Abstract
Near-infrared spectroscopy (NIRS) signals have been shown to correlate with resting-state BOLD-fMRI data across the whole brain volume, particularly at frequencies below 0.1 Hz. While the physiological origins of this correlation remain unclear, its existence may have a practical application in minimizing the background physiological noise present in BOLD-fMRI recordings. We performed simultaneous, resting-state fMRI and 28-channel NIRS in seven adult subjects in order to assess the utility of NIRS signals in the regression of physiological noise from fMRI data. We calculated the variance of the residual error in a general linear model of the baseline fMRI signal, and the reduction of this variance achieved by including NIRS signals in the model. In addition, we introduced a sequence of simulated hemodynamic response functions (HRFs) into the resting-state fMRI data of each subject in order to quantify the effectiveness of NIRS signals in optimizing the recovery of that HRF. For comparison, these calculations were also performed using a pulse and respiration RETROICOR model. Our results show that the use of 10 or more NIRS channels can reduce variance in the residual error by as much as 36 % on average across the whole cortex. However the same number of low-pass filtered white noise regressors are shown to produce a reduction of 19 %. The RETROICOR model obtained a variance reduction of 6.4 %. Our HRF simulation showed that the mean squared error (MSE) between the recovered and true HRFs is reduced by 21 % on average when 10 NIRS channels are applied and by introducing an optimized time-lag between the NIRS and fMRI time series, a single NIRS channel can provide an average MSE reduction of 14 %. The RETROICOR model did not provide a significant change in MSE. By each of the metrics calculated, NIRS recording is shown to be of significant benefit to the regression of low-frequency physiological noise from fMRI data.
Keywords: Near infrared spectroscopy, Functional magnetic resonance imaging, Low frequency oscillations, resting state
Introduction
Near-infrared spectroscopy (NIRS) uses variations in the absorption of near-infrared light to infer changes in the concentration of oxyhemoglobin (HbO) and deoxyhemoglobin (HbR) in the cerebral cortex (Obrig and Villringer, 2003; Franceschini and Boas, 2004; Gibson et al., 2005). The most common applications of NIRS employ a back-reflection geometry, such that both the source and detector of the near-infrared light are positioned on the same surface (Villringer et al., 1993). This geometry is usually necessary to ensure that an adequate amount of light can reach both the brain and then the detector, but it renders NIRS measurements highly sensitive to background hemodynamic oscillations in superficial tissues. These oscillations, due to heart rate, respiration and a variety of low frequency effects, form the background physiological noise through which changes in cortical HbO and HbR must be identified (Prince et al., 2003; Diamond et al., 2006; Lina et al., 2008).
The blood oxygen level dependent (BOLD) signal of functional magnetic resonance imaging (fMRI) occurs because of variations in the local concentration of the paramagnetic deoxyhemoglobin molecule (Huettel et al., 2004). As it is a hemodynamic measurement, BOLD-fMRI is also highly sensitive to background physiological oscillations and despite the autoregulatory actions of the brain, the effects of heart rate, respiration rate and low-frequency hemodynamic oscillations are often present in fMRI data (Hu et al., 1995).
A number of simultaneous NIRS and fMRI experiments have been performed in the study of functional activation (Toronov et al., 2001; Strangman et al., 2002; Huppert et al., 2006; Steinbrink et al., 2006). These studies have sought to investigate the relationship between the functional NIRS and BOLD signals in common regions of the brain, and have shown that there is a good correlation between the NIRS-recorded functional HbR response and the local BOLD signal. Recent studies have begun to investigate the relationship between NIRS and fMRI-BOLD during the resting state, i.e. when no stimulation paradigm is applied. Preliminary work by our group (Greve et al., 2009) and studies by Tong et al. (Tong and Frederick, 2010; Tong et al., 2011) have described a significant correlation between NIRS signals and BOLD-fMRI data across the whole brain volume during the resting state. A NIRS signal, obtained using sources and detectors placed on the scalp, appears to not only correlate with the BOLD signal present in the cortical voxels to which it is directly sensitive, but also to the BOLD signal of a large number of voxels spread throughout the brain. These resting-state correlations are particularly apparent at frequencies below 0.1 Hz (Greve et al., 2009; Tong and Frederick, 2010).
Tong and Frederick (2010) presented compelling evidence that the correlation which exists between resting-state NIRS and BOLD-fMRI is dominated by systemic oscillations in blood pressure which traverse the cerebral vasculature, rather than being suggestive of resting-state neural connectivity (Biswal et al., 1995). Systemic hemodynamic oscillations at 0.1 Hz and below are common to NIRS and BOLD-fMRI recordings and are thought to have two main sources; the variations in local vascular dilation known as vaso-motion (Gustafsson, 1993) and oscillations in arterial blood pressure known as Mayer waves (Julien, 2006). Although it was not explicitly proven, Tong and Frederick (2010) suggested that the component of the NIRS signal which correlates with BOLD-fMRI across the brain is primarily that derived from superficial tissues (i.e. the scalp) and not from the brain. While it is beyond doubt that NIRS signals contain a significant contribution from the cerebral cortex (and that this contribution will correlate with the local fMRI voxels), this contribution is invariably smaller than that of the superficial tissues (Obrig and Villringer, 2003; Boas et al., 2004). The dominance of superficial hemodynamic oscillations is partly what necessitates the detailed signal analysis and averaging common to functional NIRS experiments.
