Abstract
Changes in inter-helical hydrogen bonding are associated with the conformational dynamics of membrane proteins. The function of the protein depends on the surrounding lipid membrane. Here we review through specific examples how dynamical hydrogen bonds can ensure an elegant and efficient mechanism of long-distance intra-protein and protein-lipid coupling, contributing to the stability of discrete protein conformational substates and to rapid propagation of structural perturbations.
Keywords: membrane protein structure, hydrogen bond, membrane protein dynamics, lipid-protein interactions
I. Introduction
Membrane proteins (MPs) function in the complex environment of the lipid membrane. There, protein atoms interact not only with each other and with water, but also with atoms of the surrounding lipid molecules. Hydrogen-bonding (H bonding) interactions—which form between a hydrogen atom covalently bound to an electronegative atom, and another electronegative atom—are of paramount importance for the assembly, structure, and functioning of membrane proteins. Discussions of MP H bonds tend to focus on the energetics of H bonding in membrane protein folding [1] and structure formation [2]. Here we focus instead on the role of H bonds in MP functions. In particular, we consider how MP function depends upon networks of H bonds and their dynamics. We discuss examples, largely from our laboratories, of H-bonding networks within functionally different α-helical membrane proteins and the complexity of the networks arising from H bonds between helices and between helices and the lipid membrane. Finally, we consider the relationship between hydrogen bonding and the conformational dynamics of membrane proteins.
II. Networks of “static” hydrogen bonds in membrane protein crystal structures
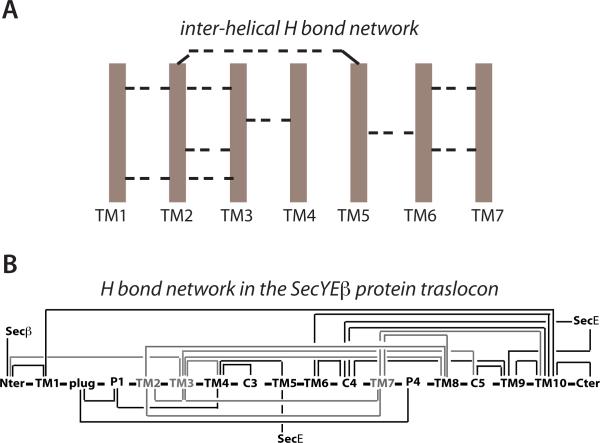
An analysis of a relatively small set of 134 TM helices in 13 α-helical protein structures indicated that almost each TM helix was connected via H bonding to the most proximal helix [3]; albeit helices with more than one H-bonding cluster were also observed, most inter-helical H bonds observed were between two amino acids from the two helices. Because each helix is likely H-bonded to a nearby helix then, even if each pair has only one H bond, a more complex picture arises in which the TM helices of the protein are interconnected via direct or indirect H bonds (Figure 1A). The dynamic breaking and forming of H bonds, allowing the TM helices to exchange H bonding partners, can only add to the complexity of this picture. Do such dynamical inter-helical H bonded networks exist? We show here that such dynamical interconnections do exist and, when present, they likely participate in controlling the conformational dynamics of TM proteins. We depict in Figure 2 examples of different classes of membrane proteins in which we have identified networks of H bonds that interconnect TM helices. These inter-helical H-bond networks ensure coupling of key structural elements to remote regions of the protein.
Figure 1.
Hydrogen bonds (H bonds) interconnect TM helices. (A) Schematic representation of TM H-bond interconnections with TM helices depicted as cartoons, and H bonds as dashed lines. One or more H bonds can interconnect pairs of TM helices. Note that TM2 is a connectivity node; it interconnects the TM1-TM2-TM3-TM4 and TM5-TM6-TM7 segments of the protein. (B) Interconnectivity map of the SecYEG protein translocon. Note that TM3 connects the gate helices TM2 and TM7 to other regions of the translocon. Panel B is modified from Bondar et al. [11].
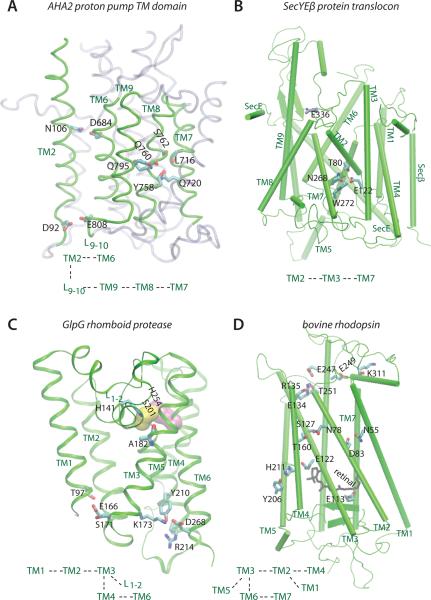
Figure 2.
