFN stimulated response elements (ISREs) in the promoters of IFN stimulated genes (ISGs) and upregulates their expression (Stark et al., 1998).
Three oligoadenylate synthetase genes (OAS1, OAS2 and OAS3) and one OAS-like (OASL) gene have been identified in the human genome (Hovnanian et al., 1998; Rebouillat et al., 1998). The transcripts of three of these genes are alternatively spliced and polymorphisms have been identified that alter splicing sites. Five isoforms have been reported for OAS1 (p42, p44, p46/p48 and p52), two for OAS2 (p69 and p71), one for OAS3 (p100) and two for OASL (p30 and p59) (Hartmann et al., 1998; Rebouillat et al., 1998; Hovnanian et al., 1999; Justesen et al., 2000; Bonnevie-Nielsen et al., 2005). In contrast, the mouse genome encodes eight OAS1 (Oas1a-Oas1h), two of OASL (Oasl1 and Oasl2), one of OAS2 (Oas2) and one of OAS3 (Oas3) gene orthologs (Rutherford et al., 1991; Justesen et al., 2000; Shibata et al., 2001; Kakuta et al., 2002). Gene duplication, rather than alternative splicing, is responsible for the multiple murine Oas1 isoforms (Perelygin et al., 2006). Enzymatically active OAS proteins play an important antiviral role. When activated by viral dsRNA, they catalyze the synthesis of short 2′-5′-oligoadenylates (2-5A) from ATP. 2-5A activates latent endonuclease RNase L which degrades both cellular and viral single-stranded RNAs (Samuel, 2001). Among the eight murine Oas1 proteins, only Oas1a and Oas1g have been reported to be active 2′-5′-oligoadenylate synthetases (Kakuta et al., 2002; Elbahesh et al., 2011). However, the inactive synthetase Oas1b mediates resistance to flavivirus-induced disease through an unknown mechanism that is independent of RNase L (Scherbik et al., 2006). The flavivirus-induced disease resistant mouse strain C3H/RV is homozygous for the dominant Oas1br allele encoding a full length Oas1b protein, while the congenic susceptible mouse strain C3H/He is homozygous for the recessive Oas1bs allele that encodes a C-terminally truncated Oas1btr protein (Perelygin et al., 2002). A previous study showed that the full length Oas1b protein, which is an inactive synthetase, can inhibit in vitro Oas1a synthetase activity in a dose-dependent manner and reduce 2-5A production in vivo in response to poly(I:C) (Elbahesh et al., 2011). A similar function was reported for Oas1d, another of the inactive Oas1 proteins (Yan et al., 2005).
Each of the human OAS genes contains an ISRE in its promoter and is induced by type I IFN (Wang and Floyd-Smith, 1997; Hartmann et al., 1998; Floyd-Smith et al., 1999; Yu et al., 1999; Rebouillat et al., 2000). Similarly, murine Oas genes with the exception of Oas1f were reported to be activated by type I IFN (Eskildsen et al., 2002; Eskildsen et al., 2003). A previous TFSEARCH analysis predicted transcription factor binding sites (TFBSs) in 500 bp promoter fragments of the murine Oas1a-h, Oas2 and Oas3 genes and identified an ISRE in only the promoters of the Oas1a, Oas1b, Oas1g and Oas2 genes (Mashimo et al., 2003). Only the ISRE in the Oas1b promoter was predicted to overlap GAS and NF-kappa B sites. These results suggested the possibility of differential regulation of Oas1a and Oas1b expression by IFN and/or viral infection but this prediction was not functionally tested.
In the present study, the Oas1a and Oas1b promoters from both C3H/RV and C3H/He mouse embryofibroblasts (MEFs) were cloned and sequenced. A GENOMATIX search of the Oas1a and Oas1b promoter sequences predicted that neither had a TATA box but that both had a canonical initiator element (INR). An inverted CCAAT element (ICE) was predicted and functional analysis showed that it may be important for the basal activities of the C3H/He and C3H/RV Oas1b promoters and the C3H/RV Oas1a promoter. The C3H/He Oas1a promoter contained a mutation in this site that was predicted to make it nonfunctional. A single ISRE as well as overlapping STAT and IRF sites were predicted in both promoters. Functional mapping of the Oas1a and Oas1b promoters by sequential 5′ deletion and TFBSs mutagenesis indicated that the ISRE as well as the overlapping STAT site are required for Oas1a promoter induction by IFN beta while Oas1b expression requires only the ISRE. Also, both STAT1 and STAT2 are required for Oas1a upregulation by IFN beta, while only STAT2 is required for Oas1b upregulation. A single nucleotide difference between the STAT sites of the Oas1b and Oas1a promoters appeared to be responsible for the differential STAT1-dependence of Oas1a and Oas1b expression.
Results
Mapping the Oas1a and Oas1b gene promoter regions required for basal promoter expression and induction by IFN beta
The Oas1a and Oas1b genes are ISGs and after treatment of C3H/He MEFs with 1000 U/ml of murine IFN beta for 3 h, Oas1a mRNA was upregulated by about 9 fold while the Oas1b mRNA was upregulated by about 7 fold (Fig. 1A). The time and the dose of IFN treatment was selected based on a previous study showing that the highest level of ISG induction was at 3h after IFN treatment of MEFs and that the levels of ISG mRNA upregulation were similar with 10, 100 and 1000 U/ml of IFN beta (Scherbik et al., 2007). The regions of the Oas1a and Oas1b proximal promoters required for basal gene expression and activation by IFN beta were then mapped using luciferase reporter assays. Two firefly luciferase reporter gene constructs, one containing a C3H/RV Oas1a gene promoter fragment [Oas1a (−1768, +28)] and the other containing a C3H/RV Oas1b gene promoter fragment [Oas1b (−1398, +51)], were first generated and then a set of 5′ sequentially deleted constructs was made for each promoter. C3H/RV MEFs were transfected with construct DNA, and 24 h later cell lysates were harvested and used to analyze basal promoter activity. To analyze the effect of IFN on Oas1 promoter activity, cells transfected with a luciferase reporter for 24 h were incubated with murine IFN beta. Initial pilot experiments showed that although luciferase activity was the highest at both 3 and 6 h after treatment, the 3h peak level was the most reproducible. At 24 and 48 h, luciferase activity was significantly decreased. Based on these preliminary results and the maximum half-life of the firefly luciferase protein of 4 h (Thompson et al., 1991; Brandes et al., 1996), assays were done in all subsequent luciferase reporter experiments after 3 h of IFN beta treatment. Low basal luciferase activities were detected for both the longest Oas1a (−1768, +28) construct (Fig. 1B) and the longest Oas1b (−1398, +51) construct (Fig. 1C). These constructs showed a modest but significant increase in promoter activity after IFN treatment. The observation that the longest Oas1a and Oas1b promoter fragments tested produced the lowest reporter activities suggested the presence of upstream transcriptional repressor elements that negatively affected the basal and IFN-induced expression levels of both promoters.
Figure 1. IFN beta-induced upregulation of Oas1a and Oas1b genes in vivo and of promoter reporter constructs in vitro.
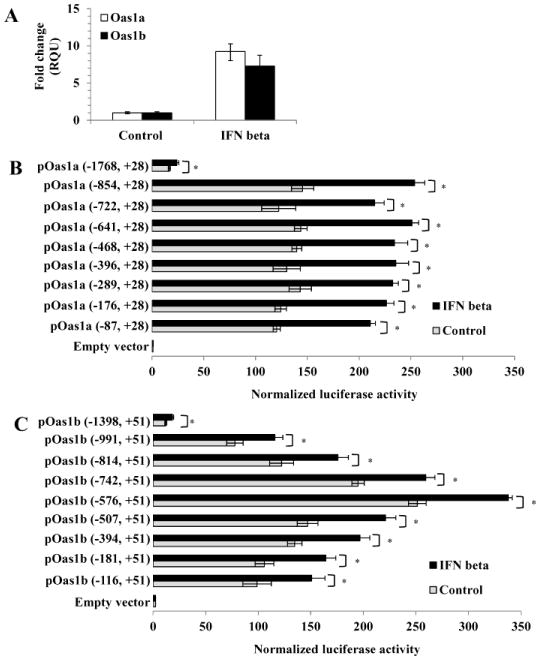
(A) C3H/He MEFs were treated with 1000 U/ml of murine IFN beta for 3h or left untreated (control). Total RNA was extracted and Oas1a and Oas1b mRNA levels were measured by real time qRT-PCR. The mRNA level for each gene was normalized to the level of GAPDH mRNA in the same sample and is shown as the fold change over the amount of mRNA in mock samples expressed in relative quantification units (RQU). Each experiment was preformed in triplicate and representative data from one of three independent experiments are shown. Luciferase reporter assays were done with (B) Oas1a and (C) Oas1b promoter fragments of different lengths. C3H/RV MEFs were co-transfected with either a luciferase reporter construct DNA or a control empty vector DNA and Renilla luciferase reporter vector DNA (transfection efficiency control). At 24 h after transfection, MEFs were either treated with 1000 U/ml of murine IFN beta for 3 h or left untreated (control). Cell lysates were prepared and luciferase activity was measured in triplicate. Firefly luciferase activity was normalized to Renilla luciferase activity for each sample. Error bars represent standard error of the mean (SEM) (n=3). Significant differences were determined with a Student's t test (*, P < 0.05).