Irrelevant of its origin, the existence of a correlation between NIRS and fMRI-BOLD in the absence of stimulation has an obvious potential application: the minimization of physiological noise in fMRI experiments. The BOLD hemodynamic response function (HRF) typically provides a peak signal change of between 1 and 5 % (Buxton et al., 1998; Friston et al., 2000), which can be significantly less than the level of physiological noise in the BOLD-fMRI signal. In order to maximize the efficiency with which the HRF can be recovered, fMRI experiments typically consist of carefully designed paradigms of stimulus repetition (Dale, 1999; Friston et al., 1999). The reliable identification of the physiological noise present in the BOLD signal allows for an increase in signal-to-noise ratio for a given experimental paradigm, or can allow the same signal-to-noise ratio to be achieved using fewer stimulations, minimizing the necessary duration of the fMRI scan. This goal is not insignificant, as fMRI experiments often require a subject to remain stationary in the scanner for periods of over an hour (Huettel et al., 2004). In addition to being expensive, this can cause serious discomfort and makes fMRI experiments less suitable for vulnerable subject groups.
The common approach to the minimization of fMRI physiological noise involves the distal measurement of hemodynamic oscillations and associated changes with a view to including these measurements in the linear model of the BOLD-fMRI signal. When applied in this way, such measurements are referred to as ‘nuisance regressors’. Measurements of heart rate and heart rate variability (Glover et al., 2000; Bianciardi et al., 2009; Chang et al., 2009) respiration rate and respiration rate variability (Birn et al., 2006, 2008) and end-tidal carbon dioxide (Wise et al., 2004; Chang and Glover, 2009) have all been investigated as nuisance regressors. The most commonly applied nuisance regression technique is known as RETROspective Image CORrection (RETROICOR) (Glover et al., 2000). This approach uses distal measurements of pulse rate (via a pulse oximeter) and respiration (via a pneumatic chest belt) to enable an image-based correction of respiration and pulsatile oscillations in the BOLD signal.
For this study, we acquired simultaneous, resting-state fMRI and 28-channel NIRS in seven healthy adults in order to thoroughly assess the utility of NIRS signals as fMRI nuisance regressors. We calculate the variance of the residual error in a general linear model of the baseline fMRI signal, and calculate the reduction in this variance that can be obtained by employing NIRS signals to regress out low frequency physiological noise. To provide a more rigorous assessment, we simulate an event-related fMRI paradigm by introducing a series of synthetic HRFs into the resting-state fMRI data of our seven subjects in order to explicitly quantify the utility of NIRS in the recovery of the HRF using a finite impulse response (FIR) model (Burock and Dale, 2000). In both cases, we compare each NIRS model with a model containing both pulse and respiration RETROICOR regressors. We also asses the utility of a single NIRS channel in groups of voxels with different NIRS-fMRI correlation in order to quantify the success of a given NIRS regressor in terms of the baseline correlation between it and the BOLD-fMRI volume. Finally, we attempt to optimize the recovery of the HRF using a single NIRS channel by introducing a voxel-specific time lag between the NIRS and BOLD-fMRI time series.
Methods
Subjects and data acquisition
Seven healthy adult subjects were recruited to this study, which was approved by the Institutional Review Board of Massachusetts General Hospital. Each subject provided written, informed consent and underwent a single scanning session consisting of three resting-state runs lasting 300 seconds.
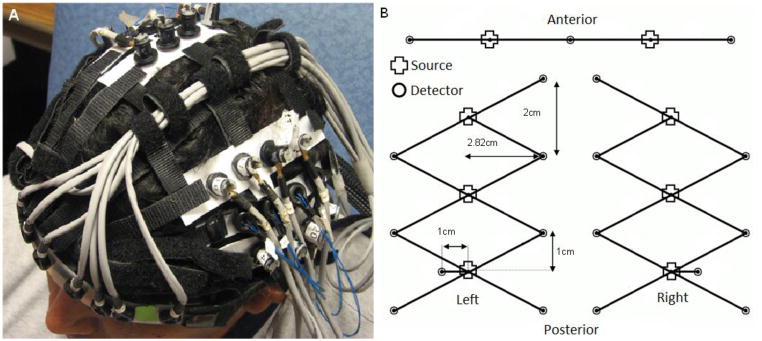
Near-infrared spectroscopy was performed using a bilaterally symmetric 28-channel probe and the TechEn CW6 continuous wave NIRS system operating at 690 and 830 nm, with a sample rate of 25 Hz (Franceschini et al., 2006). The NIRS probe covers a large proportion of the frontal and temporal lobes and is shown in position in Figure 1.
Figure 1.
The 28-channel NIRS array is shown in position on one subject in figure 1a. Figure 1b shows the schematic arrangement of the array.
All MRI data were acquired using a Siemens Tim Trio 3T system. