Abstract
Adult-onset diseases can be associated with in utero events, but mechanisms for this remain unknown1,2. The polycomb histone methyltransferase, Ezh2, stabilizes transcription by depositing repressive marks during development that persist into adulthood3–9, but its function in postnatal organ homeostasis is unknown. We show that Ezh2 stabilizes cardiac gene expression and prevents cardiac pathology by repressing the homeodomain transcription factor Six1, which functions in cardiac progenitors but is stably silenced upon cardiac differentiation10. Ezh2 deletion in cardiac progenitors caused postnatal myocardial pathology and destabilized cardiac gene expression with activation of Six1-dependent skeletal muscle genes. Six1 induced cardiomyocyte hypertrophy and skeletal muscle gene expression. Furthermore, genetically reducing Six1 levels rescued the pathology of Ezh2-deficient hearts. Thus, Ezh2-mediated repression of Six1 in differentiating cardiac progenitors is essential for stable postnatal heart gene expression and homeostasis. Our results suggest that epigenetic dysregulation in embryonic progenitor cells predisposes to adult disease and dysregulated stress responses.
Cardiac hypertrophy results from integrating numerous signalling pathways that culminate in well-described transcriptional networks11. In humans, variation in hypertrophy is induced by similar stimuli across populations. While genetic variation contributes12, epigenetic regulatory mechanisms, particularly during embryonic development are largely unexplored,1,2. The heart may be particularly sensitive to early events, as turnover of human cardiomyocytes is limited over a lifetime13. Epigenetic regulation via histone methylation stabilizes transcriptional programs in embryonic progenitors and their differentiated descendents3,4, and is likely critical for establishing and maintaining gene expression and stress responses throughout life. Polycomb complexes are candidates for stabilizing cardiac gene expression. They control cell identity and epigenetic memory in other contexts5. Ezh2, the major histone methyltransferase of the polycomb repressor complex 2 (PRC2), tri-methylates histone H3 at lysine 27 (H3K27me3). Ezh2 is essential for embryonic development5–9, but its function in organogenesis or postnatal organ maintenance is unknown.
Ezh2, and the related H3K27 methyltransferase Ezh114,15 are expressed in cardiomyocytes (Supplementary Fig. 1), and H3K27me3 levels increase in cardiac progenitor cells as they differentiate into cardiomyocytes (Supplementary Fig. 2). This suggests that PRC2 functions in the transition from cardiac progenitor to differentiated cardiomyocyte.
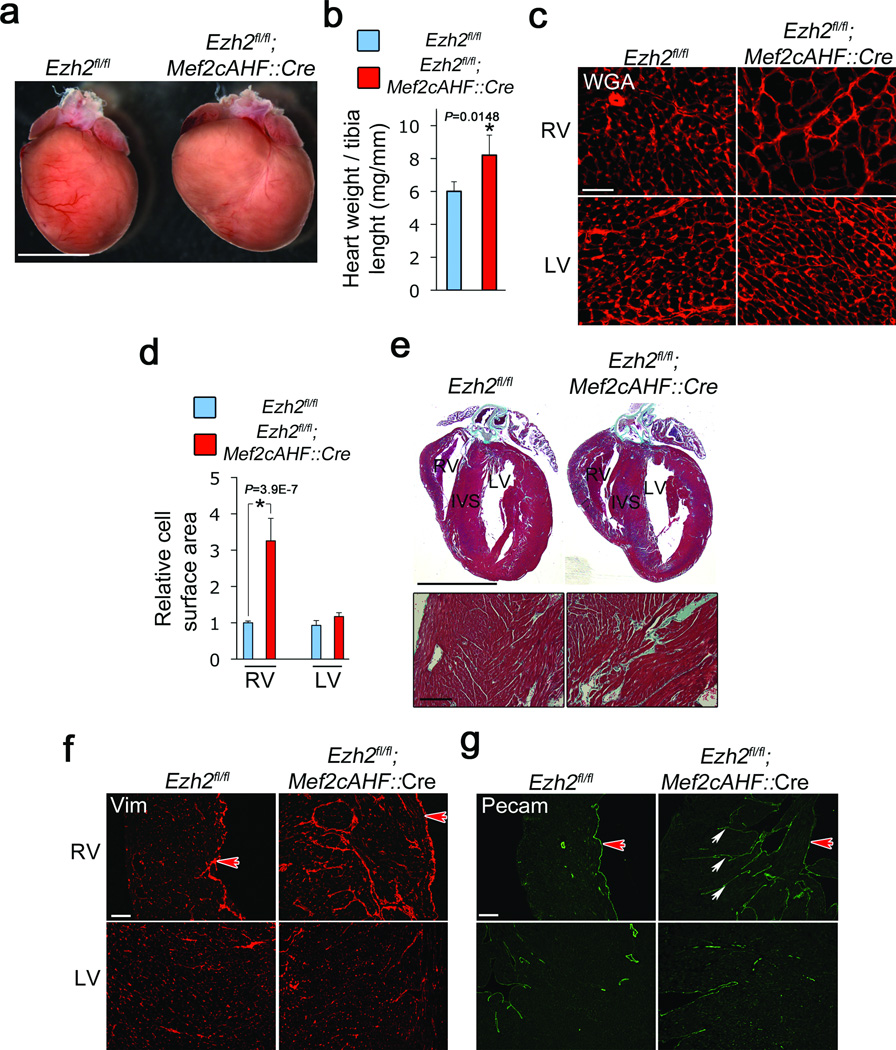
To determine the function of PRC2 in mouse heart, we inactivated Ezh2 by Cre-mediated recombination in a subpopulation of cardiac progenitors known as the anterior heart field (AHF), which contribute to the right ventricle, interventricular septum, and outflow tract16. Mice with loxP sites flanking the catalytic domain-encoding region of Ezh2 (Ezh2fl/lf)6 were crossed with mice expressing Cre recombinase under the Mef2cAHF enhancer (Mef2cAHF::Cre), which is active from embryonic day 7.5 (E7.5)16. AHF progenitors with inactivated Ezh2 failed to increase H3K27me3 levels as they differentiated into cardiomyocytes (Supplementary Fig. 2c–h), resulting in decreased Ezh2 and over 80% of right, but not left, ventricular cardiomyocytes with decreased H3K27me3 (Supplementary Fig. 3a,b), suggesting that Ezh2 is the major H3K27 histone methyltransferase in the AHF. H3K27me3 levels were not reduced in endothelial cells (Supplementary Fig. 2d). Mice with Ezh2-deficient AHF cells developed normally structured hearts (Supplementary Fig. 4). However, Ezh2-deficient hearts became enlarged after birth, with increased heart weight to tibia length ratios (Fig. 1a,b). Cardiac enlargement was restricted to the right ventricular wall, in which cardiac myocytes were massively hypertrophied, with an almost fourfold increase in cross-sectional area (Fig. 1c,d). Echocardiography confirmed right ventricular enlargement and showed mild pulmonary stenosis (Supplementary Table 1). Cardiomyocytes were already enlarged in E20 fetuses (Supplementary Fig. 5a), ruling out altered hemodynamics as a primary cause of hypertrophy. Ezh2-deficient right ventricles were fibrotic (Fig. 1e,f), and the endocardium was invaginated into the ventricular myocardium (Fig. 1g), resembling ventricular non-compaction12,17. Thus loss of Ezh2 in cardiac precursors leads to cardiac hypertrophy and fibrosis.