Promoter constructs with sequential 5′ deletions were used to determine the shortest Oas1a and Oas1b promoter fragments able to produce increased luciferase activity upon stimulation with IFN beta. Each of the 5′ deleted Oas1a promoter constructs tested produced similar significantly increased basal luciferase activities compared to the Oas1a (1768, +28) construct and the activities for each of these constructs increased upon stimulation with IFN beta (Fig. 1B). For instance, the Oas1a (−87, +28) and Oas1a (−854, +28) constructs had basal activities that were 7.1 and 8.6 fold higher, respectively, than that of the Oas1a (−1768, +28) construct and both showed a 1.75 fold increase in luciferase activity after stimulation with IFN beta. These results indicated that the TFBSs required for IFN induction of the Oas1a are located between −87 and +28.
In contrast to what was observed with the truncated Oas1a constructs, the activities of each of the 5′ deleted Oas1b constructs varied. The Oas1b (−576, +51) construct showed a 21 fold increase over the activity produced by the Oas1b (−1398, +51) construct (Fig. 1C). Constructs that were either shorter or longer than Oas1b (−576, +51) produced lower basal activities. The observation that the basal activity of the Oas1b (−116 to +51) construct was 2.5 fold lower than that of Oas1b (−576, +51) suggested that the region between −576 and −116 contains elements that enhance basal Oas1b expression. As the length of the Oas1b constructs increased beyond −576 bp, the basal activity decreased suggesting the presence of repressor elements in this region. The activity of each of the truncated Oas1b promoter constructs was induced to a similar extent above its basal level by IFN beta treatment (Fig. 1C) indicating that the elements required for the induction of the Oas1b promoter by IFN beta are located between −116 and +51 bp.
Although the results obtained were reproducible and statistically significant, basal levels were high and the maximal induction observed with IFN beta was about 2 fold. The low levels of induction by IFN are likely to be due to the use of promoter fragments with only a single ISRE in the context of a natural promoter sequence which also contains multiple additional TF binding sites that modulate both basal and IFN-induced luciferase activity. Previous studies using natural TLR9 (Guo et al., 2005) and RIG-I (Su et al., 2007) promoter sequences with single copies of an ISRE also reported a 2 fold or lower activation of the reporter gene after stimulation with IFN beta.
Analysis of the core promoter elements in the Oas1a and Oas1b promoters
Elements in the −87 to +28 C3H/RV Oas1a promoter fragment and in the −116 to +51 C3H/RV Oas1b promoter fragment were predicted with GENOMATIX and TFSEARCH programs. Neither promoter contained a TATA box but each contained an INR (Fig. 2), an element commonly found in TATA-less promoters and known to be able to direct transcriptional initiation (Smale and Baltimore, 1989; Smale, 1997). Within the INR consensus C/TC/TA+1NA/TC/TC/T, the adenine is the transcription start site (TSS) and this was used to predict the location of the Oas1a and Oas1b TSSs. The GENOMATIX program also predicted an inverted CCAAT element (ICE) at −63 to −68 in the Oas1a promoter and at −62 to −67 in the Oas1b promoter. CCAAT boxes in either the forward (CCAAT) or reverse (ICE; ATTGG) orientation are typically located about 60 to 100 nt upstream of the transcription start site (TSS) (Mantovani, 1999; Dolfini et al., 2009). To determine whether the predicted ICE plays a functional role in the Oas1a and Oas1b promoters, two nucleotides (ATTGG → ATCCG) were substituted in this element in the Oas1a (−854, +28) and Oas1b (−576, +51) constructs. A GENOMATIX search confirmed that the introduced mutations eliminated the targeted binding site and did not create a new TFBS. Mutation of ICE reduced the basal luciferase activity of the Oas1a and Oas1b promoters by 43% and 40%, respectively, but did not negatively affect induction of either promoter by IFN beta (Fig. 3A). IFN beta induced the wild type and ICE-mutated Oas1a promoters by 1.7- and 1.9 fold, respectively, and the wild type and ICE-mutated Oas1b promoter by 1.4 and 1.5 fold, respectively.
Figure 2. Predicted TFBSs in the Oas1a and Oas1b promoters.
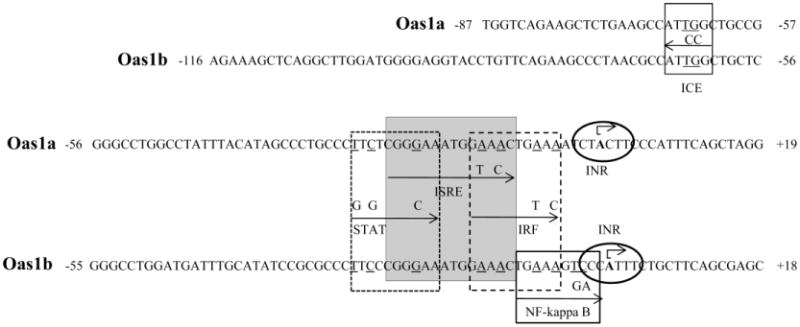
TFBSs predicted by the GENOMATIX search program. The locations of the DNA fragments with respect to the predicted TSS (+1) in an INR are indicated. Substituted nucleotides in both promoters are underlined and substitutions made in promoter fragments are shown above arrows indicating the boundaries of individual TFBSs.
Figure 3. Functional analysis of the predicted Oas1a and Oas1b ICEs.
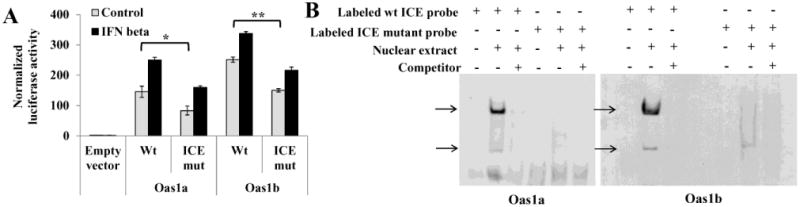
(A) Luciferase reporter assays were done with control and mutated ICE constructs. Two nts in the ICE of the Oas1a (−854, +28) and Oas1b (−576, +51) constructs were substituted as indicated in Fig. 2. At 24 h after transfection of a wild type or mutant construct, C3H/RV MEFs were either treated with murine IFN beta (1000 U/ml) for 3 h or untreated and then luciferase activities were analyzed. Firefly luciferase activity measured in triplicate was normalized to Renilla luciferase activity. Error bars represent SEM (n=3). Statistical significance was determined with a Student's t test (*, P < 0.05; **, P < 0.005). (B) EMSA. DIG-labeled Oas1b and Oas1a DNA probes containing either a wild type or mutated ICE and nuclear extracts from C3H/RV MEFs were used for the binding reactions. A 200 fold excess of unlabeled specific competitor DNA was added prior to addition of the probe in the indicated reactions. DNA-protein complexes were resolved on 6% native gels and the bound labeled probe was detected using anti-DIG antibody. The results shown are representative of at least two independent experiments.
DNA probes consisting of the −88 to −49 bp for Oas1a or −82 to −48 bp for Oas1b (Table 3) were next used in an electrophoretic mobility shift assay (EMSA) to determine whether TFs bind to the ICE sequence. Oas1a and Oas1b probes with a wild type ICE bound to two complexes in nuclear extracts from untreated C3H/RV MEFs (Fig. 3B). With both probes, the upper band was much darker than the lower band. These bands were either not detected or were much fainter when a specific unlabeled competitor DNA was included in the reaction suggesting that the binding detected was specific. Neither of the shift bands was observed when an Oas1a probe containing a mutated ICE was tested. An Oas1b probe with a mutated ICE detected a faint lower complex band but did not detect an upper complex band. The results suggest that nuclear extracts from untreated control cells contain TFs that bind to the ICE. Although the TFs binding to the ICE in the Oas1a and Oas1b promoters were not identified, both the reporter assay (Fig. 3A) and the EMSA (Fig. 3B) results suggest that the ICE element may play a role in the regulation of basal transcription from both genes.
