Abstract
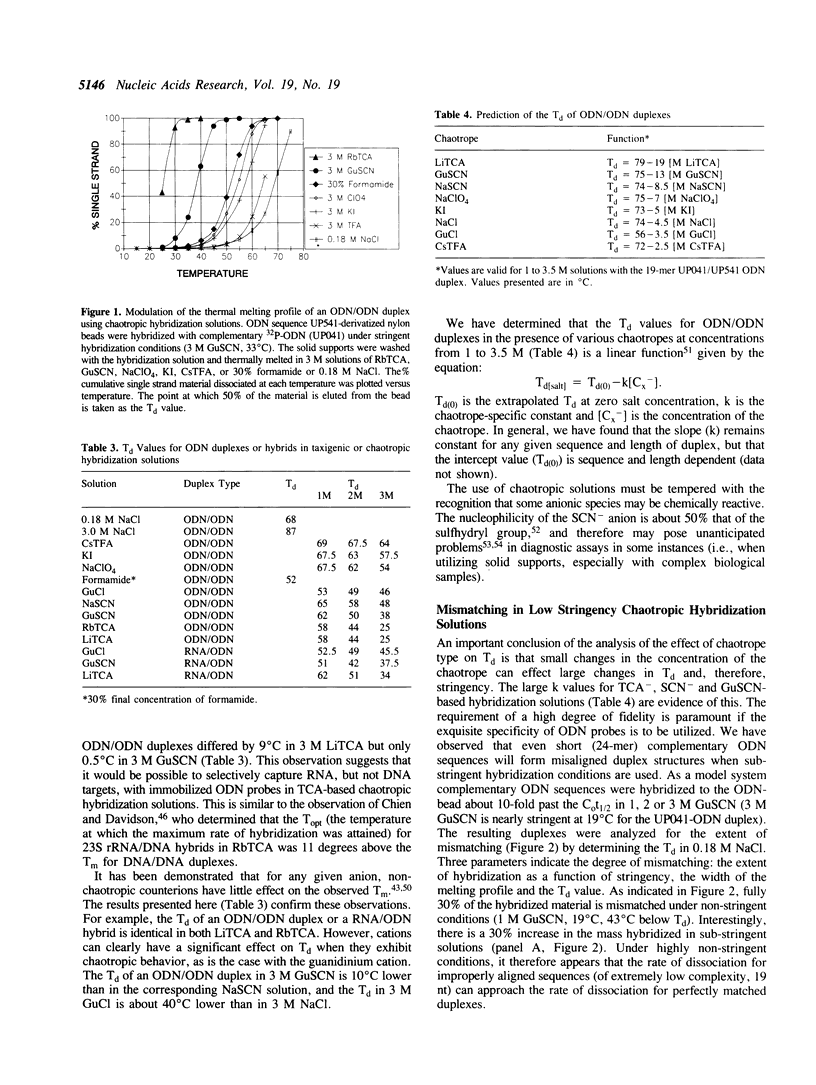
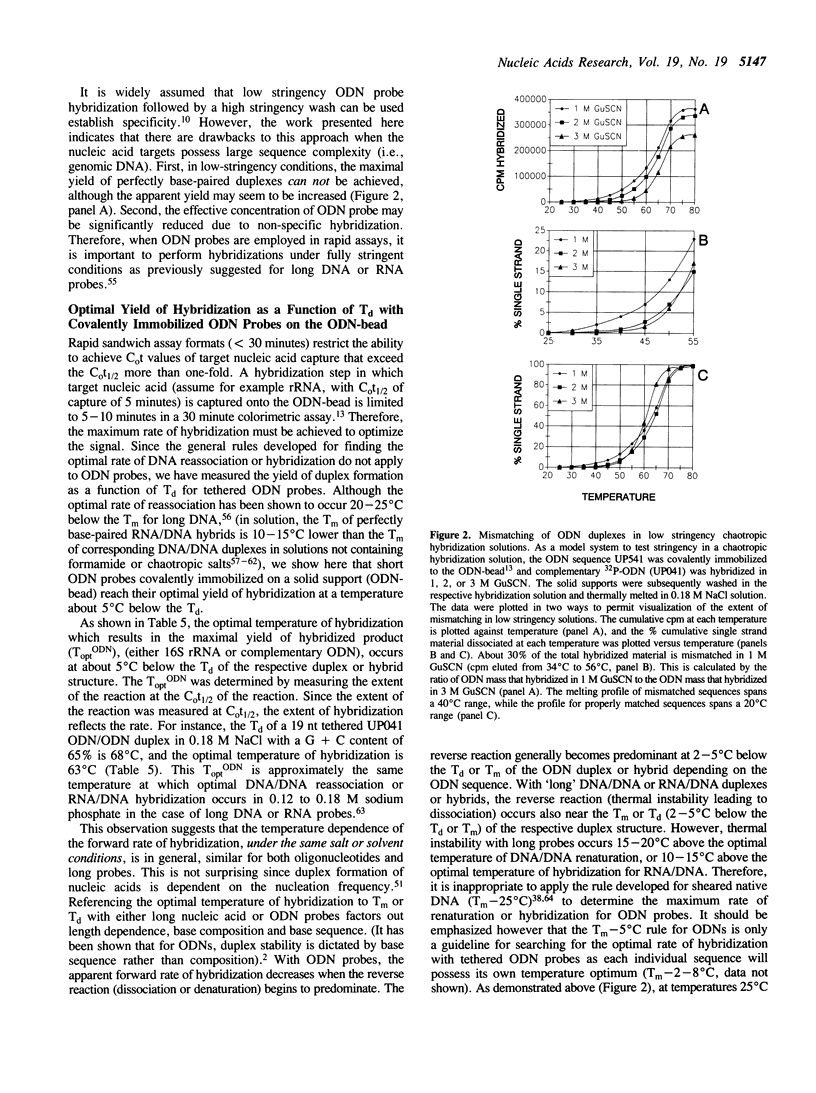
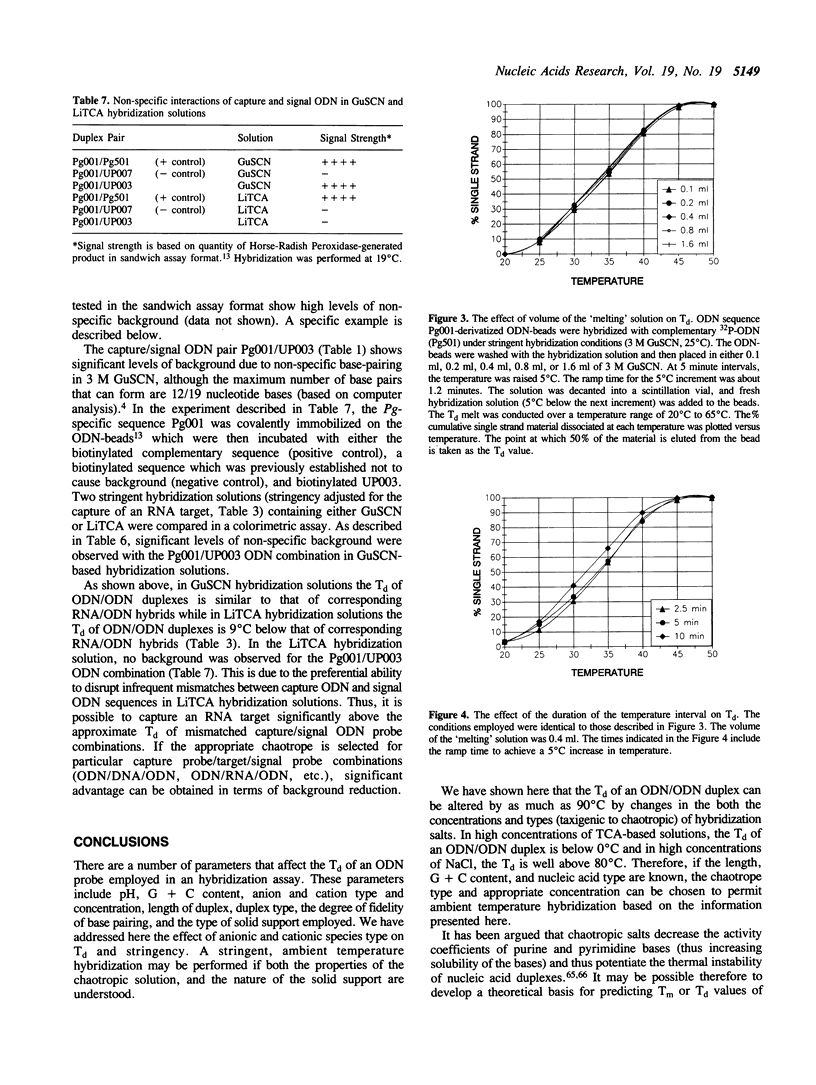
Hybridization solutions containing chaotropes may be used to modulate the thermal stability (Tm or Td) of oligodeoxynucleotide (ODN) duplexes or hybrids over a 90 degrees C range. Modulation of Td allows formulation of hybridization solutions that permit ambient temperature hybridization using most combinations of probe length, probe composition, target type, and facilitates development of convenient and rapid assay formats. The conditions required to achieve ODN duplex fidelity, and optimal yields of hybridized product, are described for trichloroacetate, thiocyanate, guanidinium salts and other chaotropic salts. The effects of different solid supports on Td are described. Also, a method is presented that uses chaotropic compounds to reduce background arising from signal ODN probes in a sandwich assay hybridization format.
Full text
PDF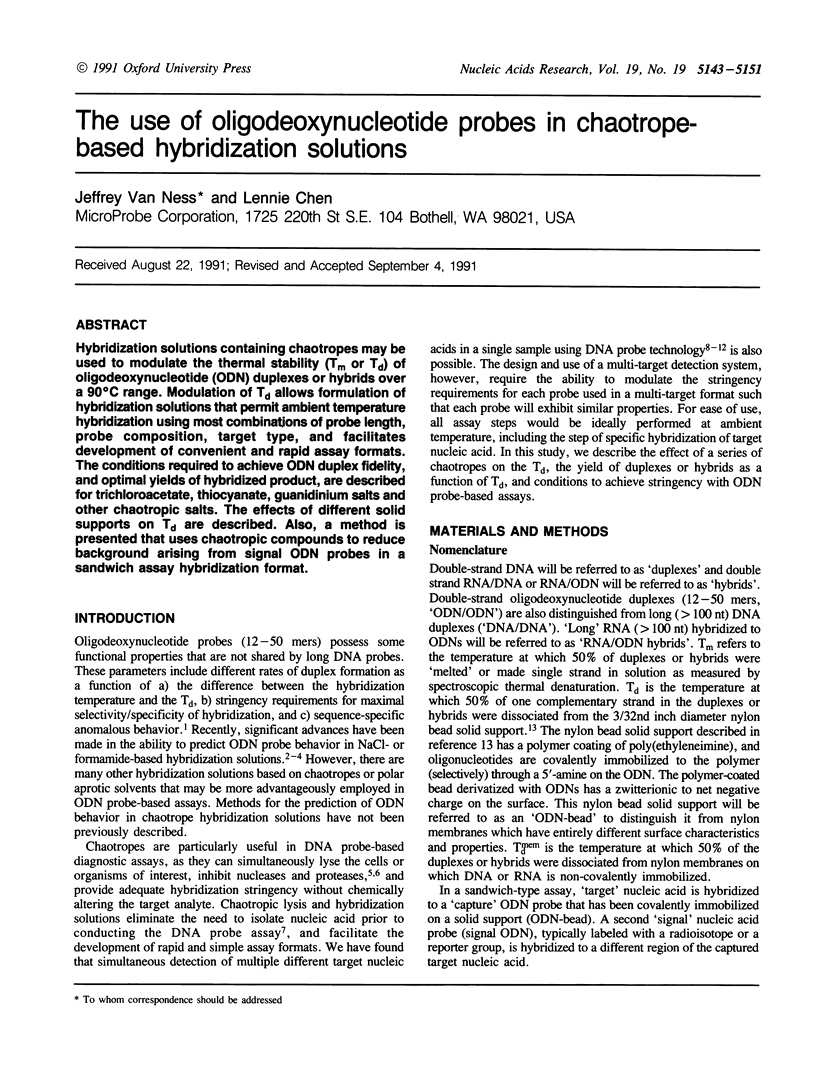




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albretsen C., Haukanes B. I., Aasland R., Kleppe K. Optimal conditions for hybridization with oligonucleotides: a study with myc-oncogene DNA probes. Anal Biochem. 1988 Apr;170(1):193–202. doi: 10.1016/0003-2697(88)90108-x. [DOI] [PubMed] [Google Scholar]
- BOLTON E. T., MCCARTHY B. J. FRACTIONATION OF COMPLEMENTARY RNA. J Mol Biol. 1964 Feb;8:201–209. doi: 10.1016/s0022-2836(64)80129-7. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Sells B. H., Purdom I. F. Kinetic complexity of RNA molecules. J Mol Biol. 1972 Jan 14;63(1):21–39. doi: 10.1016/0022-2836(72)90519-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Sells B. H., Purdom I. F. Kinetic complexity of RNA molecules. J Mol Biol. 1972 Jan 14;63(1):21–39. doi: 10.1016/0022-2836(72)90519-0. [DOI] [PubMed] [Google Scholar]