Examples of networks of H bonds that interconnect TM helices of different classes of membrane proteins. Only selected H-binding amino acids sidechains are depicted explicitly. Each panel is accompanied by a schematic representation of the inter-helical connections mediated by H bonds. (A) The AHA2 P-type plasma membrane proton pump (PDB ID: 3B8C). For simplicity, TM helices that do not participate in the H bonds depicted explicitly are shown as transparent gray ribbons. (B) The central cluster in the SecYEβ protein translocon (PDB ID: 1RHZ). E122 of TM3 mediates a cluster of H bonds that involve amino acids of the gate helices TM2 and TM7; E122 is highly conserved as Glu in archaea and eukarya, and present mostly as Gln in bacteria [11]. (C) The rhomboid intramembrane protease (PDB ID: 2IRV). The catalytic groups Ser201 and His254 are shown as yellow and purple surfaces, respectively. (D) Bovine rhodopsin with the retinal cofactor shown as black bonds (PDB ID: 1U19). E134 and R135 are part of the conserved E(D)RY motif. At 3.9 Å the distance between the E122 and H211 sidechains is somewhat long for an H bond, but in a flexible protein environment that distance could easily sample H-bonding values. The molecular graphics images in panels A–D were prepared using the VMD software [52] based on published crystal structures [4, 9, 19, 53]. The simulations of SecYEβ and GlpG were performed using the CHARMM [54] force field parameters for the protein [55] and lipid [56] atoms, and the TIP3P water model [57]. The length of the bonds involving H atoms are constrained using the SHAKE algorithm [58], the short-range interactions are cut-off at 12 Å using a switching function between 8Å and 12Å, and the Coulomb interactions are computed using the smooth particle mesh Ewald summation [59, 60]. Langevin dynamics were used to maintain the temperature constant at 300K (POPC lipids) or 310K (POPE lipids), and a Nosé-Hoover thermostat [61, 62] for keeping the pressure at 1bar. After an initial equilibration with weak harmonic constraints (2kcal mol−1 to 5 kcal mol−1) and an integration step of 1fs, we switched off all harmonic constraints and used the reversible multiple time-step algorithm [63, 64] with integration time-steps of 1fs for the bonded-forces, 2fs for the short-range non-bonded forces, and 4fs for the long-range electrostatic forces. MD simulations of GlpG were based on the crystal structure of Ben Shem et al [19]; the simulation systems comprised ~160,000 atoms (~500 lipid molecules, solvent water, and ions for charge neutrality). In the MD simulation of the SecYEG translocon we used the crystal structure of Van den Berg et al. [9] for the protein atoms, and a patch of 475 POPC lipids (217,820 atoms including solvent water and ions for charge neutrality).
The AHA2 plasma membrane proton pump is a member of the P-type ATPase family. Proteins from this family couple the hydrolysis of Adenosine Triphosphate (ATP) with large-scale conformational changes and pumping of cations across the membrane. In AHA2, the central proton donor and acceptor group on TM9 D684 is within hydrogen-bonding distance from N106 of TM2 (Figure 2A). TM2 also contains D92, one of the acidic groups proposed as putative proton release groups [4]. The close interaction between D92 and E808 (N. Bondar, work in progress) ensures coupling of TM2/TM9 to the TM7-TM8-TM9 segment of the protein. That is, the inter-helical H-bonding connections ensure that the protonation states of D684 and D92 can be relayed to remote distances in the protein, and that changes in protein structure and dynamics can be relayed to the local environment of the proton transfer groups, modulating their electrostatic environment.
SecYEβ is the protein translocation channel (translocon) found in the plasma membrane of prokaryotes. There, SecYEβ is the central component of a larger secretion machinery that ensures that newly-synthesized proteins targeted to the SecY pathway are either secreted into the periplasm or inserted into the plasma membrane. The selection process appears to be based on biophysical principles of partitioning between the membrane and the translocon [5–7]. Release of TM helices into the lipid membrane is thought to occur via the lateral opening of helices TM2 and TM7 (Figure 2B) [8–10]. We have found that SecYEβ has a remarkable network of inter-helical H bonds: no fewer than 70 H bonds that interconnect different structural elements of the translocon, and each of the TM helices has at least one inter-helical H bond (Figure 1B) [11]. Importantly, the gate helices TM2 and TM7 are interconnected via H bonding with each other, and with TM3 (Figures 1B, 2B). The presence of this central cluster of H bonds, and the extensive inter-helical H bonding of the translocon, couple the heart of the translocon to the remaining parts of the machinery.
The GlpG rhomboid protease from E. coli is a model system for understanding intramembrane proteolysis, a fascinating process in which a membrane-embedded protein cleaves other TM segments [12–18]. Although the function of GlpG is completely different from that of SecY, there is a remarkable symmetry in their mechanism of action: whereas SecY must open a lateral helical gate to release TM substrates into the surrounding lipid membrane, in GlpG the lateral gate helix TM5 opens towards the membrane to admit the TM substrates for docking and cleavage. That is, both SecY and GlpG are helix-gated membrane proteins.
The flexible TM5 gate helix of GlpG is not involved in inter-helical H bonding, but it has been noted that it connects to TM2 via hydrophobic interactions [19–22]. On the other hand, TM2 is part of an H-bonding cluster with TM1 and TM3; TM3 is further connected to TM4 and TM6, which carry the catalytic groups, and to loop L12 (Figure 2C)—a loop that may play important structural [23] and lipid-sensing roles [22]. TM3 of GlpG could thus be seen as having a similar role as that of TM3 in SecY (Figure 1B), serving as a key node in the network of H bonds that couple different regions of the protein.