Table 3.
Sequences of DNA probes used in EMSA.
| Gene | TFBS | Location | Sequence |
|---|---|---|---|
| Oas1a | ISRE | −31 to +7 | GCCCTTCTCGGGAAATGGAAACTGAAAATCTACTTCCC |
| Oas1a | ICE | −83 to −49 | CAGAAGCTCTGAAGCCATTGGCTGCCGGGGCCTGG |
| Oas1a | mutICE | −83 to −49 | CAGAAGCTCTGAAGCCATCCGCTGCCGGGGCCTGG |
| Oas1b | ISRE | −33 to +8 | CGCCCTTCCCGGGAAATGGAAACTGAAAGTCCCATTTCTGC |
| Oas1b | ICE | −82 to −48 | CAGAAGCCCTAACGCCATTGGCTGCTCGGGCCTGG |
| Oas1b | mutICE | −82 to −48 | CAGAAGCCCTAACGCCATCCGCTGCTCGGGCCTGG |
Comparison of the sequences of the Oas1a and Oas1b promoters in resistant C3H/RV and susceptible C3H/He MEFs
To determine whether the sequences of the Oas1a or Oas1b promoters differ between congenic flavivirus disease susceptible C3H/He and flavivirus disease resistant C3H/RV mice, Oas1a (−1768, +28) and Oas1b (−1398, +51) DNA fragments were amplified by PCR from MEFs from each mouse strain, cloned and sequenced. Although complete identity was observed between the C3H/RV and C57BL/6J (NT_078458.6) Oas1a and Oas1b promoter sequences, the C3H/He Oas1a promoter sequence differed by T to C substitutions at positions −66 and −298 and the C3H/He Oas1b promoter sequence differed by a G to C substitution at position +15 and a 4 nt deletion between −327 to −323. GENOMATIX and TFSEARCH TFBS searches of the C3H/He promoter sequences predicted that only the T to C substitution at −66 in the Oas1a promoter was located in a TFBS. This substitution changed the ICE binding site from ATTGG to ATCGG. Based on the observation that mutation of the ICE from ATTGG to ATCCG significantly decreased the basal activities of both the C3H/RV Oas1a and Oas1b promoters, the single nt substitution in the C3H/He Oas1a promoter producing a ATCGG ICE sequence could be expected to reduce the basal expression level of the Oas1a gene in C3H/He MEFs.
Analysis of transcription factors binding to the Oas1a and Oas1b promoters
Both the previously published search (Mashimo et al., 2003) and the TFSEARCH and GENOMATX TFBS searches done in this study predicted canonical ISREs with identical sequences (5′ GGGAAATGGAAACT 3′) between −22 to −9 bp in the Oas1a promoter and between −23 to −10 bp in the Oas1b promoter (Fig. 2B). To determine whether the components of the ISGF3 complex bind to the predicted Oas1a and Oas1b ISREs, −31 to +6 bp Oas1a and −32 to +5 bp Oas1b DNA fragments were used as DNA probes in EMSAs with nuclear extracts from untreated or murine IFN beta-treated MEFs. Two bands were detected with the Oas1a probe and three with the Oas1b probe using nuclear extracts from IFN-treated cells (Fig. 4A). However, the upper complex band was not observed with either probe when the reaction was done with nuclear extracts from untreated cells. To determine whether the STAT1 and STAT2 transcription factors were present in one or more of these bands, nuclear extracts were incubated with either anti-STAT1 or anti-STAT2 antibody prior to addition of the probe. With the Oas1a probe, the upper band was not detected when STAT1 antibody was present, and a supershift band (indicated by an arrow) was observed when STAT2 antibody was present. With the Oas1b probe, a decrease in the intensity of the upper band was observed when STAT1 antibody was added and a supershift band was detected when the STAT2 antibody was added. The addition of a nonspecific rabbit IgG did not alter the band patterns of either probe. The results suggested that both STAT1 and STAT2 were present in the upper complex band detected with both probes. In order to confirm the validity of the supershift data the binding of STAT1 and STAT2 to the ISREs in the Oas1a and Oas1b promoters in cells stimulated with IFN beta was analyzed by ChIP (Fig. 4B and C). Pilot ChIP experiments showed that the levels of STAT1 and STAT2 bound to these ISG promoters was higher at 30 m than at 3 h after treatment with IFN beta. C3H/He cells were left untreated or treated with 1000 U/ml of IFN beta for 30 m and then treated with formaldehyde. Crosslinked DNA-protein complexes were immunoprecipitated using anti-STAT1, anti-STAT2 or a nonspecific IgG antibody. The crosslinks were reversed and the immunoprecipitated DNA was purified and quantified by real time qPCR. The results showed that both STAT1 and STAT2 are present on the Oas1a and Oas1b promoters after stimulation of the C3H/He cells by IFN beta. Although one of the two anti-IRF-9 antibodies (Santa Cruz Biotechnology) tested detected IRF-9 in Western blots, neither of these antibodies was found to be suitable for EMSA nor ChIP, so that the interaction of IRF-9 with the ISREs of these promoters would not be directly tested.
Figure 4. Binding of STAT1 and STAT2 to the Oas1a and Oas1b promoters in vitro and in vivo.
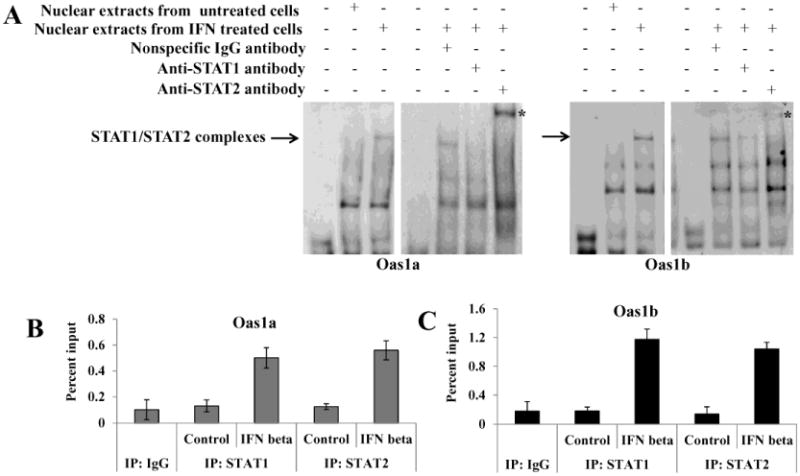
(A) DIG-labeled Oas1b and Oas1a DNA probes and nuclear extracts from untreated or murine IFN beta (1000 U/ml) treated C3H/RV MEFs were used for EMSA. Anti-STAT1, anti-STAT2 or a nonspecific IgG antibody was added to the nuclear extract prior to addition of the probe. DNA-protein complexes were resolved on 6% native gels and the labeled probe was detected with anti-DIG antibody. The results shown are representative of at least two independent experiments. ChIP analysis of the binding of STAT1 or STAT2 to the (B) Oas1a or (C) Oas1b promoter in control or 30 min IFN beta treated C3H/He cells. The amounts of precipitated Oas1a and Oas1b promoter DNA were quantified by real-time qPCR using promoter-specific primers and fluorogenic TaqMan FAM/MGB probes. Nonspecific IgG antibody was used as a negative control and averaged values are shown. The bars represent standard deviation (SD) (n=3).
Functional analysis of TFBSs predicted in the Oas1a and Oas1b promoters
The GENOMATIX search done in the present study predicted a STAT site overlapping the 5′ end and an IRF overlapping the 3′ end of both the Oas1a and Oas1b ISREs (Fig. 2). These predictions suggested that the sequences located immediately upstream and downstream of the ISRE might be important for stabilizing the binding of STAT1 and IRF-9 to the core ISRE and/or for facilitating the binding of additional STAT and IRF factors. To functionally test these predictions, the ISRE and overlapping STAT and IRF TFBSs were individually mutated in the Oas1a (−854, +28) and Oas1b (−576, +51) constructs as indicated in Fig. 2. The luciferase activity of each mutated reporter construct after stimulation with murine IFN beta was compared to that of the wild type construct. A GENOMATIX search confirmed that the introduced mutations eliminated the targeted binding site but did not create a new TFBS. Mutation of the ISRE (3′ GAAA to 3′ GTAC) in the Oas1a (Fig. 5A) and Oas1b (Fig. 5B) constructs reduced the basal activities of both promoters by more than 80% suggesting that the ISRE plays a role in modulating the basal expression of Oas1a and Oas1b. Low levels of IFN were previously reported to be secreted by unstimulated, cultured MEFs (Takaoka and Yanai, 2006) and this could explain the negative effect of the ISRE mutation on basal expression. No increase in activity above basal levels was observed for either the Oas1a or Oas1b promoter with a mutated ISRE after stimulation with IFN beta confirming that the ISRE is required for IFN beta-induction of both genes.