- Blüthmann H., Brück D., Hübner L., Schöffski A. Reassociation of nucleic acids in solutions containing formamide. Biochem Biophys Res Commun. 1973 Jan 4;50(1):91–97. doi: 10.1016/0006-291x(73)91068-1. [DOI] [PubMed] [Google Scholar]
- Bonner J., Kung G., Bekhor I. A method for the hybridization of nucleic acid molecules at low temperature. Biochemistry. 1967 Dec;6(12):3650–3653. doi: 10.1021/bi00864a005. [DOI] [PubMed] [Google Scholar]
- Bonner J., Kung G., Bekhor I. A method for the hybridization of nucleic acid molecules at low temperature. Biochemistry. 1967 Dec;6(12):3650–3653. doi: 10.1021/bi00864a005. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Bauer W. R. The early melting of closed duplex DNA: analysis by banding in buoyant neutral rubidium trichloroacetate. Nucleic Acids Res. 1980 Mar 11;8(5):1145–1165. doi: 10.1093/nar/8.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Bauer W. R. The properties of native and denatured DNA in buoyant rubidium trichloroacetate at neutral pH. Nucleic Acids Res. 1977 Jun;4(6):1891–1909. doi: 10.1093/nar/4.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAMBERLIN M., BERG P. MECHANISM OF RNA POLYMERASE ACTION: FORMATION OF DNA-RNA HYBRIDS WITH SINGLE-STRANDED TEMPLATES. J Mol Biol. 1964 Feb;8:297–313. doi: 10.1016/s0022-2836(64)80139-x. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y. H., Davidson N. RNA:DNA hybrids are more stable than DNA:DNA duplexes in concentrated perchlorate and trichloroacetate solutions. Nucleic Acids Res. 1978 May;5(5):1627–1637. doi: 10.1093/nar/5.5.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- DUGGAN E. L. Deformation of DNA. III. The effect of glycol and glycerol on the ultraviolet absorbances of DNA. Renaturation by dilution. Biochem Biophys Res Commun. 1961 Nov 1;6:93–99. doi: 10.1016/0006-291x(61)90391-6. [DOI] [PubMed] [Google Scholar]