Bovine rhodopsin is a prototype for the rhodopsin family of G-Protein-Coupled Receptors (GPCRs). In the absence of light, the protein is covalently bound to the 11-cis retinal cofactor via a protonated Schiff base; absorption of light by the retinal chromophore triggers conformational changes of the protein that ultimately lead to the active state of the receptor, in which binding to the G protein occurs. More than 70% of the GPCRs of the rhodopsin family contain the sequence (D/E)R(Y/W) on TM3 [24]. Amino acids E134 and R135 of this conserved motif form the so-called ionic lock with TM6 amino acids E247 and T251 (Figure 2D). The ionic lock stabilizes the inactive conformation of the GPCR [24, 25], and controls the energetics of the transition between the active and inactive states of bovine rhodopsin [26]. TM7 connects to TM6 [26], which connects to TM3 that is part of a network involving TM5, TM2, TM1, and TM4 (Figure 2D). It thus appears that TM3, which harbors not only amino acids from the ionic lock, but also the E113 counterion of the protonated retinal Schiff base, is interconnected to other regions of the protein via H bonding (Figure 2D). Changes in H bonding between TM3 and TM5 have been associated with the formation of the active state of rhodopsin [27].
The above discussion of H-bond networks that interconnect critical regions of the protein to other structural elements is largely based on the analysis of static crystal structures. Although the H bonds observed in crystal structures are undoubtedly important in the function of the protein in the native membrane environment, the dynamics of complex H-bonded networks and how these networks may respond to perturbations (such as mutations or variations in the lipid membrane environment) cannot be determined from crystallographic structures without the help of molecular dynamics (MD) simulations. In what follows, we will use the results from MD simulations of the SecYEβ protein translocon and the GlpG rhomboid protease to illustrate the complex dynamics of inter-helical networks of H bonds, the dependence of the H-bond dynamics on the lipid membrane environment, and how extensive H-bonding networks can control the conformational states of membrane proteins. We begin with a brief introduction to MD simulations.
III. Hydrogen bond interactions can be extracted from MD simulations
Membrane proteins in their native membrane environments are not static as observed in crystal structures. Rather, they are dynamic. MD simulations are crucial for understanding H-bond dynamics, because they allow us to extend the observational range of the experiments by reconstructing the physiological lipid membrane environment of solvated membrane proteins, and consequently to investigate dynamics in atomic detail. A molecular mechanics (classical) MD simulation consists of solving numerically the classical equations that describe the motions of all particles of the system—the protein, lipid, and water atoms, and ions [28]. One obtains from simulations trajectories that describe the evolution in time of the coordinates of all the atoms of the system (generally at room temperature) for a finite period of time. The resulting MD trajectories can be used to dissect interactions between atoms. Currently, typical all-atom simulation times are 50–100 nsec long, but μsec times can be achieved for small systems at elevated temperatures using modest processors [29]. Emerging processor technology [30] will soon make it possible to achieve routinely 10 μsec time scales and beyond for membrane proteins [31, 32] and msec time scales for small soluble proteins [33, 34].
Starting with the crystallographic coordinates, the first step in the simulation of a membrane protein is to insert it into a lipid bilayer composed of the lipids of interest. To perform the insertion, the centers of mass of the bilayer system and the protein are made to coincide by aligning the protein's transmembrane principal normal to the bilayer, followed by removal of lipids that overlap the protein. The system must then be equilibrated. In short, the protein is first restrained to allow the bilayer to relax around it. Then the restraints are slowly removed until the system runs freely. Besides the composition, the temperature and pressure of system are held constant using standard algorithms. The system is allowed to run until it is well equilibrated as judged by the stability of the simulation box with periodic boundary conditions. To illustrate the kind of information on H bond dynamics that can be gleaned from MD simulations, we consider two examples from our laboratories: Prolonged MD simulations of the SecYEG protein translocon [11] and of the GlpG intramembrane protease [22]. Those papers should be consulted for the technical details of the simulations. A brief description of the protocol used for MD simulations of GlpG and SecY is given in the legend of Figure 2.
