Abstract
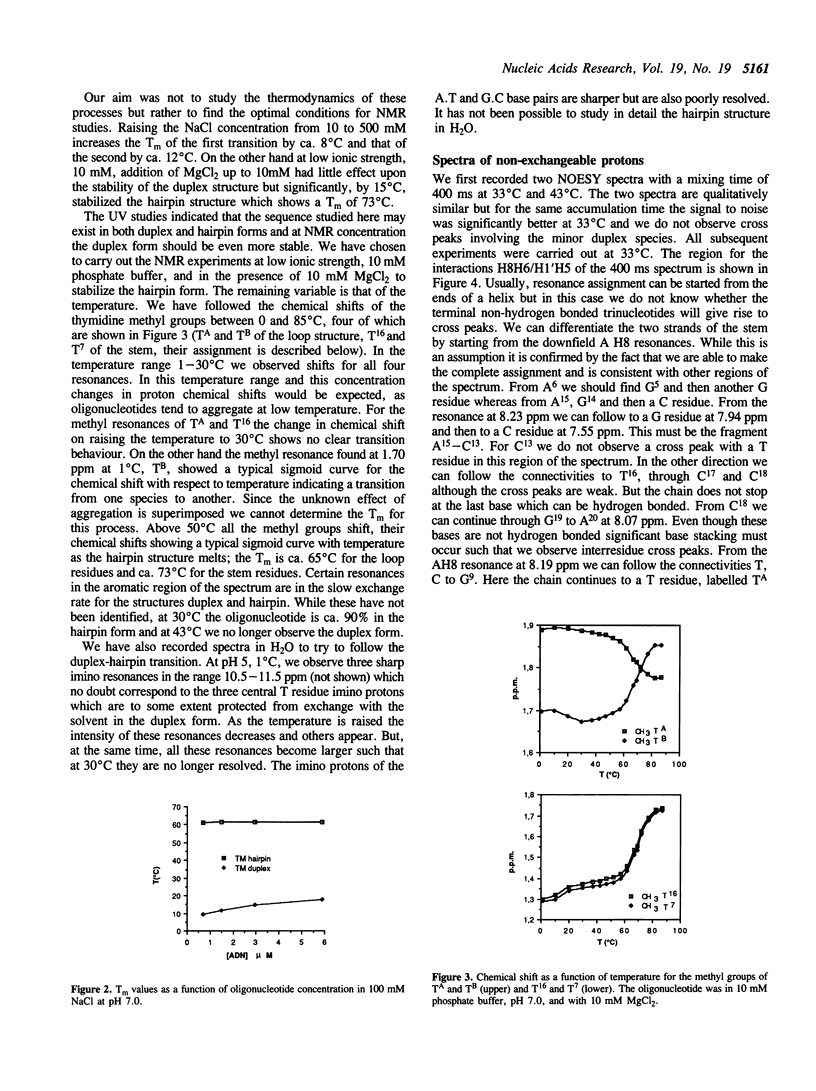
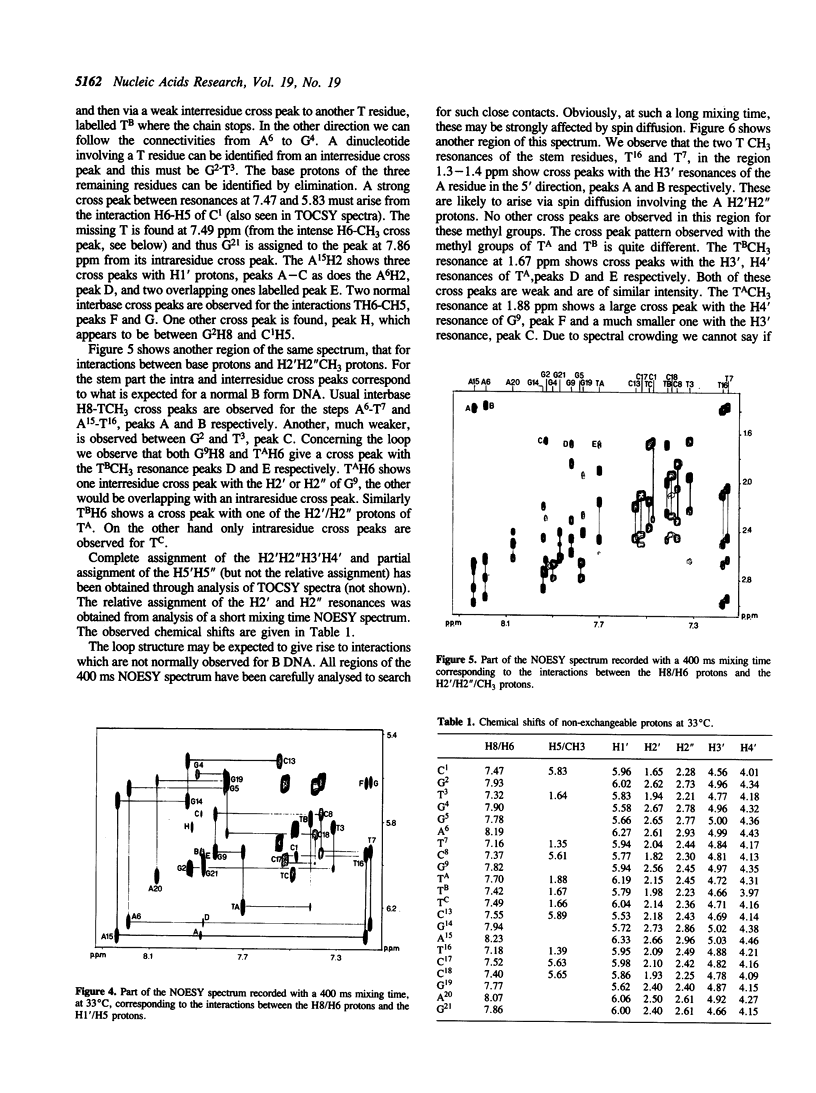
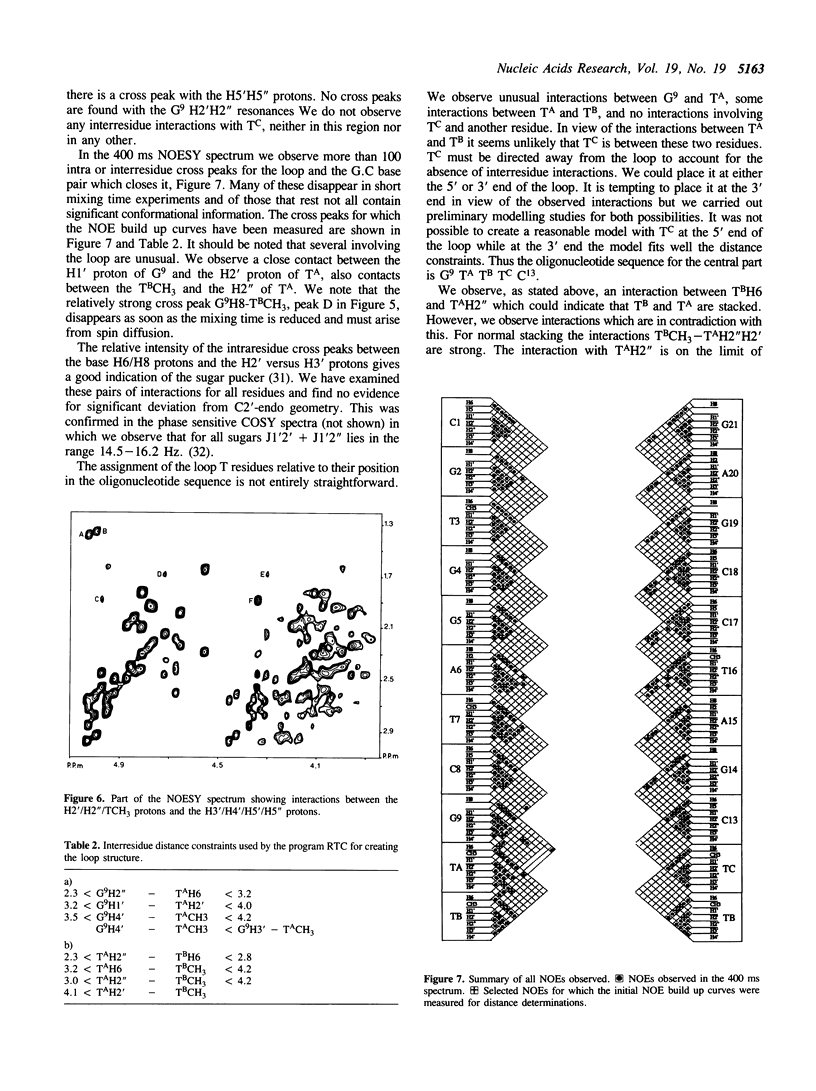
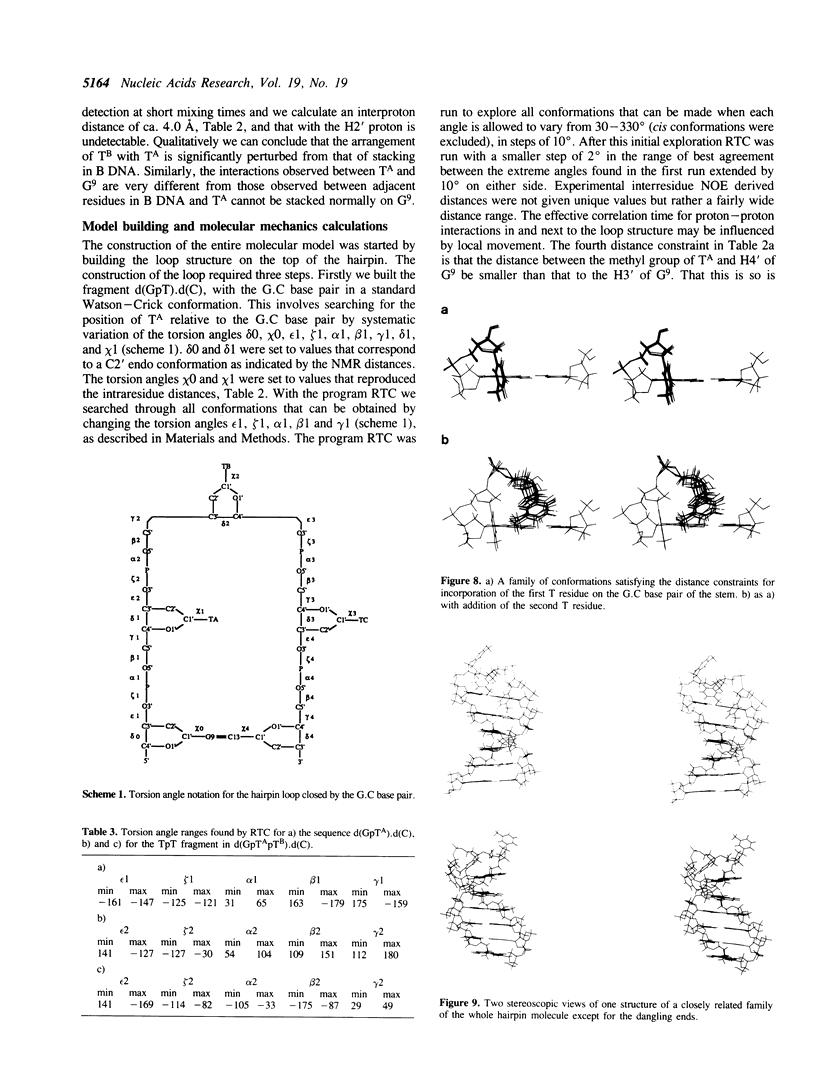
We have determined by two-dimensional nuclear magnetic resonance studies and molecular mechanics calculations the three-dimensional solution structure of a 21 residue oligonucleotide capable of forming a hairpin structure with a loop of three thymidine residues. This structure is in equilibrium with a duplex form. At 33 degrees C, low ionic strength and in the presence of MgCl2 the hairpin form dominates in solution. Six Watson-Crick base pairs are formed topped by the loop structure. The residues 1-3 and 18-21 are not complementary and form dangling ends. Distance constraints have been derived from nuclear Overhauser enhancement measurements. These, together with molecular mechanics calculations, have been used to determine the structure. We do not observe stacking of thymidine residues either over the 3' or the 5' end of the stem.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benham C. J. Stable cruciform formation at inverted repeat sequences in supercoiled DNA. Biopolymers. 1982 Mar;21(3):679–696. doi: 10.1002/bip.360210314. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R., Grzeskowiak K., Dickerson R. E. Structure of a T4 hairpin loop on a Z-DNA stem and comparison with A-RNA and B-DNA loops. J Mol Biol. 1990 Jan 5;211(1):189–210. doi: 10.1016/0022-2836(90)90020-M. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Fazakerley G. V., Guschlbauer W., Kaplan B. E., Sowers L. C. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990 May 20;213(2):303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Sowers L. C., Eritja R., Kaplan B., Goodman M. F., Cognet J. A., Le Bret M., Guschlbauer W., Fazakerley G. V. Abasic frameshift in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Biochemistry. 1989 Mar 7;28(5):2018–2026. doi: 10.1021/bi00431a009. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Sowers L. C., Eritja R., Kaplan B., Goodman M. F., Cognet J. A., LeBret M., Guschlbauer W., Fazakerley G. V. An abasic site in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Nucleic Acids Res. 1987 Oct 12;15(19):8003–8022. doi: 10.1093/nar/15.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Quignard E., Teoule R., Guy A., Guschlbauer W. A two-dimensional 1H-NMR study of the dam methylase site: comparison between the hemimethylated GATC sequence, its unmethylated analogue and a hemimethylated CATG sequence. The sequence dependence of methylation upon base-pair lifetimes. Eur J Biochem. 1987 Sep 15;167(3):397–404. doi: 10.1111/j.1432-1033.1987.tb13351.x. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., Téoule R., Guy A., Fritzsche H., Guschlbauer W. NMR studies on oligodeoxyribonucleotides containing the dam methylation site GATC. Comparison between d(GGATCC) and d(GGm6ATCC). Biochemistry. 1985 Aug 13;24(17):4540–4548. doi: 10.1021/bi00338a009. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985 Jul-Aug;67(7-8):685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Müller U. R., Fitch W. M. Evolutionary selection for perfect hairpin structures in viral DNAs. Nature. 1982 Aug 5;298(5874):582–585. doi: 10.1038/298582a0. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van Beuzekom A. A., Altona C. Conformational and model-building studies of the hairpin form of the mismatched DNA octamer d(m5C-G-m5C-G-T-G-m5C-G). J Biomol Struct Dyn. 1987 Jun;4(6):965–987. doi: 10.1080/07391102.1987.10507692. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res. 1986 May 27;14(10):4187–4196. doi: 10.1093/nar/14.10.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pieters J. M., de Vroom E., van der Marel G. A., van Boom J. H., Altona C. Conformational consequences of the incorporation of arabinofuranosylcytidine in DNA. An NMR study of the DNA fragments d(CGCTAGCG) and d(CGaCTAGCG) in solution. Eur J Biochem. 1989 Sep 15;184(2):415–425. doi: 10.1111/j.1432-1033.1989.tb15033.x. [DOI] [PubMed] [Google Scholar]