- Davis K. A., Hatefi Y. Kinetics of the resolution of complex I (reduced diphosphopyridine nucleotide-coenzyme Q reductase) of the mitochondrial electron transport system by chaotropic agents. Biochemistry. 1969 Aug;8(8):3355–3361. doi: 10.1021/bi00836a033. [DOI] [PubMed] [Google Scholar]
- Davis K. A., Hatefi Y. Succinate dehydrogenase. I. Purification, molecular properties, and substructure. Biochemistry. 1971 Jun 22;10(13):2509–2516. doi: 10.1021/bi00789a014. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Dix K., Watanabe S. M., McArdle S., Lee D. I., Randolph C., Moncla B., Schwartz D. E. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J Clin Microbiol. 1990 Feb;28(2):319–323. doi: 10.1128/jcm.28.2.319-323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn A. R., Hassell J. A. A novel method to map transcripts: evidence for homology between an adenovirus mRNA and discrete multiple regions of the viral genome. Cell. 1977 Sep;12(1):23–36. doi: 10.1016/0092-8674(77)90182-9. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R., Feix G. RNA-RNA hybridization in aqueous solutions containing formamide. Anal Biochem. 1972 Dec;50(2):467–476. doi: 10.1016/0003-2697(72)90056-5. [DOI] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P., HERSKOVITS T. T. Nonaqueous solutions of DNA. Reversible and irreversible denaturation in methanol. Arch Biochem Biophys. 1961 Oct;95:114–129. doi: 10.1016/0003-9861(61)90116-3. [DOI] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P., HOLTZER A. Application of light scattering to biological systems: deoxyribonucleic acid and the muscle proteins. Adv Biol Med Phys. 1958;6:431–551. doi: 10.1016/b978-1-4832-3112-9.50014-1. [DOI] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W. E., Pettijohn D. E., Van Ness J. One strand equivalent of the Escherichia coli genome is transcribed: complexity and abundance classes of mRNA. Science. 1977 Aug 5;197(4303):582–585. doi: 10.1126/science.327551. [DOI] [PubMed] [Google Scholar]