Figure 5. Effect of mutation of the ISRE or overlapping/adjacent TFBSs on Oas1a and Oas1b promoter activity.
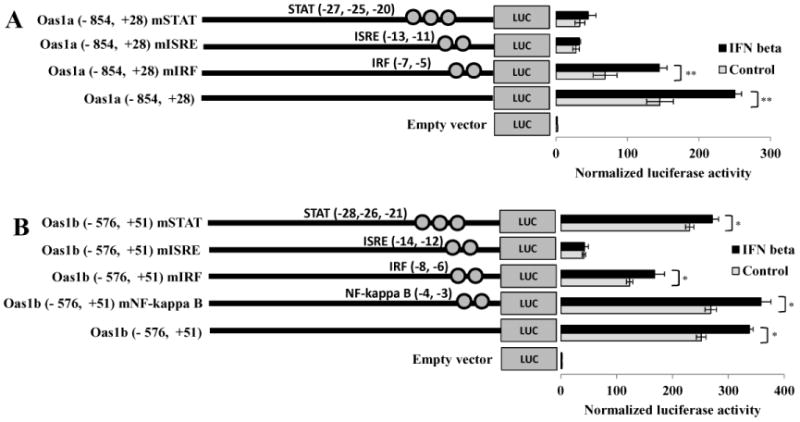
Individual TFBSs were mutated in reporter constructs as indicated in Fig. 2. The luciferase activities of control and mutated (A) Oas1a and (B) Oas1b reporter constructs were assayed. Each construct was transfected into C3H/RV cells and 24 h after transfection, cells were treated with 1000 U/ml of murine IFN beta for 3 h or left untreated. Luciferase activity was measured in triplicate and normalized to Renilla luciferase activity. The values shown are the average of three independent experiments. Error bars represent SEM (n=3). Statistical significance was determined with a Student's t test (*, P < 0.05; **, P < 0.005)
The predicted IRF site overlapping the 3′ end of the ISRE was mutated at the −5 and −7 positions in the Oas1a promoter and at the −6 and −8 positions in the Oas1b promoter (Fig. 2). Although mutation of the IRF site resulted in about a 50% reduction in basal Oas1a and Oas1b promoter activity, both mutated promoters were efficiently upregulated by IFN (Fig. 5). IFN beta induced the wild type Oas1a construct activity by 1.7 fold and that of the Oas1a IRF mutant construct by 2.1 fold. Both the wild type and mutant IRF Oas1b constructs showed a 1.3 fold induction after IFN treatment. These results indicate that the IRF binding site overlapping the ISRE in both promoters positively regulates basal promoter activity, but does not contribute to the induction of these promoters by IFN beta.
Mutations were next introduced into the STAT site overlapping the 5′ end of the Oas1a and Oas1b ISREs. Substitutions were introduced at positions −20, −25 and −27 in the Oas1a promoter and at positions −21, −26 and −28 in the Oas1b promoter (Fig. 2). Similar to what was observed for the Oas1a construct with a mutated ISRE, the Oas1a mutant STAT construct showed a more than an 80% reduction in basal activity but was not upregulated by IFN beta. In contrast, mutation of the STAT site in the Oas1b promoter only minimally affected basal or IFN beta-induced activities. These results suggest that the STAT site overlapping the Oas1a ISRE is required for the induction of this promoter by IFN beta, while the STAT site overlapping the Oas1b ISRE is not.
The TFBS searches done by Mashimo et al. (2003) and in the present study predicted an NF-kappa B site immediately downstream of the Oas1b ISRE but not of the Oas1a ISRE. The sequence of this site differs from the NF-kappa B consensus GGGA/GNNC/TC/TCC (Grilli et al., 1993) at the two underlined positions (Fig. 2). Type 1 IFN binding to the IFN alpha/beta receptor has been reported to also activate the phosphatidylinositol 3 kinase (PI3K) pathway (Kaur et al., 2005) which was shown to play a role in activating NF-kappa B (Chang et al., 2006). The Oas1b NF-kappa B site was mutated as indicated in Fig. 2. The luciferase activity observed for this mutant construct was the same as that for the wild type construct with and without IFN treatment (Fig. 5B). The results indicate that the NF-kappa B site does not modulate the induction of the Oas1b promoter by IFN beta.
IFN beta-induction of the Oas1a and Oas1b genes in the absence of STAT1 or STAT2
As one means of determining whether STAT1 is required for the activation of the Oas1a and Oas1b genes by IFN beta, the expression of these genes was analyzed by quantitative real time RT-PCR in wild type, STAT1-/- and STAT2-/- MEFs. In wild type MEFs, Oas1a and Oas1b gene expression was efficiently induced by IFN beta (Fig. 6A). Basal levels of Oas1a mRNA were not detectable in either untreated STAT1-/- or STAT2-/- MEFs and minimal upregulation of Oas1a expression was observed after incubation of these cells with IFN beta. Basal levels of Oas1b were detected in both STAT1-/- and STAT2-/- MEFs. Although IFN beta did not upregulate Oas1b expression in STAT2-/- MEFs, it efficiently upregulated Oas1b expression in STAT1-/- MEFs. To confirm these observations, the binding of STAT1 and STAT2 to the Oas1a and Oas1b promoters in control, STAT-/- and STAT2-/- MEFs was analyzed by ChIP. Both STAT1 and STAT2 bound the Oas1a and Oas1b promoters in IFN beta-treated control 129 MEFs but neither bound to either promoter in the STAT2-/- MEFs (Fig. 6B and C). In contrast, STAT2 bound to the Oas1b promoter but not to the Oas1a promoter in STAT1-/- MEFs confirming that Oas1a promoter induction by IFN beta occurs in a STAT1- and STAT2-dependent manner, while the induction of Oas1b by IFN beta is STAT2-dependent but can be STAT1-independent.
Figure 6. Analysis of the induction of Oas1a and Oas1b by IFN beta in wild type, STAT1-/- and STAT2-/- MEFs.
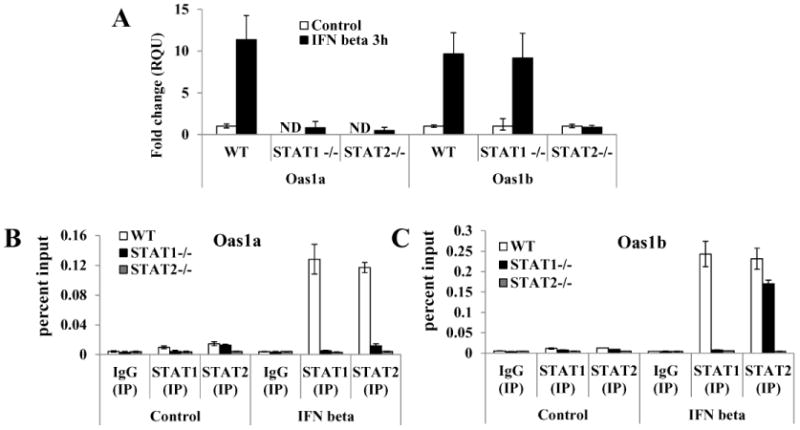
MEFs were treated with 1000 U/ml of murine IFN beta for 3 h or left untreated. Total RNA was collected and fold induction of Oas1a and Oas1b mRNA expression levels was measured by real-time qRT-PCR. The mRNA level of each gene was normalized to the level of GAPDH mRNA in the same sample. The fold change over the amount of mRNA in mock samples is expressed in RQU. Each experiment was preformed in triplicate and representative data from one of three independent experiments are shown. ND - not detected. ChIP analysis of the binding of STAT1 or STAT2 to the (B) Oas1a or (C) Oas1b promoter in control 129, STAT1-/- and STAT2-/- MEFs mock treated or treated with 1000 U/ml of IFN beta for 30 min. The amounts of precipitated Oas1a and Oas1b promoter DNA were quantified by real-time qPCR using promoter specific primers and fluorogenic TaqMan FAM/MGB probes. Nonspecific IgG antibody was used as a negative control and averaged values are shown. The bars represent SD (n=3).