IV. The dynamics of inter-helical H-bonding networks can be very complex
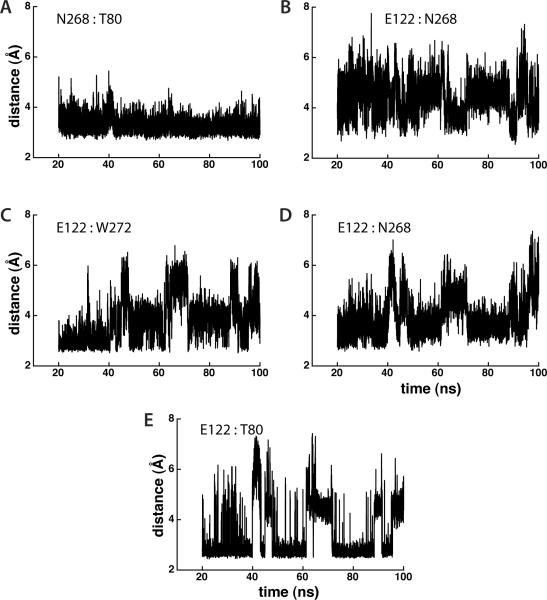
In the crystal structure of the M. jannaschii SecY protein translocon [9], amino acids T80, E122 and N268 of the central H-bonding cluster are interconnected via H-bonds characterized by distances of 2.6Å to 3.2 Å (Figure 2B). The static distance between W272 and E122 (4.4 Å) is somewhat long for an H bond. The MD simulations of the translocon at room temperature revealed a much more complex picture [11]: only one of the H bonds of the TM2-TM3-TM7 cluster, formed by sidechains of T80 and N268, is stable throughout the entire simulation (Figure 3A). The dynamics of the other sidechain:sidechain distances have a stable pattern in which H bonds are broken and reformed (Figure 3B–D). The breaking and re-forming of an H bond within this cluster can take between picoseconds to nanoseconds and tens of nanoseconds. W272 H bonds transiently to E122 (Figure 3C). E122 appears as a key player in this cluster; it can have two H bonds with N268 (one for each carboxylic oxygen atom), and it also H bonds with T80 and W272. The complexity of the interactions mediated by E122 is reflected in the existence of two or even three sub-conformers of SecY characterized by different distances between E122 and N268 (Figure 4B), E122 and W272 (Figure 4C), and between E122 and T80 (Figure 4E). Because the breaking and forming of the H bonds is very fast – that is, the energy barriers separating the sub-conformers are small and can be overcome easily – the overall structure of the protein remains stable; indeed, in MD simulations at room temperature the root-mean squared distances of the TM region of SecY relative to the starting crystal-structure coordinates was stable at ~2Å [11].
Figure 3.
The dynamics of inter-helical H bonding can be highly complex. Illustration of the dynamics of the inter-helical H bonds in the central cluster (TM2-TM3-TM7) of the SecY protein translocon. The dynamics of the H bonds are monitored here by the time-dependent distances between the heavy atoms of the H bond donor and acceptor groups. All distances are reported in Å. Panels B and D illustrate H bonding of N268 to the two carboxyl oxygen atoms of E122. Distances ≤ 3.5Å are considered here as H-bonding distances. We used for this analysis the last 80ns of the trajectory. See Figure 2B for a molecular picture of the central H-bonding cluster. The complex dynamics of the inter-helical H bonds in wild-type SecY was discussed in [11]. The protonation state of His amino depends on the local electrostatic environment [65]. In the MD simulation of the wild-type translocon from Bondar et al. [11], we modeled all His amino acids in the Nδ1 tautomeric state; that simulation was prolonged to 49.3ns. The time-series of H bonds presented here are from a new ≈100ns trajectory in which the His amino acids were modeled in the Nε2 tautomeric state; test distance analyses from the prolonged MD trajectory of the SecY translocon confirm the complex dynamics of the inter-helical H bonds observed in [11].
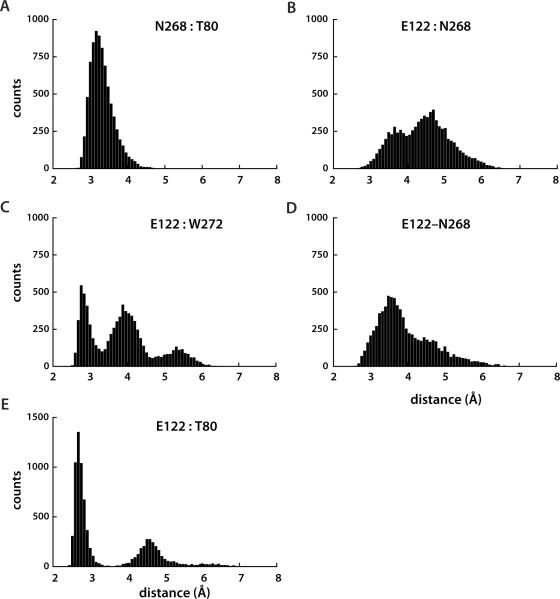
Figure 4.
Distribution of sidechain distances in the TM2-TM3-TM7 cluster of the SecY translocon. Histograms were calculated using the time-series presented in Figure 3, for the range of values between 2Å and 10Å, with a bin size of 0.08Å (i.e., 100 bins). All distances are reported in Å. Note that the distribution of the N268:T80 distances is slightly skewed towards non-H-bond distances; the skew towards large distances is more pronounced for the E122:N268 distance in panel D.
The central role of E122 is supported by our observations that this amino acid is highly conserved as Glu in archaea, as Gln or Glu in eukarya, and largely as Gln in bacteria [11]. Within the data set used for the sequence analyses, E122 is highly conserved. E122 is never replaced by an Asp sidechain, suggesting that a long sidechain at position 122 is required for mediating inter-helical H bonds. The replacement of E122 by Gln in bacterial SecY is accompanied by the absence of the N268 sidechain [11]. One would thus expect that the dynamics of the TM2-TM3-TM7 helices in bacterial SecY to be different from that of the archaeal SecY discussed here.