- Hanstein W. G., Davis K. A., Hatefi Y. Water structure and the chaotropic properties of haloacetates. Arch Biochem Biophys. 1971 Dec;147(2):534–544. doi: 10.1016/0003-9861(71)90411-5. [DOI] [PubMed] [Google Scholar]
- Hatefi Y., Hanstein W. G. Solubilization of particulate proteins and nonelectrolytes by chaotropic agents. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1129–1136. doi: 10.1073/pnas.62.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I., Nishimura Y., Naraoka T., Watanabe K., Arata Y., Miura K. Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: d(GCGAAAGC). Nucleic Acids Res. 1989 Mar 25;17(6):2223–2231. doi: 10.1093/nar/17.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton J. R. Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res. 1977 Oct;4(10):3537–3555. doi: 10.1093/nar/4.10.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVINE L., GORDON J. A., JENCKS W. P. The relationship of structure to the effectiveness of denaturing agents for deoxyribonucleic acid. Biochemistry. 1963 Jan-Feb;2:168–175. doi: 10.1021/bi00901a030. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANDELL J. D., HERSHEY A. D. A fractionating column for analysis of nucleic acids. Anal Biochem. 1960 Jun;1:66–77. doi: 10.1016/0003-2697(60)90020-8. [DOI] [PubMed] [Google Scholar]
- MARMUR J., TS'O P. O. Denaturation of deoxyribonucleic acid by formamide. Biochim Biophys Acta. 1961 Jul 22;51:32–36. doi: 10.1016/0006-3002(61)91013-7. [DOI] [PubMed] [Google Scholar]