Figure 1.
Ezh2 limits cardiac growth and fibrosis. (a) Adult mice in which Ezh2 was deleted in second heart field progenitors (Ezh2fl/fl; Mef2cAHF::Cre) present hypertrophied right ventricle, with increased (b) heart weight to tibia length ratio (mg/mm), as compared to controls (Ezh2fl/fl). Ezh2fl/fl (blue bars) and Ezh2fl/fl; Mef2cAHF::Cre (red bars). (c) Wheat germ agglutinin (WGA) staining of heart sections in control (Ezh2fl/fl) or Ezh2-deficient (Ezh2fl/fl; Mef2cAHF::Cre) right ventricle (RV) and left ventricle (LV). (d) Cardiomyocyte cell surface area in Ezh2fl/fl (blue bars) or Ezh2fl/fl; Mef2cAHF::Cre (red bars) hearts. (e) Tissue disorganization and fibrosis, as revealed with Masson’s trichrome staining. In b and d, bars represent the mean +/− S.D. of measurements from at least three hearts per genotype. * indicates P < 0.05. (f) Vimentin staining of RV and LV of Ezh2fl/fl or Ezh2fl/fl; Mef2cAHF::Cre hearts. (g) Pecam staining showing invaginations of endocardium into myocardium (arrows). In f,g, the red arrow points to endocardial lining of the RV lumen. Reference bars in (a) and (e) (upper panel) = 0.5 cm; (e) (lower panel) = 100 µm; (c) = 25 µm; (f) and (g) = 100 µm.
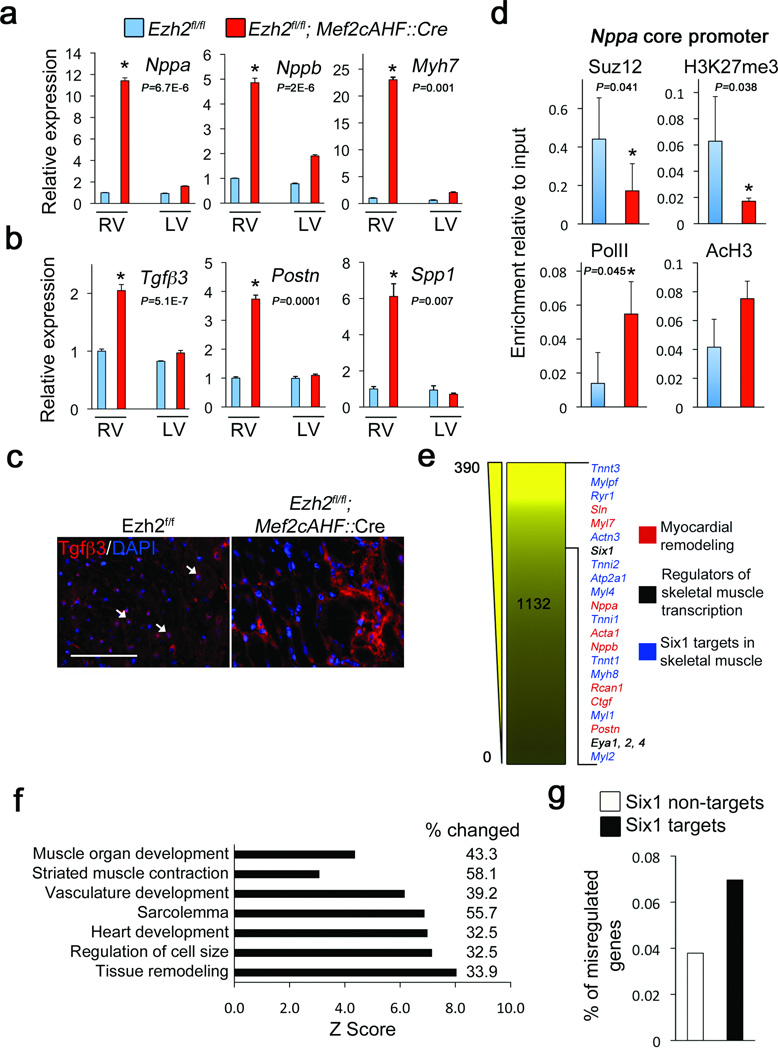
We examined altered gene expression in Ezh2-deficient adult hearts. The hypertrophy-associated genes18,19 Nppa, Nppb, and Myh7, as well as the profibrotic genes Tgfβ3, Postn and Spp1 were highly upregulated (Fig. 2a,b). Immunofluorescence detected low levels of Tgfβ3 in the perinuclear region of wild-type cardiac myocytes, while it was enriched in the extracellular region in Ezh2-deficient hearts (Fig. 2c). Thus, Ezh2 might suppress gene expression that promotes cardiac hypertrophy and fibrosis.