Discussion
The mouse genome contains 8 duplications of the Oas1 gene. As an active 2-5 A synthetase murine Oas1a functions as a broad spectrum antiviral protein through its ability to activate RNase L (Kakuta et al., 2002). Oas1b is an inactive synthetase that can function in a dominant negative manner to suppress Oas1a synthetase activity (Elbahesh and Brinton, 2011). Oas1b also provides flavivirus-specific antiviral activity through an as yet uncharacterized mechanism that does not involve the RNase L pathway (Scherbik et al., 2006). Based on TFBS differences identified in the Oas1a and Oas1b promoters, a previous study predicted that these two genes may be differentially activated by IFN and/or virus infection but this hypothesis was not functionally tested (Mashimo et al., 2003). In the present study, a firefly luciferase reporter assay and truncated and mutated promoter constructs were used to map the Oas1a and Oas1b promoter regions required for basal activity and IFN beta induction. For both genes, the lowest basal promoter activity was observed with the longest promoter fragment tested but activity increased significantly with 5′ truncation suggesting the presence of binding sites for transcriptional repressors in the upstream region. Previous studies reported that deletion of the 5′ upstream region was a prerequisite for mapping the activation sites in the proximal promoters of the interleukin 19 and fucosyltransferase VI genes (Chen et al., 2006; Higai et al., 2008). Upstream sites that repress the transcriptional activation by TFs binding to TFBSs in the proximal promoter region were also previously identified in the promoters of genes for which transcription is known to be tightly regulated (Garban and Bonavida, 2001; Lindas and Tomkinson, 2007). Since activation of the RNase L pathway by 2-5A leads to degradation of not only viral but also cellular ssRNAs, it would be expected that the expression of the Oas1 genes would be tightly regulated.
The core promoters of the Oas1a and Oas1b genes contain an INR element and an inverted ICE. Mutation of the ICE element reduced the basal activities of the Oas1a and Oas1b promoters by about 40% but did not affect the induction of either promoter by IFN beta suggesting that ICE plays a role in basal promoter activity but not in IFN-mediated induction. The EMSA results showed that TFs in control cell nuclear extracts bound to a probe with a wild type but not a mutated ICE sequence. Although the identity of the TFs was not determined, NF-Y, also termed CBF (CCAAT binding factor), was previously shown to be the primary TF binding to the CCAAT element in either orientation (Mantovani, 1999). NF-Y is a ubiquitous transcription factor composed of three subunits that can increase the DNA binding affinity of factors bound to neighboring TFBSs (Reith et al., 1994; Wright et al., 1995; Jackson et al., 1998) and interact with p300/CBP, GCN5 and PCAF complexes that mediate promoter acetylation (Currie, 1998; Jin and Scotto, 1998; Li et al., 1998; Salsi et al., 2003). NF-Y has also been reported to recruit RNA polymerase II to a promoter (Kabe et al., 2005). The mutation identified in the ICE of the C3H/He Oas1a promoter suggests that the basal levels of Oas1a expression would be reduced in the cells of these mice.
Deletion analysis of the Oas1b promoter showed that the regulation of this gene is more complex than that of the Oas1a gene. The mapping data indicated that the region of the Oas1b promoter between −576 and −116 contains elements that enhance basal Oas1b expression. TFBSs predicted by TFSEARCH and GENOMATIX in this region of the Oas1b promoter that were not found in the Oas1a promoter include C/EBPb, GATA-1, AML-1, CdxA, HSF2, IK-2, Lyf-1, and MZF-1. C/EBPb (also known as CCAAT/enhancer-binding protein beta) was previously shown to positively regulate gene induction by recruiting transcriptional coactivators as well as basal transcription factors (Kowenz-Leutz and Leutz, 1999).
Upon type I IFN stimulation, the transcription factor complex ISGF3, composed of STAT1, STAT2 and IRF-9, binds to ISREs in ISG promoters with the consensus sequence 5′ A/GNGAAANNGAAACT 3′ (Darnell et al., 1994). The Oas1a and Oas1b promoters were each predicted to have a single canonical ISRE by a previous TFSEARCH (Mashimo et al., 2003) and also by both the GENOMATIX and TFSEARCH database searches done in the present study. Since the structure of ISGF3/DNA complex has not yet been solved is not clear exactly how the ISGF3 complex components bind to the ISRE. It was previously reported that STAT1 recognizes the 5′ GAAA sequence and IRF-9 recognizes the 3′ GAAA sequence in the ISRE. STAT2 does not stably bind to DNA but interacts with both STAT1 and IRF-9 and is essential for the transcriptional activity of the ISGF3 complex (Bluyssen et al., 1995; Qureshi et al., 1995). The substitutions introduced into the ISRE 3′ GAAA in both the Oas1a and Oas1b promoters abolished IFN beta induction indicating that this region is required for IFN beta induction of these ISGs. A GAS site was previously predicted to overlap the 5′ end of the Oas1b ISRE but not that of the Oas1a (Mashimo et al., 2003). GAS elements were originally identified in the promoters of IFN-gamma responsive genes. Upon IFN-gamma stimulation, a STAT1 homodimer, also referred to as the gamma interferon activation factor (GAF), was shown to bind to GAS elements with the consensus sequence TTCN2-4GAA (Decker et al., 1997). Although other STATs can bind to this consensus sequence, the DNA binding specificities of different STAT proteins depends on the length of the spacer sequence and specific nucleotides in both the binding site and immediately adjacent to it (Darnell, 1997; Decker et al., 1997; Ehret et al., 2001). The GENOMATIX search done in this study predicted a STAT binding site overlapping the 5′GAAA motif of the ISRE in both the Oas1a and Oas1b promoters. Substitution of the G in this ISRE motif was reported to prevent the binding of STAT1 but not of the IRF-9/STAT2 complex (Bluyssen and Levy, 1997). Mutation of the Oas1a STAT site including the G mentioned above abolished IFN beta-induced promoter activity suggesting that the binding of STAT1 is crucial for the induction of this promoter by IFN beta. In contrast, mutation of the same nucleotides in the Oas1b STAT site did not significantly reduce upregulation by IFN beta. Consistent with these observations, the Oas1b gene but not the Oas1a gene was efficiently upregulated in IFN-treated STAT1-/- MEFs confirming that the induction of Oas1b by IFN beta does not require STAT1.
Although the sequences of the Oas1a and Oas1b ISREs are identical, the STAT sites overlapping the 5′ ISRE GAAA motif differ by one nt, Oas1a (TTCTCGGGAA) and Oas1b (TTCCCGGGAA). A previous report proposed that the presence of an A, G or T at this position promotes STAT2 binding (Tenoever et al., 2007). However, the finding in the present study that IFN beta activation of the Oas1a promoter with a T in this position is STAT1-dependent while activation of the Oas1b promoter with a C in this position is STAT1-independent suggests that this site is important for stabilizing STAT1 binding and that the substitution at this site is responsible for the differential STAT1 dependence between these two promoters. Although other ISGs, such as Adar1, APOBEC3G and Irf7, were previously reported to be upregulated by type I IFN in a STAT1-independent manner, the TF complexes involved in the STAT1-independent activation of ISGs by type I IFN have not yet been well characterized (Ousman et al., 2005; Sarkis et al., 2006; George et al., 2008). Expression of a hybrid IRF-9/STAT2 protein efficiently activated the transcription of an ISRE-dependent luciferase reporter in the absence of type I IFN stimulation (Kraus et al., 2003). Also, an IRF-9/STAT2 complex was sufficient to induce the expression of a reporter construct containing the RIG-G gene promoter upon stimulation with IFN alpha (Lou et al., 2009). However, it is currently not known whether additional factors are also involved in STAT1-independent upregulation of ISGs by type I IFN.
The GENOMATIX search predicted an IRF binding site overlapping the 3′GAAA motif of the ISRE in both the Oas1a and Oas1b promoters and an NF-kappa B binding site immediately downstream of the Oas1b ISRE. Although these sites were not found to be involved in gene activation by IFN, they may be involved in the upregulation of these genes in virus-infected cells. It was previously reported that IRF-3, IRF-7 and NF-kappa B transcription factors are activated during viral infections and mediate induction of type I IFN as well as upregulate a subset of ISGs independently of IFN (Nakaya et al., 2001; Grandvaux et al., 2002; Peters et al., 2002; Barnes et al., 2004; Elco et al., 2005; Andersen et al., 2008; Basagoudanavar et al., 2011).