- Martinson H. G. The nucleic acid-hydroxylapatite interaction. I. Stabilization of native double-stranded deoxyribonucleic acid by hydroxylapatite. Biochemistry. 1973 Jan 2;12(1):139–145. doi: 10.1021/bi00725a023. [DOI] [PubMed] [Google Scholar]
- Matthews J. A., Kricka L. J. Analytical strategies for the use of DNA probes. Anal Biochem. 1988 Feb 15;169(1):1–25. doi: 10.1016/0003-2697(88)90251-5. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Melli M., Whitfield C., Rao K. V., Richardson M., Bishop J. O. DNA-RNA hybridization in vast DNA excess. Nat New Biol. 1971 May 5;231(18):8–12. [PubMed] [Google Scholar]
- NYGAARD A. P., HALL B. D. FORMATION AND PROPERTIES OF RNA-DNA COMPLEXES. J Mol Biol. 1964 Jul;9:125–142. doi: 10.1016/s0022-2836(64)80095-4. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Olsen G. J., Woese C. R. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986 May 9;45(3):325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Ranki M., Palva A., Virtanen M., Laaksonen M., Söderlund H. Sandwich hybridization as a convenient method for the detection of nucleic acids in crude samples. Gene. 1983 Jan-Feb;21(1-2):77–85. doi: 10.1016/0378-1119(83)90149-x. [DOI] [PubMed] [Google Scholar]
- Robinson D. R., Grant M. E. The effects of aqueous salt solutions on the activity coefficients of purine and pyrimidine bases and their relation to the denaturation of deoxyribonucleic acid by salts. J Biol Chem. 1966 Sep 10;241(17):4030–4042. [PubMed] [Google Scholar]
- Roychoudhury R., Wu R. Terminal transferase-catalyzed addition of nucleotides to the 3' termini of DNA. Methods Enzymol. 1980;65(1):43–62. doi: 10.1016/s0076-6879(80)65009-5. [DOI] [PubMed] [Google Scholar]