Figure 2.
Ezh2 represses expression of fetal genes, profibrosis factors and Six1. (a) QRTPCR showing relative expression of Nppa, Nppb and Myh7 mRNA. Ezh2fl/fl (blue bars) and Ezh2fl/fl; Mef2cAHF::Cre (red bars). (b) QRTPCR showing expression of Tgfβ3, Postn and Spp1. (c) Immunofluorescence showing low levels of perinuclear Tgfβ3 in control cardiomyocytes (arrows), and strong extracellular staining in the extracellular region in Ezh2fl/fl; Mef2cAHF::Cre right ventricle (RV). Nuclei were counterstained with DAPI. (d) Chromatin immunoprecipitation for Suz12, H3K27me3, RNA PolII, and acetylated histone H3 (AcH3) on the Nppa core promoter in Ezh2-deficient Hearts (red bars), as compared with controls (blue bars). e, Heat map of 1132 upregulated genes identified by RNA-seq comparing Ezh2-deficient hearts to control hearts. Numbers at left indicate fold increase. Indicated genes were amongst the most upregulated. (f) Analysis of functional categories identified a high percentage of genes regulating skeletal muscle and tissue remodelling. (g) RNA-seq analyses showed a higher percentage of deregulated genes targeted by Six1, as compared to deregulated genes not targeted by Six1, in Ezh2-deficient hearts. Reference bars = 25 µm. Bars in panels (a), (b) and (d) represent the mean +/− S.D. from at least three biological replicates. * Indicates P < 0.05.
If Ezh2 function were epigenetic, a sustained requirement for its activity would be anticipated. To address this, Ezh2 was deleted in differentiating ventricular myocytes using ventricular myocyte-specific Nkx2.5::Cre transgenic mice20,21, which express Cre only after cardiomyocytes begin to differentiate. Ezh2fl/fl;Nkx2.5::Cre mice had mild cardiomyocyte hypertrophy and increased Myh7 mRNA levels (Supplementary Fig. 6b,e), perhaps due to reduced Nkx2.5::Cre-mediated recombination efficiency20,21 (Supplementary Fig. 3c,d), or to a critical window of activity of Ezh2 during early cardiac progenitor differentiation. Thus, Ezh2 functions in cardiomyocyte progenitors, with long-term consequences.
We hypothesized Ezh2-mediated H3K27me3 directly represses fetal genes, and reactivation in Ezh2-deficient hearts results from loss of epigenetic repression. At the Nppa promoter19, occupancy of the PRC2 core subunit suppressor of zeste 12 homolog (Suz12) and H3K27me3 was decreased in Ezh2 mutant hearts, and acetylated histone H3 (AcH3, activating mark) and RNA polymerase II (PolII) were increased (Fig. 2d, Supplementary Fig. 7). These results suggest Ezh2 directly represses fetal gene expression to maintain cardiac homeostasis.
The global gene expression profile of adult Ezh2-deficient right ventricles was determined by RNA sequencing (RNA-seq) and DNA microarrays. With a stringent P<0.001 and fold-change cutoff of >1.4, 1,132 genes were upregulated, and 1,800 downregulated (Supplementary Table 2). Consistent with hypertrophy and fibrosis, myocardial remodelling-associated genes (e.g., Nppa, Nppb) and atrial-specific genes (Sln, Myl7), were upregulated in Ezh2-deficienct hearts (Fig. 2e). Genes involved in signalling pathways mediating myocardial remodelling were also misregulated (Supplementary Table 3). Gene Ontology classification showed surprisingly that many misregulated genes encode skeletal muscle-specific contractile proteins and developmental regulators (Fig. 2f), including some that are normally induced in cardiac hypertrophy (e.g., Acta1). Thus, Ezh2 is required to stabilize postnatal cardiac gene expression.
In pluripotent cells, Polycomb complexes repress transcription factors regulating cell differentiation and development5. We hypothesized Ezh2-mediated repression of key transcription factors is essential to stabilize cardiac gene expression. Our analyses showed increased expression of the homeodomain transcription factor Six122 and its coactivators, Eya1, Eya2, and Eya423,24 (Fig. 2e; Supplementary Table 2). Six1 functions in non-cardiac cell types, including skeletal muscle. Indeed, many (P<2×10−8) genes misregulated in Ezh2-deficient hearts are direct Six1 targets in differentiating skeletal myoblasts (Fig. 2e)25. Six1 targets were preferentially misregulated over non-Six1 targets (Fig. 2g). Levels of Six1, Eya1, and some skeletal muscle genes were also increased in Ezh2fl/fl;Nkx2.5::Cre hearts (Supplementary Fig. 6e). Six1 functions in early cardiac progenitors, but not in differentiating cardiomyocytes10 and remains silent in developing and postnatal heart (Fig. 3b,c; 6d; Supplementary Fig. 8). Upon loss of Ezh2, Six1 might activate transcription of a skeletal muscle genes subset in cardiomyocytes, contributing to instability of cardiac gene expression.