Low levels of IFN produced by uninfected cells are required for maintaining basal levels of ISGs (Taniguchi and Takaoka, 2001). Low level constitutive expression of ISGs in uninfected cells is thought to be crucial for mounting an effective antiviral response in the initial stages of a virus infection (Basagoudanavar et al., 2011). However, the data obtained in this study indicated that basal expression of neither Oas1a nor Oas1b is solely dependent on type I IFN. Many viruses antagonize type I IFN signaling and the induction of antiviral ISGs by blocking the activation of the Jak-STAT signaling pathway (Randall and Goodbourn, 2008; Diamond, 2009). The observation made in this study that Oas1b and in previous studies, that some other ISGs can be induced by type I IFN in a STAT1-independent manner indicates that an effective antiviral response can still be mounted in infected cells when STAT1 activation/translocation is blocked. The importance of an alternative STAT1-independent response in providing host protection was reported for dengue virus (another flavivirus) in a mouse model (Perry et al., 2011). However, other types of viruses, such as measles virus and lymphocytic choriomeningitis virus, have been reported to enhance their survival by suppressing dendritic cell development and expansion through a STAT2-dependent but STAT-1-independent pathway (Hahm et al., 2005).
Materials and Methods
Cells
SV40-T antigen transformed C3H/He and C3H/RV MEFs were grown in minimal essential medium (MEM) supplemented with 5% fetal bovine serum (FBS) and 10 μg/ml gentamicin. Transformed 129/SvEv, STAT1-/- and STAT2-/- MEF lines (provided by Christian Schindler, Columbia University, New York, NY) were grown in MEM supplemented with 10% FCS and 1% PenStrep (Gibco).
Quantification of mRNA levels
Real time quantitative reverse transcription-PCR (qRT-PCR) analyses of mouse Oas1a and Oas1b gene expression were performed with a 50 μl reaction mixture containing 500 ng of total cellular RNA, the primer pair (1 μM), and the probe (0.2 μM) in an Applied Biosystems 7500 Sequence Detection System. Applied Biosystems Assays-on-Demand 20× primer and fluorogenic TaqMan FAM/MGB probe mixes Mn00836412_m1 for Oas1a and Mn00449297_m1 for Oas1b were used. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an endogenous control for each sample and was detected using TaqMan mouse GAPDH primers and probe (Applied Biosystems). One-step RT-PCR was performed for each target gene and for the endogenous control in a singleplex format using 200 ng of RNA and the TaqMan One-Step RT-PCR Master Mix Reagent Kit (Applied Biosystems). The cycling parameters were as follows: reverse transcription at 48°C for 30 m, AmpliTaq activation at 95°C for 10 m, denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 m (cycle repeated 40 times). The values were normalized to those for GAPDH and presented as the relative fold change compared to the uninfected calibrator sample in relative quantification (RQ) units (Scherbik et al., 2007).
Cloning the Oas1a and Oas1b gene promoter regions
A DNA fragment consisting of 1768 bp upstream and 28 bp downstream of the Oas1a gene TSS and a fragment consisting of 1398 bp upstream and 51 bp downstream of the Oas1b TSS were amplified from C3H/He and C3H/RV genomic DNA using primers designed from the GenBank C57BL/6J mouse genomic sequence [(NCBI) Genbank ID: NT_078458.6]. The amplified PCR products were cloned into TopoXL vector (Invitrogen) and sequenced. The C3H/RV DNA fragments were then subcloned into the pGL4.17 firefly luciferase reporter vector (Promega) using NheI and BgLII cloning sites to generate the Oas1a (−1768, +28) and Oas1b (−1398, +51) reporter constructs. These two constructs were subsequently used as templates in additional PCR reactions to amplify DNA fragments containing sequential 5′ deletions. Each of the deleted PCR products was then cloned into the pGL4.17 firefly luciferase reporter vector to generate eight additional Oas1a reporter constructs [Oas1a (−854, +28), Oas1a (−722, +28), Oas1a (−641, +28), Oas1a (−468, +28), Oas1a (−396, +28), Oas1a (−289, +28), Oas1a (−176, +28) and Oas1a (−87, +28)] and eight additional Oas1b reporter constructs [Oas1b (−991, +51), Oas1b (−814, +51), Oas1b (−742, +51), Oas1b (−576, +51) Oas1b (−507, +51), Oas1b (−394, +51), Oas1b (−181, +51), and Oas1b (−116, +51)]. The primers used for these PCR reactions are listed in Table 1.
Table 1.
Primers used to generate the Oas1a and Oas1b promoter deletion reporter constructs.
| Construct | Forward primer | Reverse primer |
|---|---|---|
| Oas1a (−854 to +28) | AGCTAGCTAGGCTCGGGGACCAGTAGTAG1 | AAGATCTGCTAAGTCTGGAGCTTCCTGG1 |
| Oas1a (−722 to +28) | AGCTAGCGCCCATGAGTGATCCTCCA | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−641 to +28) | AGCTAGCTCAACCTTGGGAATGTCCTGG | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−468to +28) | AGCTAGCGATGTGAGTAAGAGAGGGGGC | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−396 to +28) | AGCTAGCAAAGAAAGAAAGAAAGGAAGAAAG | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−289 to +28) | AGCTAGCTGGAAGCCACAGCCACCTTCTGCAG | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−176 to +28) | AGCTAGCAGAAGAAACCCCAAGAAAGCCAG | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1a (−87 to +28) | AGCTAGCTGGTCAGAAGCTCTGAAGCC | AAGATCTGCTAAGTCTGGAGCTTCCTGG |
| Oas1b (−1398 to +51) | AGCTAGCTTTTTCCCCCTCACACTCTG | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−991 to +51) | AGCTAGCCCAATCCTGCTCTTGCAGAAGGC | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−814 to +51) | AGCTAGCGTGTGTGTGTGTGTGTGTTTGGGAC | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−742 to +51) | AGCTAGCACAGCCCGAGCTCTTAAACTCTGAG | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−507 to +51) | AGCTAGCAGACCCCACCCTCACCCAGAC | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−394 to +51) | AGCTAGCACCTGCAAGTCCAGAGGTAAAGG | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−181 to +51) | AGCTAGCCAGAAGAAATCCCGAGAAAG | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
| Oas1b (−116 to +51) | AGCTAGCGTACCTGTTCAGAAGCCCTAACGCC | AAGATCTCCTCTGCAGCCAGCAGGTCCT |
Underlined sequences indicate an NheI restriction site included in all forward primers and a BglII restriction site included in the reverse primers.
Prediction and mutation of TFBSs
The TFSEARCH (version 1.3) and GENONATIX programs were used to predict TFBSs in the Oas1a and Oas1b promoter fragment sequences. Specific mutations in predicted IRF, ISRE, STAT and ICE binding sites were introduced into the promoter sequence in the Oas1a (−854, +28) construct to generate the Oas1a (−854, +28) mIRF, Oas1a (−854, +28) mISRE, Oas1a (−854, +28) mSTAT and Oas1a (−854, +28) mICE reporter constructs, respectively. Specific mutations in the predicted NF-kappa B, IRF, ISRE, STAT and ICE binding sites were introduced into the Oas1b (−576, +51) construct to generate the Oas1b (−576, +51) mNF-kappa B, Oas1b (−576, +51) mIRF, Oas1b (−576, +51) mISRE, Oas1b (−576, +51) mSTAT Oas1b (−576, +51) mICE constructs, respectively. The mutations were introduced using the primers shown in Table 2 and a QuikChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol. The mutant promoter sequences were analyzed using GENOMATIX software to ensure that the substitutions introduced eliminated the targeted binding site but did not create a new TFBS. The sequences of all the constructs generated were verified by DNA sequencing.
Table 2.