The complex pattern of H bonding dynamics of the TM2-TM3-TM7 cluster in SecY could not have been foreseen from the crystal structure alone. Furthermore, it appears that the dynamics of the inter-helical H bonds can depend on the lipid interactions.
In extensive MD simulations of GlpG, we observed that the dynamics of the H-bonded network that interconnects helices TM1, TM2, and TM3 (Figure 2C) is different in lipid membranes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) compared to 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE): the TM2:TM3 H bond mediated by E166 and S171 is stable at 2.6 ± 0.1Å in POPE, whereas in POPC it breaks and reforms rapidly throughout the 35ns MD, being present only for ~50% of the time.
The observation that the dynamics of the inter-helical H bonds can depend on the composition of the lipid membrane is important, because there is increasing evidence from experiments on various membrane proteins that changes in the lipid membrane composition can affect protein function, or that a protein can function only if certain lipids are present. For example, the E. coli GlpG rhomboid protease is active when reconstituted in PE lipids, but not in PC [35]. Proper functioning of the secondary multidrug transporter LmrP requires H bonding between a surface-exposed Asp and the PE lipid membrane, and is incompatible with PC lipids [36]. Anionic lipids enhance significantly binding of the SecY translocation channel to the SecA motor of the translocase machinery [37], and direct lipid:protein interactions are certainly involved in the recognition of TM helices by the translocon [5–7]. The energetics of the transition between the active and inactive states of visual rhodopsin depends on the composition of the lipid membrane [38]. Lipids can have conserved binding sites on the protein surface [39], and bind tightly to these specific sites [40].
The mechanisms by which the lipid membrane composition affects the functioning of the membrane protein are not yet entirely clear. Because changes in inter-helical H bonding can be associated with protein conformational transitions, differences in the dynamics of inter-helical H bonds [22] could contribute to the observed effects of the lipid membrane composition on membrane protein function. The question then is why would the dynamics of inter-helical H bonds—or of the entire protein—depend on the lipid membrane? The location of the GlpG inter-helical H bonds relatively close to the helix termini—and thus close to the lipid headgroup region—could be used as an argument to suggest that simple electrostatic effects are important. That is, different lipid headgroups would create a different electrostatic environment for the inter-helical H bonds. Although it certainly is true that different lipid headgroups would provide different electrostatic environments, the effects of the lipid membrane composition on protein dynamics, and in particular on its H-bonding interactions, can be rather complex.
V. The lipid membrane affects membrane protein H-bonding interactions
Several excellent reviews of the mechanisms by which lipids influence protein function have been published in the last several years [41–43]. These reviews noted, for example, that different lipid membrane compositions could imply differences in the macroscopic properties of the membrane (viscosity, phase transition, lateral pressure), but also differences in specific protein:protein and protein:lipid interactions [41]. The MD simulations reviewed by Jensen and Mouritsen [42] illustrate at atomic detail how changes in the lipid headgroups influence the formation of a water wire inside the GlpF channel.
The close coupling between GlpG and the lipid membrane that is necessary for docking and cleaving the TM substrate makes GlpG a challenging model system for understanding the general physical principles of how lipids modulate protein function. We use here GlpG to illustrate how complex the molecular picture of protein:lipid interactions can be when one accounts for dynamics.
Crystal structure analyses in which electron densities for detergent and/or lipids could be observed have provided valuable glimpses into the possible interactions of GlpG with the membrane [44]. The electron densities observed by Wang et al. [44] indicated that the membrane is very thin close to the protease. Vinothkumar [45] has shown in atomic detail how lipids adapt to the surface features of GlpG to match its varying hydrophobic thickness. Indeed, significant nonuniform thinning of the membrane close to the protease was revealed by detailed MD simulations of GlpG in hydrated lipid membranes [22].
The thinning of the membrane (Figure 5) occurs as the lipid molecules mold to the small hydrophobic thickness and rather unusual shape of GlpG (Figures 2C, 5). Although thinning of the membrane is observed with both POPC and POPE lipids [22], there are significant differences in how POPC and POPE interact with GlpG, and the H-bond dynamics of GlpG depends on the lipid membrane environment. The difference comes from the fact that the critical loop L12 and the cap loop thought to control access to the catalytic site contain polar amino acids located at the lipid headgroup interface.
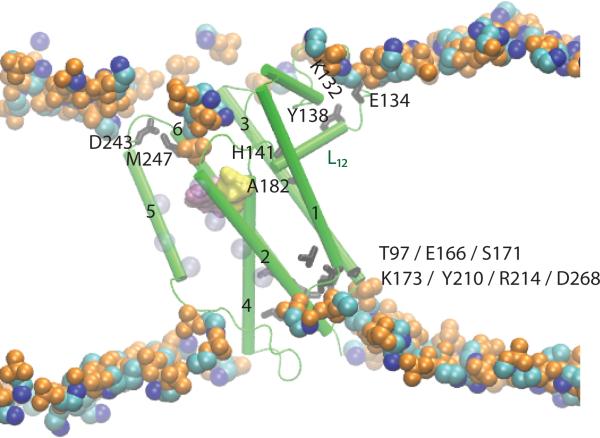
Figure 5.