- Rychlik W., Rhoads R. E. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELA M., ANFINSEN C. B., HARRINGTON W. F. The correlation of ribonuclease activity with specific aspects of tertiary structure. Biochim Biophys Acta. 1957 Dec;26(3):502–512. doi: 10.1016/0006-3002(57)90096-3. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Suggs S. V., Wallace R. B., Hirose T., Kawashima E. H., Itakura K. Use of synthetic oligonucleotides as hybridization probes: isolation of cloned cDNA sequences for human beta 2-microglobulin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6613–6617. doi: 10.1073/pnas.78.11.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TS'O P. O., HELMKAMP G. K., SANDER C. Interaction of nucleosides and related compounds with nucleic acids as indicated by the change of helix-coil transition temperature. Proc Natl Acad Sci U S A. 1962 Apr 15;48:686–698. doi: 10.1073/pnas.48.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Gillespie D. Molecular hybridization with RNA probes in concentrated solutions of guanidine thiocyanate. Anal Biochem. 1987 Jun;163(2):281–291. doi: 10.1016/0003-2697(87)90225-9. [DOI] [PubMed] [Google Scholar]
- Van Ness J., Kalbfleisch S., Petrie C. R., Reed M. W., Tabone J. C., Vermeulen N. M. A versatile solid support system for oligodeoxynucleotide probe-based hybridization assays. Nucleic Acids Res. 1991 Jun 25;19(12):3345–3350. doi: 10.1093/nar/19.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. RNA-DNA hybridization in solution without DNA reannealing. Biochem Biophys Res Commun. 1977 Apr 25;75(4):1127–1132. doi: 10.1016/0006-291x(77)91500-5. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]
- Woods D. E., Markham A. F., Ricker A. T., Goldberger G., Colten H. R. Isolation of cDNA clones for the human complement protein factor B, a class III major histocompatibility complex gene product. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5661–5665. doi: 10.1073/pnas.79.18.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]