Primers used for site directed mutageneis.
| Construct | Primer | Primer sequence |
|---|---|---|
| Oas1a (−854, +28) mIRF | F1 | GCCCTTCTCGGGAAATGGAAACTGTACATCTACTTCCC2 |
| R | GGGAAGTAGATGTACAGTTTCCATTTCCCGAGAAGGGC | |
| Oas1a (−854, +28) mISRE | F | GCCCTTCTCGGGAAATGGTACCTGAAAATCTACTTCCC |
| R | GGGAAGTAGATTTTCAGGTACCATTTCCCGAGAAGGGC | |
| Oas1a (−854, +28) mSTAT | F | CATAGCCCTGCCCGTGTCGGCAAATGGAAACTG |
| R | CAGTTTCCATTTGCCGACACGGGCAGGGCTATG | |
| Oas1a (−854, +28) mICE | F | CAGAAGCTCTGAAGCCATCCGCTGCCGGGGCCTGG |
| R | CCAGGCCCCGGCAGCGGATGGCTTCAGAGCTTCTG | |
| Oas1b (−576, +51) mNF-kappa B | F | CCCGGGAAATGGAAACTGAAAGGACCATTTCTGCTTCAGCG |
| R | CGCTGAAGCAGAAATGGTCCTTTCAGTTTCCATTTCCCGGG | |
| Oas1b (−576, +51) mIRF | F | CCCGGGAAATGGAAACTGTACGTCCCATTTCTGCTTCAGCG |
| R | CGCTGAAGCAGAAATGGGACGTACAGTTTCCATTTCCCGGG | |
| Oas1b (−576, +51) mISRE | F | GCCCTTCCCGGGAAATGGTACCTGAAAGTCCC |
| R | GGGACTTTCAGGTACCATTTCCCGGGAAGGGC | |
| Oas1b (−576, +51) mSTAT | F | GCATATCCGCGCCCGTGCCGGCAAATGGAAACTGAAAGTCCC |
| R | GGGACTTTCAGTTTCCATTTGCCGGCACGGGCGCGGATATGC | |
| Oas1b (−576, +51) mICE | F | CAGAAGCCCTAACGCCATCCGCTGCTCGGGCCTGG |
| R | CCAGGCCCGAGCAGCGGATGGCGTTAGGGCTTCTG |
F, forward primer; R, reverse primer
Underlined letters indicate mutated nts.
Luciferase reporter assay
The pGL4.17 firefly luciferase reporter plasmid DNA (1 μg) containing either an Oas1a or Oas1b promoter fragment and 20 ng of a pGL4.74 Renilla luciferase reporter construct DNA (Promega) in a solution consisting of 3 μl of FuGENE 6 (Roche Applied Science) and 97 μl of serum free media were added to C3H/RV MEF cultures (50-70% confluency) and the cells were incubated at 37°C for 24 h. Murine recombinant IFN beta (1000 U/ml) (PBL Interferon Source) was added to the cell cultures 21 h after transfection. After a 3 h incubation, cells were harvested using the lysis buffer supplied with the Dual Luciferase Reporter Assay System (Promega). Firefly and Renilla luciferase activities were separately measured according to the manufacturer's protocol using a luminometer (LMax II384, Molecular Devices). The firefly luciferase activity was normalized to the Renilla luciferase activity in each sample.
Electrophoretic mobility shift assay (EMSA)
The Oas1a and Oas1b DNA probes (Table 3) were labeled using a DIG gel shift 2nd generation kit (Roche Applied Science) according to the manufacturer's instructions. Nuclear extracts (2 μg) prepared from C3H/RV MEFs that were either untreated or treated with 1000 U/ml of murine IFN beta for 1 h were used for EMSA. The specificity of the binding detected was analyzed by addition of a 200 fold excess of unlabeled probe to one of the reactions. For supershift assays, nuclear extracts were incubated with anti-STAT1 (Santa Cruz Biotechnology Inc.), anti-STAT2 (provided by Christian Schindler, Colombia University, New York, NY) or a nonspecific IgG antibody (Zymed) for 30 m at room temperature. Binding reactions were prepared according to the DIG gel shift 2nd generation kit protocol. DNA-protein complexes were resolved on a 6% native polyacrylamide gel and labeled probe was detected using the anti-DIG antibody supplied with the kit.
ChIP
Confluent C3H/He monolayers were incubated with 1000 U/ml of murine IFN beta for 30 min or left untreated. Chromatin was crosslinked in situ with 1% formaldehyde for 10 min at ∼25°C. After washing with ice cold phosphate-buffered saline (PBS), cells were incubated with 1 ml of cell lysis buffer [5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP40 and Complete EDTA-free Protease Inhibitors (Roche Applied Sciences)] for 10 m and the nuclei were pelleted at 2000 rpm at 4°C. Nuclei were then incubated with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0, and Complete EDTA-free Protease Inhibitors) for 20 m on ice. The crosslinked chromatin was sonicated to an average size of 500–1,000 base pairs. Sonicated chromatin obtained from 1 × 106 cells was used for each immunoprecipitation reaction. Samples were first precleared with 60 μl of salmon sperm DNA-coated agarose beads (Millipore) and then incubated with 5 μg of anti-STAT1 (Santa Cruz Biotechnology), anti-STAT2 (provided by Christian Schindler, Columbia University, New York, NY) or rabbit nonspecific IgG antibody overnight at 4°C. After incubation with 60 μl salmon sperm-coated agarose beads, the samples were washed sequentially for 5 m at 4°C with each of the following buffers: low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.0 and 150 mM NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.0 and 500 mM NaCl) and LiCl buffer (0.25 M LiCl, 1% NP40, 1% DOC, 1 mM EDTA and 10 mM Tris, pH 8.0). Finally, the samples were washed twice with TE buffer (1 mM EDTA and 10 mM Tris, pH 8.0), and then eluted with 500 μl of SDS elution buffer (1% SDS, 0.1 M NaHCO3). Samples were incubated with 20 μl of 5 M NaCl at 65°C overnight to reverse the crosslinks, then 10 μl of 500 mM EDTA, 20 μl of Tris pH 6.5, and 2 μl of Proteinase K (10 mg/mL) was added and the samples were incubated for 1 h at 45°C. Imput and immunoprecipitated DNA were isolated using a phenol:chloroform:isoamyl alcohol mixture (USB Biochemicals) according to the manufacturer's protocol and analyzed by real time quantitative PCR (qPCR) using probes and primers designed to span the proximal Oas1a and Oas1b promoters. The sequences of the primers were: Oas1a forward primer 5′-GGATCCTAAGAAAGCTCAGACTTCA-3′, Oas1a reverse primer 5′- CCCGGCAGCCAATGG-3′, Oas1b forward primer 5′- GAAGCCCTAACGCCATTGG-3′, Oas1b reverse primer 5′-AGGGCGCGGATATGCA-3′. The sequence of the FAM-MGB Oas1a probe was 5′-TGGAAGTGTGGGAAAGGTCTTT-3′ and that of the Oas1b probe was 5′-CGGGCCTGGATGAT-3′. To generate standard curves, known amounts of Oas1a, Oas1b and Irf7 promoter DNA in a TOPO-XL vector (Invitrogen) were titrated and assayed by real time qPCR using a FastStart Universal Probe Master (ROX) kit (Roche Applied Science) according to the manufacturer's protocol. The standard curves were independently generated at the same time as the immunoprecipitated DNA samples were assayed by qPCR and used to quantify the immunoprecipitated DNA.
Highlights.
>5′ STAT and 3′ IRF binding sites overlap the Oas1a and Oas1b ISREs. >The overlapping IRF binding sites are not required for gene induction by IFN beta. >The ISRE and STAT sites are required for IFN beta induction of Oas1a. >The ISRE but not the STAT site is required for IFN beta induction of Oas1b. >Oas1a requires STAT1 and STAT2 while Oas1b needs only STAT2 for IFN beta induction.