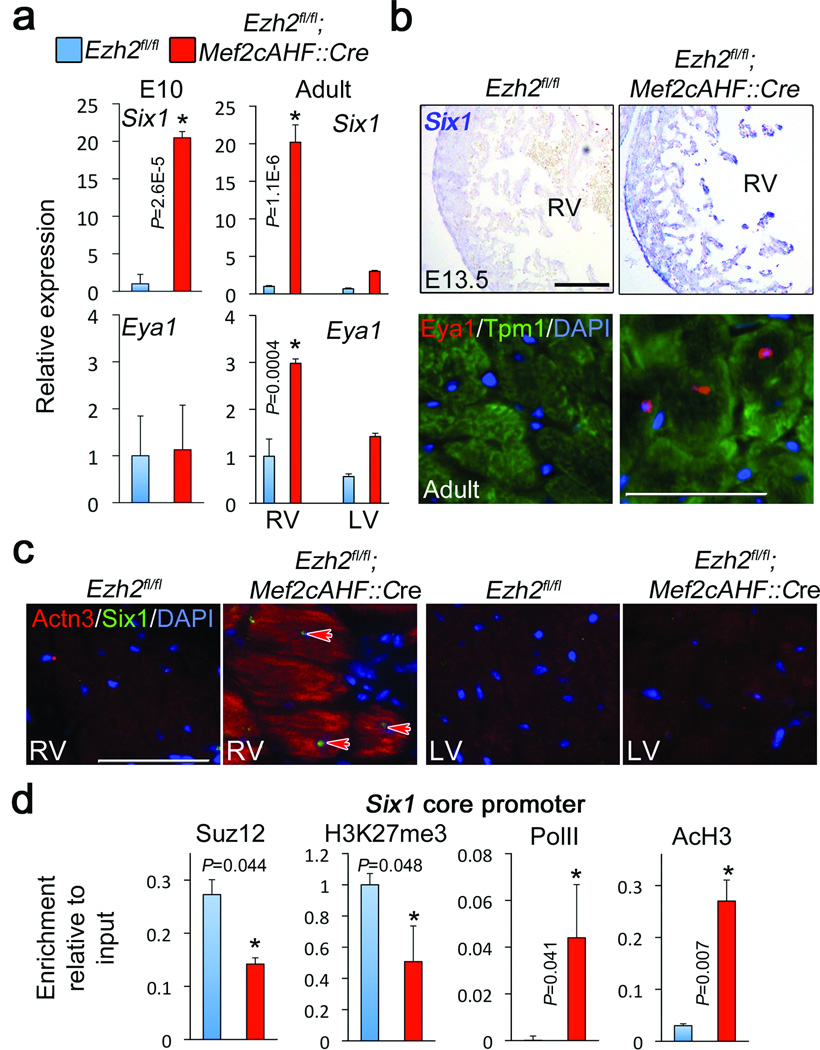
Figure 3.
Six1 is epigenetically repressed by PRC2. (a) QRTPCR showing expression of Six1 and Eya1 in embryonic and adult myocardium. (b) Upper panel: In situ hybridization showing Six1 mRNA in E13.5 Ezh2-deficient (Ezh2fl/fl; Mef2cAHF::Cre) right ventricle (RV). Reference bar = 200 µm. Lower panel. Immunofluorescence for Eya1. Cardiomyocytes were co-stained with an anti tropomyosin (Tpm1) antibody, and nuclei with DAPI. (c) Immunofluorescence for Six1 and alpha-actinin 3 (Actn3) in control (Ezh2fl/fl) and Ezh2fl/fl; Mef2cAHF::Cre adult hearts. Only Ezh2-defficient right ventricular (RV) cardiomyocytes co-expressed Six1 and Actn3. Arrows point to Six1-possitive cardiomyocyte nuclei. LV: left ventricle. Reference bar = 25 µm. (d) Chromatin immunoprecipitation for Suz12, H3K27me3, RNA PolII, and acetylated histone H3 (AcH3) on the Six1 core promoter in Ezh2-deficient Hearts (red bars), and controls (blue bars). Bars in panels (a) and (d) represent the mean +/− S.D. of at least three biological replicates. * Indicates P < 0.05.
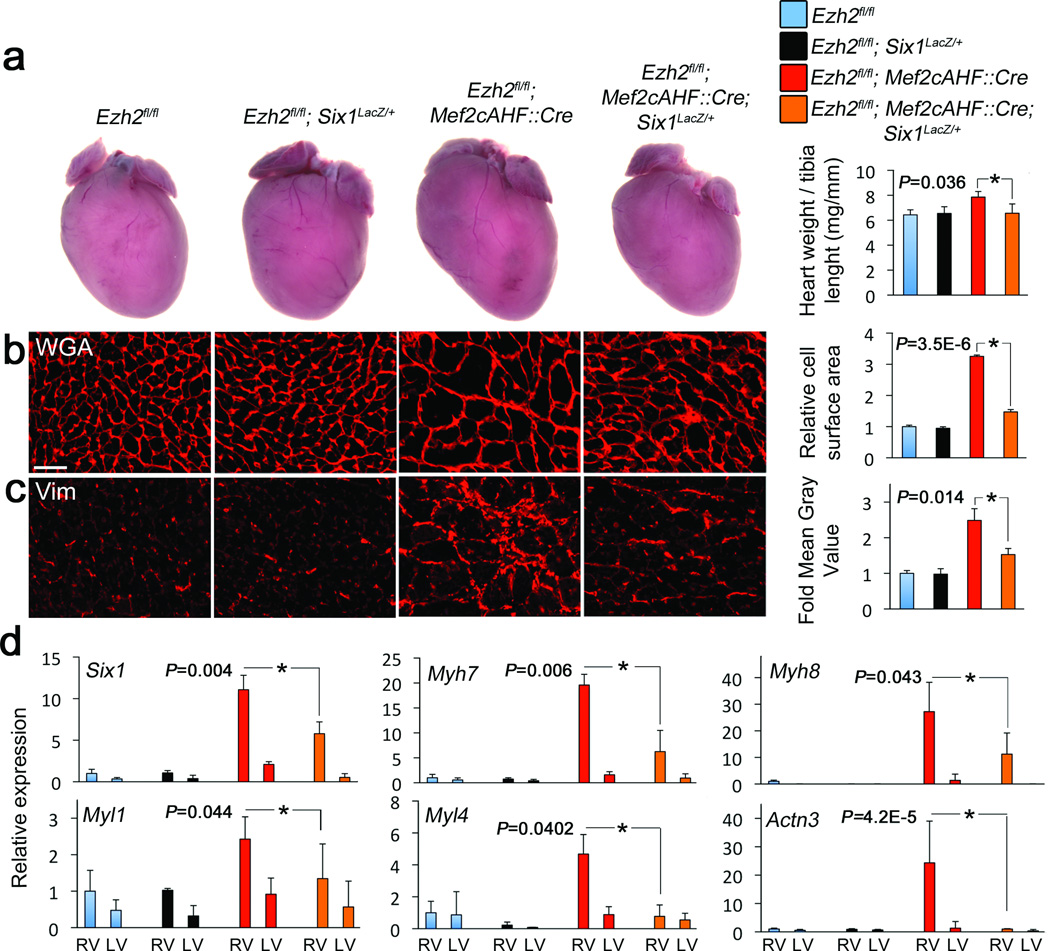
Figure 6.
Ezh2 limits cardiac hypertrophy and fibrosis by repressing Six1. (a) Whole heart images and heart mass, as represented by heart weight to tibia length ratio of Ezh2 floxed (Ezh2fl/fl), Six1 heterozygous (Ezh2fl/fl; Six1LacZ/+), Ezh2-deficient mice (Ezh2fl/fl; Mef2c::Cre), and Ezh2-deficient mice with decreased levels of Six1 (Ezh2fl/fl; Mef2c::Cre;Six1LacZ/+) (b), Heart sections from mice with the same genotypes as in (a) stained with wheat germ agglutinin (WGA) and (c) vimentin. Graphs show the respective quantitations. (d) QRTPCR for Six1, Myh7, Myh8, Myl1, Myl4 and Actn3. Reference bars = 10 µm. Bars represent the mean +/− S.D. from at least three hearts per genotype. * indicates P < 0.05.
We addressed the relevance of increased Six1 expression in deregulating gene expression in Ezh2-deficient hearts. Quantitative real time PCR (QRTPCR) confirmed Six1 de-repression in early developing ventricular myocardium (E10) and exclusively in the adult Ezh2-deficient right ventricle (Fig. 3a), consistent with specific repression by Ezh2. At E13.5, in newborn, and adult hearts, Six1 expression was increased in Ezh2-deficient right ventricular myocardium (Fig. 3b,c; Supplementary Fig. 8), and Eya1 was derepressed exclusively in adult cardiomyocytes (Fig. 3b). Furthermore, upon loss of Ezh2, Six1 was expressed in nuclei of adult cardiomyocytes that concomitantly expressed the skeletal muscle-specific Six1 target Actn3 (Fig. 3c).
To determine if Six1 and Eya1 are targets for PRC2-mediated repression, we performed chromatin immunoprecipitation. In Ezh2-deficient adult hearts, H3K27me3 and Suz12 were depleted at the Six1, but not Eya1, promoter (Fig. 3d, not shown). PolII and acetylated histone H3 (AcH3) were enriched, consistent with Six1 derepression. Thus, PRC2 epigenetically represses Six1 expression.
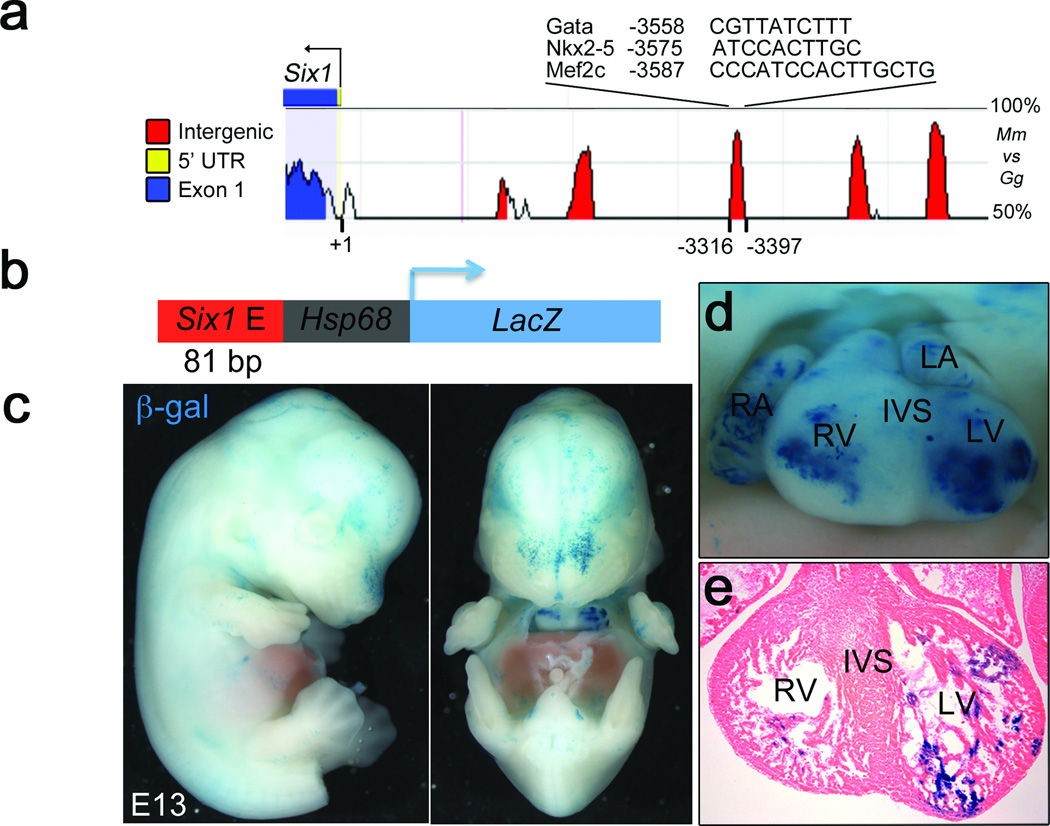
Six1 expression is rapidly extinguished upon differentiation of cardiac progenitors10, but its derepression in Ezh2fl/fl;Mef2cAHF::Cre hearts suggests an expression potential in differentiated myocardium. Computational analysis of the Six1 locus identified a highly conserved sequence in the Six1 5' non-coding region with putative binding motifs for cardiac development regulators Nkx2-5, GATA4 and Mef2c (Fig. 4a). To test its potential to drive cardiac expression, this conserved element was fused to a LacZ-reporter driven by the Hsp68 promoter, and the resulting reporter construct (Fig. 4b) was injected in mouse zygote pronuclei. About 50% of transient transgenic embryos (6/12) showed β-galactosidase activity in the myocardium (Fig. 4c–e), suggesting that this enhancer is active in the heart when not in its endogenous repressed genomic location. This agrees with suppression of a cardiac expression-potential of the Six1 locus in differentiating cardiac progenitors.