Lipid and intra-protein H bond coupling in GlpG. The active site groups S201 and H254 are shown as yellow and purple surfaces, respectively. Loop L12 H bonds to the POPE lipid membrane via E134, and to the protein via the L12-H141:TM3-A182 H bond. TM3 further connects via H bonding to TM1, TM2, TM4, and TM6 (see also Figure 2C). TM2 is connected to the gate helix TM5 via hydrophobic interactions; these interactions are illustrated schematically by the transparent blue van der Waals spheres on TM2 and TM5, which represent the Cα atoms of amino acids L161, W157, and F153 TM2, and L229, F232, W236 on TM5. The sidechain:backbone interaction between D243 and M247 is significantly more stable in POPE than POPC lipids [22]. In a POPC lipid bilayer the H bond between the Y138 sidechain and the K132 backbone is broken as water molecules penetrate deeper into the lipid membrane. Figure 5 is modified from Bondar et al. [22].
E134 of L12 H bonds tightly to a POPE lipid (Figure 5A); as a result of the E134:POPE H bond, water molecules cannot penetrate into the lipid bilayer. In POPC, however, the E134:lipid headgroup interaction is water-mediated, and water molecules move deeper into the membrane, where they replace the Y138:K132 H bond with protein:water interactions. As a result, L12 is locally less structured in POPC than in POPE. A dependence on the lipid membrane environment was also observed for lipid H bonding of other amino acids located on the GlpG surface [22].
The H-bonding connectivity between L12 and TM3 implies that the structure and dynamics of the loop is coupled to that of TM3, and can affect the dynamics of the H-bonding network mediated by TM3 (Figure 2C). This was indeed observed in the MD simulations [22]. Because TM3 H bonds to TM2, which contributes to the substrate-docking site, the extensive coupling via H bonds mediated by TM3 and the sensitivity of L12 to the lipid environment could represent (or be part of) the mechanism by which the protein is tightly coupled to the lipid membrane. That is, the protein has at least one structural element that can H bond to both lipid and protein groups. H bonding to the lipid ensures that the local protein structure and dynamics depends on the composition of the lipid membrane; the intra-protein H bond couples the structure and dynamics at the protein:lipid interface to remote regions of the protein.
Tight H-bond-mediated coupling between the protein and the surrounding lipid membrane was also observed in the case of the SecY protein translocon [11]. Amino acid E336 is located on the cytoplasmic tip of TM8 (Figure 2B), where it participates in an H-bond cluster that includes TM2, TM3, and TM8 [11]. It has been shown by experiments that mutating to Arg the corresponding E382 in yeast increases the translocation of protein segments with more positively charged ends [46]. Direct changes in the electrostatic interactions between SecY and the translocating protein segments could contribute to the observed changes in peptide translocation [46]. But simulations on the E336R mutant of the M. jannaschii translocon indicated that R336 H bonds to a lipid headgroup instead of participating in the TM2-TM3-TM8 H bond network; the structure and internal solvation of the mutant are different from those of the wild-type translocon [11]. The presence of the extensive H-bonded network interconnecting the TM helices and the loops of the translocon (Figure 1B) likely explains why the protein structure and water interactions change when an H bond from an inter-helical H bond cluster is replaced with a lipid H bond.
VI. Conclusions
Inter-helical H bonding of TM membrane proteins can ensure an elegant and efficient mechanism for long-distance coupling within the protein. The coupling can be easily extended to the surrounding lipid membrane via a structural element that H bonds both to the membrane and to the protein.
The H bonds interconnecting TM helices can have a complex dynamics that can be assessed with MD simulations, but not from visual inspection of a static crystal structure. Inter-helical H bonds can be very stable, or can have a stable pattern of breaking and reforming with timescales ranging from picoseconds to nanoseconds and tens of nanoseconds (Figure 3). An important question that emerges from the observation of such H-bonding patterns is how the breaking and reforming of H bonds on the nanosecond timescale relate to the large-scale, slow global structural rearrangements that may be required for protein function. Based on the analysis discussed here, we suggest that the clusters of H bonds that inter-connect TM helices of membrane proteins contribute significantly to controlling protein conformation.
In the absence of perturbations, the clusters of H bonds help stabilize stabilizing protein conformation. Although an H bond may break and reform rapidly, without a high energetic cost, during the time that that particular H bond is broken the H-bonding partners H bond with other groups within the cluster. For example, while T80 and N268 are engaged in a stable interaction (Figures 3A,4A) E122 interacts mostly with N268 at time ~65ns (Figure 3B), and with T80 at time ~80ns (Figure 3E). The overall protein structure is maintained.
Conformational dynamics is essential for enzyme function. Changes in the preferred geometry occur along the enzyme reaction cycle - for example, upon binding of a ligand. Importantly, the enzyme can sample conformations similar to those in the ligand-bound state even in the absence of the ligand; binding of the ligand would then simply shift the enzyme's conformational equilibrium towards the active, ligand-bound form [47]. The slow collective motion of lid opening may be facilitated by fast ps-ns dynamics at local hinge sites of the enzyme [48]. As discussed below, we think that networks of inter-helical H bonds with distinct conformational modes (as the example in Figure 4) may presage shifts in the population of the conformational states sampled along the protein functional cycle.