Acknowledgments
This work was supported by Public Health Service research grant AI045135 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank B. Stockman for technical assistance. We thank Jerry Boss's lab at Emory University School of Medicine for technical advice on ChIP assays and Christian Schindler, Columbia University for providing STAT1 and STAT2 knockout MEFs and STAT2 antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen J, VanScoy S, Cheng TF, Gomez D, Reich NC. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 2008;9:168–175. doi: 10.1038/sj.gene.6364449. [DOI] [PubMed] [Google Scholar]
- Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- Basagoudanavar SH, Thapa RJ, Nogusa S, Wang J, Beg AA, Balachandran S. Distinct roles for the NF-kappa B RelA subunit during antiviral innate immune responses. J Virol. 2011;85:2599–2610. doi: 10.1128/JVI.02213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluyssen HA, Levy DE. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J Biol Chem. 1997;272:4600–4605. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- Bluyssen HA, Muzaffar R, Vlieststra RJ, van der Made AC, Leung S, Stark GR, Kerr IM, Trapman J, Levy DE. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc Natl Acad Sci U S A. 1995;92:5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes C, Plautz JD, Stanewsky R, Jamison CF, Straume M, Wood KV, Kay SA, Hall JC. Novel features of drosophila period Transcription revealed by real-time luciferase reporting. Neuron. 1996;16:687–692. doi: 10.1016/s0896-6273(00)80088-4. [DOI] [PubMed] [Google Scholar]
- Chang TH, Liao CL, Lin YL. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 2006;8:157–171. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Chen PJ, Wei CC, Wang C, Chen FW, Hsu YH, Chang MS. Promoter analysis of interleukin 19. Biochem Biophys Res Commun. 2006;344:713–720. doi: 10.1016/j.bbrc.2006.03.200. [DOI] [PubMed] [Google Scholar]
- Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- Diamond MS. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J Interferon Cytokine Res. 2009;29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8:4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology. 2011;409:262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Williams BR, Sen GC. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol. 2005;79:3920–3929. doi: 10.1128/JVI.79.7.3920-3929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Hartmann R, Kjeldgaard NO, Justesen J. Gene structure of the murine 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2002;59:1212–1222. doi: 10.1007/s00018-002-8499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Justesen J, Schierup MH, Hartmann R. Characterization of the 2′-5′-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res. 2003;31:3166–3173. doi: 10.1093/nar/gkg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd-Smith G, Wang Q, Sen GC. Transcriptional induction of the p69 isoform of 2′,5′-oligoadenylate synthetase by interferon-beta and interferon-gamma involves three regulatory elements and interferon-stimulated gene factor 3. Exp Cell Res. 1999;246:138–147. doi: 10.1006/excr.1998.4296. [DOI] [PubMed] [Google Scholar]
- Garban HJ, Bonavida B. Nitric oxide inhibits the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J Immunol. 2001;167:75–81. doi: 10.4049/jimmunol.167.1.75. [DOI] [PubMed] [Google Scholar]
- George CX, Das S, Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008;380:338–343. doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilli M, Chiu JJ, Lenardo MJ. NF-kappa B and Rel: participants in a multiform transcriptional regulatory system. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- Guo Z, Garg S, Hill KM, Jayashankar L, Mooney MR, Hoelscher M, Katz JM, Boss JM, Sambhara S. A distal regulatory region is required for constitutive and IFN-beta-induced expression of murine TLR9 gene. J Immunol. 2005;175:7407–7418. doi: 10.4049/jimmunol.175.11.7407. [DOI] [PubMed] [Google Scholar]
- Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Hartmann R, Olsen HS, Widder S, Jorgensen R, Justesen J. p59OASL, a 2′-5′ oligoadenylate synthetase like protein: a novel human gene related to the 2′-5′ oligoadenylate synthetase family. Nucleic Acids Res. 1998;26:4121–4128. doi: 10.1093/nar/26.18.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higai K, Miyazaki N, Azuma Y, Matsumoto K. Transcriptional regulation of the fucosyltransferase VI gene in hepatocellular carcinoma cells. Glycoconj J. 2008;25:225–235. doi: 10.1007/s10719-008-9114-z. [DOI] [PubMed] [Google Scholar]
- Hovnanian A, Rebouillat D, Levy ER, Mattei MG, Hovanessian AG. The human 2′,5′-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2′,5′-OAS locus. Genomics. 1999;56:362–363. doi: 10.1006/geno.1998.5737. [DOI] [PubMed] [Google Scholar]
- Hovnanian A, Rebouillat D, Mattei MG, Levy ER, Marie I, Monaco AP, Hovanessian AG. The human 2′,5′-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics. 1998;52:267–277. doi: 10.1006/geno.1998.5443. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Ericsson J, Mantovani R, Edwards PA. Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein. J Lipid Res. 1998;39:767–776. [PubMed] [Google Scholar]
- Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Shibata S, Iwakura Y. Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J Interferon Cytokine Res. 2002;22:981–993. doi: 10.1089/10799900260286696. [DOI] [PubMed] [Google Scholar]
- Kaur S, Uddin S, Platanias LC. The PI3′ kinase pathway in interferon signaling. J Interferon Cytokine Res. 2005;25:780–787. doi: 10.1089/jir.2005.25.780. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. 2003;278:13033–13038. doi: 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas AC, Tomkinson B. Characterization of the promoter of the gene encoding human tripeptidyl-peptidase II and identification of upstream silencer elements. Gene. 2007;393:62–69. doi: 10.1016/j.gene.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lou YJ, Pan XR, Jia PM, Li D, Xiao S, Zhang ZL, Chen SJ, Chen Z, Tong JH. IRF-9/STAT2 functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Glaser P, Lucas M, Simon-Chazottes D, Ceccaldi PE, Montagutelli X, Despres P, Guenet JL. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics. 2003;82:537–552. doi: 10.1016/s0888-7543(03)00176-9. [DOI] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Sato M, Hata N, Asagiri M, Suemori H, Noguchi S, Tanaka N, Taniguchi T. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- Ousman SS, Wang J, Campbell IL. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79:7514–7527. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci U S A. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Zharkikh AA, Scherbik SV, Brinton MA. The mammalian 2′-5′ oligoadenylate synthetase gene family: evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J Mol Evol. 2006;63:562–576. doi: 10.1007/s00239-006-0073-3. [DOI] [PubMed] [Google Scholar]
- Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KL, Smith HL, Stark GR, Sen GC. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc Natl Acad Sci U S A. 2002;99:6322–6327. doi: 10.1073/pnas.092133199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi SA, Salditt-Georgieff M, Darnell JE., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci U S A. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rebouillat D, Hovnanian A, David G, Hovanessian AG, Williams BR. Characterization of the gene encoding the 100-kDa form of human 2′,5′ oligoadenylate synthetase. Genomics. 2000;70:232–240. doi: 10.1006/geno.2000.6382. [DOI] [PubMed] [Google Scholar]
- Rebouillat D, Marie I, Hovanessian AG. Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2′-5′ oligoadenylate synthetase. Eur J Biochem. 1998;257:319–330. doi: 10.1046/j.1432-1327.1998.2570319.x. [DOI] [PubMed] [Google Scholar]
- Reith W, Siegrist CA, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci U S A. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford MN, Kumar A, Nissim A, Chebath J, Williams BR. The murine 2-5A synthetase locus: three distinct transcripts from two linked genes. Nucleic Acids Res. 1991;19:1917–1924. doi: 10.1093/nar/19.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi V, Caretti G, Wasner M, Reinhard W, Haugwitz U, Engeland K, Mantovani R. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J Biol Chem. 2003;278:6642–6650. doi: 10.1074/jbc.M210065200. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177:4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbik SV, Stockman BM, Brinton MA. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J Virol. 2007;81:12005–12018. doi: 10.1128/JVI.01359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Kakuta S, Hamada K, Sokawa Y, Iwakura Y. Cloning of a novel 2′,5′-oligoadenylate synthetase-like molecule, Oasl5 in mice. Gene. 2001;271:261–271. doi: 10.1016/s0378-1119(01)00508-x. [DOI] [PubMed] [Google Scholar]
- Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Su ZZ, Sarkar D, Emdad L, Barral PM, Fisher PB. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007;213:502–510. doi: 10.1002/jcp.21128. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- Wang Q, Floyd-Smith G. The p69/71 2-5A synthetase promoter contains multiple regulatory elements required for interferon-alpha-induced expression. DNA Cell Biol. 1997;16:1385–1394. doi: 10.1089/dna.1997.16.1385. [DOI] [PubMed] [Google Scholar]
- Wright KL, Moore TL, Vilen BJ, Brown AM, Ting JP. Major histocompatibility complex class II-associated invariant chain gene expression is up-regulated by cooperative interactions of Sp1 and NF-Y. J Biol Chem. 1995;270:20978–20986. doi: 10.1074/jbc.270.36.20978. [DOI] [PubMed] [Google Scholar]
- Yan W, Ma L, Stein P, Pangas SA, Burns KH, Bai Y, Schultz RM, Matzuk MM. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol Cell Biol. 2005;25:4615–4624. doi: 10.1128/MCB.25.11.4615-4624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Wang Q, Floyd-Smith G. Transcriptional induction of p69 2′-5′-oligoadenylate synthetase by interferon-alpha is stimulated by 12-O-tetradecanoyl phorbol-13-acetate through IRF/ISRE binding motifs. Gene. 1999;237:177–184. doi: 10.1016/s0378-1119(99)00284-x. [DOI] [PubMed] [Google Scholar]