Figure 4.
Six1 has differentiated myocardium expression potential. (a) rVista plot of the percentage of conservation between the mouse (Mm) and chicken (Gg) Six1 upstream regulatory region. The indicated peak represents a highly conserved element including potential Nkx2-5, Mef2c and Gata binding motifs. (b) A LacZ reporter driven by the Hsp68 promoter and a 80bp DNA fragment including such a conserved element was injected into mouse blastocysts. (c) Lateral and frontal views of whole mount transient transgenic Six1-hsp68-lacZ embryos at E11.5 that were stained for β-galactosidase (β-gal) activity. (d) Close up of the heart from the same embryo. (e) Section from heart in g showing b-gal staining in ventricular myocardium. RA, LA, RV and LV: right and left atria, and ventricles, respectively.
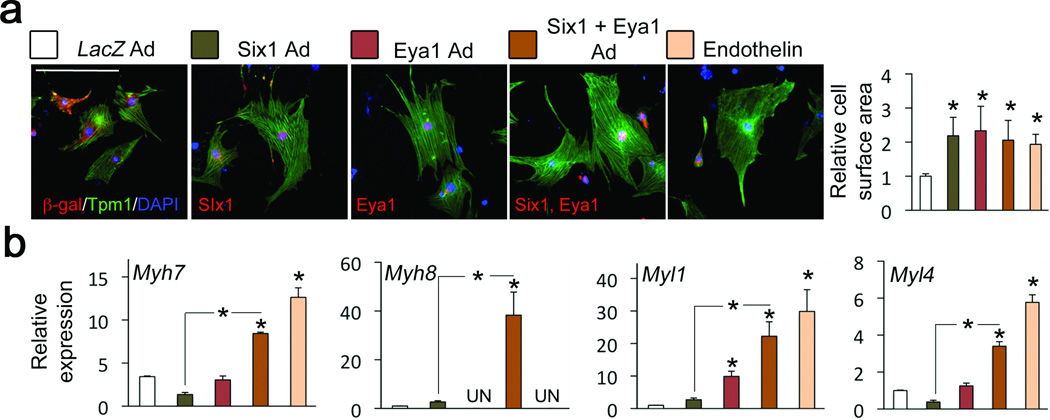
We investigated the effect of persistent Six1 and Eya1 expression in cardiomyocytes on hypertrophy. Overexpression of Six1, with or without Eya1, in cultured neonatal mouse cardiomyocytes resulted in hypertrophy comparable to that induced by the potent hypertrophic agonist, Endothelin-1 (Fig. 5a). In addition, Six1 and Eya1 synergistically activated fetal cardiac and skeletal muscle gene expression (Fig. 5b; Supplementary Fig. 9).
Figure 5.
Six1 induces cardiac hypertrophy and skeletal muscle gene expression in cardiomyocytes. (a) Cardiac myocytes infected with adenoviruses (ad) expressing control cDNA (lacZ), Six1, or Eya1, or treated with Endothelin-1. Expression of LacZ, Six1 and Eya, was confirmed with staining for β-galactosidase (β -gal), and V5-tagged Six1 and Eya1. Cardiomyocytes were stained for tropomyosin, and nuclei counterstained with DAPI. Graph shows average cell surface area. Bars represent the mean +/− S.D. from at least 100 myocytes measured. Reference bars = 10 µm. (b) QRTPCR of skeletal muscle genes on RNA from infected cardiomyocytes. Bars in (a,b) represent the mean +/− S.D. from at least three independent infections. * Indicates P < 0.05. LV: left ventricle
To determine if Six1 derepression causes the phenotype of mice with Ezh2-deficient hearts, we reduced Six1 levels. Ezh2fl/fl;Mef2cAHF::Cre mice were bred onto a heterozygous Six1-LacZ knock-in background23, and cardiomyocyte hypertrophy and fibrosis were analyzed in adult mice. Reducing levels of Six1 by 50% in Ezh2-deficient hearts significantly normalized heart size, as measured by heart weight to tibia length ratios (Fig. 6a). Furthermore, cardiomyocyte size and the levels of Tgfβ3 and fibrosis were almost completely rescued (Fig. 6b,c; Supplementary Fig. 10). Skeletal muscle genes, the hypertrophy marker Myh7, and the profibrosis factors Tgfβ3 and Postn, also returned to normal expression levels (Fig. 6d; Supplementary Fig. 10). Residual Six1 (Fig. 6d) might account for the persistent phenotype. Evidence points to a direct effect of Six1 misregulation in myocardium and rule out secondary effects due to outflow tract pressure overload. Six1 and its target Actn3 are coexpressed in Ezh2-deficient cardiomyocytes (Fig. 3c), and Six1 occupied the Nppa, Myl1 and Tgfβ3 locus in Ezh2-deficient hearts (Supplementary Fig. 11). In addition, mild thickening of the pulmonary valve leaflets in Ezh2fl/fl;Mef2cAHF::Cre adult hearts (Supplementary Fig. 12) was not present in already hypertrophied fetal hearts (Supplementary Fig. 5b), and was not rescued upon decreasing Six1 levels (Supplementary Fig. 12), despite a normalization of hypertrophy and fibrosis (Fig. 6b–c). Thus, myocardial deregulation of Six1 alone is the major cause of myocardial hypertrophy and fibrosis in the Ezh2-deficient heart.