SecY helices TM2 and TM7 are expected to undergo motions that would allow opening of a lateral gate towards the lipid membrane [8–10]. One would thus expect that at least some of the inter-helical H bonds of TM2 and TM7 (Figures 1B, 2B&3) would break when the translocon opens towards the membrane. TM3, which H bonds to the both TM2 and TM7, H bonds with additional regions of the translocon (Figure 1B); the extensive H bonds of TM3 would indicate that large-scale motions of TM3 are unlikely to accompany lateral opening of the translocon. Since the H bonds between TM3 and TM7 are relatively weak (Figures 3B–D, 4B–D), and in the closed state of the translocon TM3-E122, TM7-N268 and TM7-W272 side-chains already sample conformations in which the E122:N267 and E122:W272 are too long for TM3:TM7 inter-helical H bonds, one could expect that lateral opening of the translocon could involve breaking of the TM3:TM7 H bond. Breaking of the TM3:TM7 H bond would mean enhanced dynamics of TM7, and thus a de-stabilization of the TM2:TM7 H bond (Figure 4A) with a shift towards a conformer in which TM7 is free of H bonds with TM2 and TM3, while the TM3:TM2 H bond (Figure 4E) may still be present.
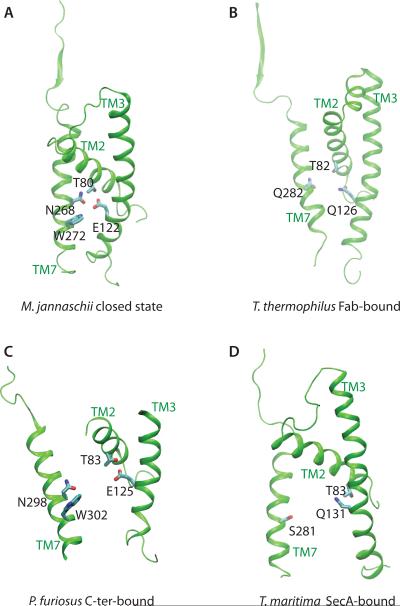
Our proposal that the pattern of H bonding in the central TM2-TM3-TM7 predicts qualitatively conformational changes associated with translocon opening appears to be supported by inspection of crystal structures thought to represent snapshots of the translocon along its opening path (Figure 6)—the bacterial T. maritima translocon in its open SecA-bound state [49], the Fab-bound pre-open T. thermophilus translocon [50], and the P. furiosus translocon structure solved from a crystal in which the C-terminal α-helical region of one SecY copy is bound to the cytoplasmic region of another SecY copy [51]. In these three structures binding of the ligand (SecA, Fab segment, or C-terminal region of another translocon) is associated with changes in how the TM2, TM3, and TM7 helices interact with each other: TM7 is away from TM2 and TM3, but TM2 remains relatively close to TM3. The distance between the groups corresponding to the M. jannaschii T80:E122 H bond are 3.4Å in T. maritima (T83:Q131), 4.7 Å in T. thermophilus (T82:Q126), and 3.3 Å in P. furiosus (T83:E125).
Figure 6.
H bonding in the central TM2-TM3-TM7 cluster of SecY is coupled to SecY's conformation. The TM2, TM3, and TM7 helices with selected amino acid residues are depicted for the M. jannaschii SecY translocon in the closed state [9] (A), and for the structures thought to represent SecY open to various extents: the Fab-bound T. thermophilus [50] (B), the archaeal P. furiousus bound to the C-terminal fragment of another SecY copy [51] (C), and the SecA-bound T. maritima translocon [49] (D). Note that in the various open structures TM2 and TM3 remain relatively close to each other, whereas TM7 moves away from TM2/TM3. M. jannaschii E122 is conserved as Glu in the archaeal P. furiosus SecY (panel C), but replaced by a Gln in the bacterial T. thermophilus and T. maritima translocons (panels B&D). S281 of T. maritima SecY is part of an array of Ser/Thr groups along TM7 [11]. In the closed state of the T. maritima SecY, the short sidechain of S281 and the presence of a Gln instead of a Glu at position 131 could make H bonding to T83/Q131 weaker than in the corresponding M. jannaschii cluster. That is, the extent to which the translocon opens and the kinetics of translocon opening may be different in archaeal vs. bacterial translocons.
The dynamics of the inter-helical H bonds appear to be coupled to the overall conformational dynamics of the protein. Marginally stable H bonds (that is, bonds that rapidly break and reform) could contribute to the structural and dynamical stability of the protein in the absence of perturbations. Once the membrane protein is perturbed, however, these H bonds may be rapidly rearranged to help stabilize a new conformation. The perturbation could be binding of a substrate, mutation, or changes in the lipid membrane composition.