- Pieters J. M., de Vroom E., van der Marel G. A., van Boom J. H., Koning T. M., Kaptein R., Altona C. Hairpin structures in DNA containing arabinofuranosylcytosine. A combination of nuclear magnetic resonance and molecular dynamics. Biochemistry. 1990 Jan 23;29(3):788–799. doi: 10.1021/bi00455a029. [DOI] [PubMed] [Google Scholar]
- Quignard E., Fazakerley G. V., Teoule R., Guy A., Guschlbauer W. Consequences of methylation on the amino group of adenine. A proton two-dimensional NMR study of d(GGATATCC) and d(GGm6ATATCC). Eur J Biochem. 1985 Oct 1;152(1):99–105. doi: 10.1111/j.1432-1033.1985.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Quignard E., Téoule R., Guy A., Fazakerley G. V. An NMR study of A-T base pair opening rates in oligonucleotides. Influence of sequence and of adenine methylation. Nucleic Acids Res. 1985 Nov 11;13(21):7829–7836. doi: 10.1093/nar/13.21.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J Biomol Struct Dyn. 1987 Feb;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res. 1985 Sep 11;13(17):6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- Wemmer D. E., Chou S. H., Hare D. R., Reid B. R. Duplex-hairpin transitions in DNA: NMR studies on CGCGTATACGCG. Nucleic Acids Res. 1985 May 24;13(10):3755–3772. doi: 10.1093/nar/13.10.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]