We uncovered a finely regulated epigenetic regulatory mechanism in which Ezh2-mediated stable repression of Six1 in differentiating embryonic cardiac progenitors is essential for normal cardiac growth and stress-responsiveness in the adult. Ezh2 modulates a feed-forward pathway repressing fetal gene expression, which is reinforced by repression of Six1. (Supplementary Fig. 13). This represents a novel pathway for regulating cardiac growth: neither Ezh2 nor Six1 is upregulated in hearts subjected to hypertrophic stimuli (Supplementary Fig. 14). Repressing Six1 is required to stabilize cardiac gene expression by preventing activation of skeletal muscle genes in cardiac muscle. Ezh2 loss is likely insufficient to activate this program merely acting as a “brake”, but reactivation of Six1 (the “gas pedal”) and expression of its targets results in pathologic cardiac remodelling with hypertrophy and fibrosis. Mutations in EYA4 cause cardiomyopathy in humans24, strengthening the idea that control of Six-Eya function is critical for normal cardiac homeostasis. Our results suggest Ezh2-mediated Six1 repression in cardiac progenitors stabilizes postnatal cardiac gene expression, illustrating the importance of epigenetic regulation early in development and its implications in postnatal organ homeostasis.
METHODS
Mice
The following mice strains were used: Ezh2fl/fl, Mef2cAHF::Cre, Nkx2-5::Cre, and Six1Lacz/+. All animal experiments were conducted following guidelines established and approved by the UCSF Institutional Animal Care and Use Committee.
Plasmids
Six1 and Eya1 expression vectors were constructed by cloning PCR-amplified full-length cDNAs into pCDNA3.1/V5-His TOPO (Invitrogen).
Cardiac hypertrophy induction
Isoproterenol (60 mg/kg body weight) was administered for 14 days in 9 weeks old mice using ALZET osmotic minipumps, which were surgically implanted subdermally. As control mice were implanted with PBS-filled minipumps. 5 mice per genotype were treated.
Protein analysis
Immunostaining and in situ hybridization procedures were carried out by standard protocols. For immunostaining, cryosections were incubated with the next primary antibodies: anti-trimethyl-Histone H3 (Lys 27) (Millipore 07-449, 1:3000), histone H3 trimethyl Lys9 (39161 Active Motif, 1:1000), alpha tropomyosin (CH1 monoclonal, Hybridoma bank, 1:100), cardiac troponin t (Thermo Scientific,1:1000) Pab to Vimentin (GP53 Progen, 1:500), Tgfβ3 (sc-83 Santa Cruz Biotechnology, 1:100), Alpha-actinin 3 (Epitomics, 1:100), Six125 (1:100), Isl1 (Developmental Studies Hybridoma Bank, 1:250), Ezh2 (Active Motif, 1:200), and Ezh1 (Abcam, 1:200). Cultured myocytes were incubated with the next primary antibodies: β-galactosidase (Abcam, 1:100) and anti-V5 (R960-25 Invitrogen, 1:100). After incubation with primary antibodies, the sections were washed 3× with PBS, incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen, 1:500), washed and mounted with Prolong gold with 4,6-diamidino-2-phenylindole (Invitrogen) to counterstain nuclei. Fluorescent signal for vimentin and Tgfβ3 was quantified as Mean Gray Value with ImageJ. Multiple sections from at least three hearts per genotype were analyzed. The next antibodies were used for Western blot: anti-Ezh2 (Cell Signalling, 1:1000) and anti-GAPDH (Santa Cruz Biotechnology, 1:1000).
Gene expression analysis
RNA was isolated from the right ventricle and interventricular septum and treated with DNaseI. cDNA was prepared with the SuperScript III First Strand Synthesis Kit (Invitrogen). 10 pg of cDNA were used for quantitative real-time amplification with the next TaqMan probes. Nppa Mm01255747_g1, Nppb Mm00435304_g1, Myh7 Mm00600555_m1, Tgfβ3 Mm01307950_m1, Postn NM_015784.2, Spp1 Mm00436767_m1, Six1 Mm00808212_m1, Eya1 Mm00438796_m1, Myh8 Mm01329494_m1, Myl1 Mm00659043_m1, Myl2 Mm00440384_m1, Myl4 Mm00440377_m1, Mylpf Mm00443940_m1, Tnni1 Mm00502426_m1, Actn3 Mm00496495_m1 and Tnni2 Mm00437157_g1. In situ hybridization on whole mount embryos and on 10 um paraffin sections was performed as previously reported. RNA antisense probes were labeled with digoxigenin-coupled UTP. After hybridization, samples were washed and incubated with alkaline phosphatase-coupled anti-digoxigenin antisera (Roche). Signal was developed with BM purple (Roche).