Highlights
-
-
Networks of inter helical hydrogen bonds can be present in membrane proteins
-
-
these inter helical hydrogen bonds have a complex dynamics
-
-
lipid hydrogen bonding change the local structure and dynamics of membrane proteins
-
-
intra protein and lipid:protein hydrogen bonding couple the protein to lipids
VI. Acknowledgements
This research was supported in part by grants GM-74637 from the National Institute of General Medical Sciences and GM-86685 from NIGMS and the National Institute of Neurological Disorders and Stroke (S.H.W.), a Marie Curie International Reintegration Award IRG-276920 (A.-N.B), and an allocation of computer time from the National Science Foundation through TeraGrid resources. We are indebted to our collaborators Prof. Douglas J. Tobias, Dr. Coral del Val, and Dr. Alfredo J. Freites, for many valuable discussions on the protein translocon and the GlpG intramembrane protease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bowie JU. Membrane protein folding: how important are hydrogen bonds? Curr. Opin. Struct. Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].White SH. How hydrogen bonds shape membrane protein structure. Adv. Protein Chem. 2005;72:157–172. doi: 10.1016/S0065-3233(05)72006-4. [DOI] [PubMed] [Google Scholar]
- [3].Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: Polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- [4].Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- [5].White SH, Von Heijne G. How translocons select transmembrane helices. Ann. Rev. Biophys. Biophys. Chem. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- [6].Jaud S, Fernández-Vidal M, Nillson I, Meindl-Beinker NM, Hübner NC, Tobias DJ, von Heijne G, White SH. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA. 2009;106:11588–11593. doi: 10.1073/pnas.0900638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Öjemalm K, Higuchi T, Jiang Y, Langel Ü, Nilsson I, White SH, Suga H, von Heijne G. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E359–E364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- [9].Van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- [10].Du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM. The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bondar A-N, del Val C, Freites JA, Tobias DJ, White SH. Dynamics of SecY translocons with translocation-defective mutations. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brown MS, Goldstein JL. The SREBP Pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- [13].McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- [14].Struhl G, Greenwald I. Presenilin is required for activity and nuclear access to Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- [15].Urban S, Lee JR, Freeman M. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- [16].Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- [17].Wolfe MS, Kopan R. Intramembrane proteolysis: Theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- [18].Lal M, Caplan M. Regulated intramembrane proteolysis: Signaling pathways and biological functions. Physiology. 2011;26:34–44. doi: 10.1152/physiol.00028.2010. [DOI] [PubMed] [Google Scholar]
- [19].Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–183. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- [21].Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nature Struct. Mol. Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- [22].Bondar A-N, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- [25].Kim H-J, Altenbach C, Thurmond RL, Khorana HG, Hubbell WL. Structure and function in rhodopsin: Rhodopsin mutants with a neutral amino acid at E134 have a partially activated conformation in the dark state. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vogel R, Mahalingam M, Lüdeke S, Huber T, Siebert F, Sakmar TP. Functional role of the “ionic Lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. Journal of Molecular Biology. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- [27].Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, Smith SO. Changes in interhelical hydrogen bonding upon rhodopsin activation. J. Mol. Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- [28].Leach AR. Molecular Modelling: Principles and Applications. 2nd ed. Pearson Education Ltd.; Harlow: 2001. [Google Scholar]
- [29].Ulmschneider JP, Smith JC, White SH, Ulmschneider MB. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J. Am. Chem. Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, Ho CR, Lerardi DJ, Kolossváry I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan Y, Spengler J, Theobald M, Towles B, Wang SC, Anton a special-purpose machine for molecular dynamics simulation. Proc. 34th Annu. Internat. Sym. Computer Architect. 2007 [Google Scholar]
- [31].Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- [34].Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How Fast-Folding Proteins Fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- [35].Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hakizimana P, Masureel M, Gbaguidi B, Ruysschaert J-M, Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for protein gradient sensing? J. Biol. Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- [37].Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- [39].Adamian L, Naveed H, Liang J. Lipid-binding surfaces of membrane proteins: Evidence from evolutionary and structural analysis. Biochim. Biophys. Acta. 2011;1808:1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim. Biophys. Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- [41].Lee AG. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- [42].Jensen MØ, Mouritsen OG. Lipids do influence protein function: The hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- [43].Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: an energetic perspective. Annu.Rev.Biophys.Biomol.Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- [44].Wang Y, Ha Y. Open-cap conformation of intramembrane protease GIpG. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vinothkumar KR. Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 2011;407:232–247. doi: 10.1016/j.jmb.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J. Biol. Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- [47].Hanson JA, Duderstadt K, Watkins LP, Bhattacharyya S, Brokaw J, Chu J-W, Yang H. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature Letters. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- [49].Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [53].Okada T, Sugihara M, Bondar A-N, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- [54].Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- [55].MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [56].Feller SE, MacKerell AD., Jr. An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
- [57].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- [58].Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- [59].Darden T, York D, Pedersen L. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- [60].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- [61].Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- [62].Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- [63].Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991;6:121–142. [Google Scholar]
- [64].Tuckerman M, Berne BJ. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- [65].Baran KL, Chimenti MS, Schlessman JL, Fitch CA, Herbst KJ, Garcia-Moreno BE. Electrostatic effects in a network of polar and ionizable groups in Staphylococcal nuclease. J. Mol. Biol. 2008;379:1045–1062. doi: 10.1016/j.jmb.2008.04.021. [DOI] [PubMed] [Google Scholar]