Cell measurements
Cryosections were fixed in 4% PFA for 5 min, incubated with 5 ug/ul wheat germ agglutinin, Alexa Fluor® 594 conjugate (W11262 Invitrogen) for 10 min and washed 3× ten min each with PBS. Sections were then mounted with Prolong gold with 4,6-diamidino-2-phenylindole (Invitrogen). Micrographs were taken from right and left ventricles. The cross sectional area of myocytes was measured using ImageJ. At least 100 cardiomyocytes were measured per image. Multiple sections obtained from at least three hearts per genotype were analyzed. The same method was used to measure alpha tropomyosin-stained cardiomyocytes in culture. At least 50 cells were measured from three biological replicates.
Microarray analysis and RNA-seq
RNA was isolated from the right ventricle and interventricular septum of five Ezh2fl/fl and five Ezh2fl/fl;Mef2cAHF::Cre mice. Affymetrix mouse Gene ST 1.0 arrays were hybridized and scanned according to the manufacturer's recommendations. Linear models were fitted for each gene on Ezh2fl/fl;Mef2cAHF::Cre vs Ezh2fl/fl samples to derive the mutant effect using the limma package in R/Bioconductor. Moderated t-statistics and the associated P-values were calculated, adjusting for multiple testing by controlling for false discovery rate with the Benjamini-Hochberg method and for family-wise error rate using the Bonferroni correction (adjP). Genes with adjP<0.05 were considered differentially expressed. RNAseq was performed as previously described in RNA from the right ventricle of five hearts per genotype26. Gene ontology analysis was performed using GO Elite.
Chromatin immunoprecipitation
Chromatin was isolated from the right ventricle and the interventricular septum with the Imprint Chromatin Immunoprecipitation Kit (Sigma), following manufacturer instructions. The immunoprecipitated chromatin, input and IgG-bound chromatin were analized by qPCR using TaqMan probes for the Nppa core promoter, Nppa distal promoter, Six1 core promoter, Eya1 core promoter, and the Srf core promoter (Supplementary Table. 4)
Cardiomyocyte culture and infection
Newborn mouse cardiomyocytes were isolated and maintained using the Neonatal Cardiomycyte Isolation System (Cellutron) as recommended by the manufacturer.
Cardiomyocytes were seeded in 8 well chamberslides 48 h before infection. Cells were infected with 5.5×108 IFU of Six1-encoding adenovirus and 1.2×108 IFU of Eya1-encoding adenovirus per well. Cells were fixed and stained 48 h post infection.
For induction of cadiomyocyte hypertrophy, 0.2 mM of Endothelin 1 (Sigma) was added to the culture media. Cells were exposed for 48 h.
Adenoviruses
For generation of adenoviral vectors, Six1 and Eya1 cDNAs were cloned into pAd/CMV/V5-DEST (Invitrogen).
Viral particles were generated using the ViraPower Adenoviral Expression System. Viral particles were tittered using the AdEasy Viral Titer Kit (Agilent Technologies).
Statistical analysis
Data are presented as mean +/− S.D. The P-value was determined by two-tailed T-test. A P-value <= 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Blais for Six1 antiserum, T. Sukonnik for in situ hybridization, J. Wythe for Cre/mT/mG embryos, J. Wylie for technical assistance, D. Miguel-Perez for animal husbandry, A. Holloway and A. Williams (Gladstone Bioinformatics core) for RNA-seq and microarray data analysis, L. Ta (Gladstone Genomics Core) for microarray experiments, C. Miller (Gladstone Histology Core) for histology, Bruneau lab members for helpful discussions, and Gary Howard for editorial assistance. This work was supported by a fellowship from the California Institute for Regenerative Medicine (P.D.-O.), a grant from the NIH (U01HL098179), the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association Established Investigator Award (B.G.B.), and by William H. Younger, Jr. (B.G.B.). Animal work was supported by an NIH/NCRR grant (C06 RR018928) to the J. David Gladstone Institutes.
Footnotes
Accession codes.
Microarray data can be accessed at GEO. Accession ID: GSE30076
RNA-Seq data can be accessed at GEO. Accession ID: GSE34274
AUTHOR CONTRIBUTIONS
P.D.-O. designed the study with B.G.B. and performed most of the experiments; Y.H. performed echocardiography; X.L. provided essential information prior to publication; X.L. and A.T. provided mouse lines; D.C. generated and analyzed RNAseq data with C.E.S. and J.G.S.; P.D.-O. and B.G.B. wrote the paper with input from all the authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nature reviews. Endocrinology. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 2.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 3.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell stem cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 7.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu CR, et al. Chromatin "prepattern" and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes & development. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo C, et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. The Journal of clinical investigation. 2011;121:1585–1595. doi: 10.1172/JCI44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nature reviews. Molecular cell biology. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annual review of genomics and human genetics. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 13.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:437–449. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Weiford BC, Subbarao VD, Mulhern KM. Noncompaction of the ventricular myocardium. Circulation. 2004;109:2965–2971. doi: 10.1161/01.CIR.0000132478.60674.D0. [DOI] [PubMed] [Google Scholar]
- 18.Swynghedauw B. Phenotypic plasticity of adult myocardium: molecular mechanisms. The Journal of experimental biology. 2006;209:2320–2327. doi: 10.1242/jeb.02084. [DOI] [PubMed] [Google Scholar]
- 19.Houweling AC, van Borren MM, Moorman AF, Christoffels VM. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res. 2005;67:583–593. doi: 10.1016/j.cardiores.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 20.McFadden DG, et al. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi JK, et al. Chromatin remodeling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cellular and molecular life sciences : CMLS. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 24.Schonberger J, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005 doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic acids research. 2010;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Current protocols in molecular biology / edited by Frederick M. Ausubel … [et al.] 2011 doi: 10.1002/0471142727.mb0412s94. Chapter 4, Unit4 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.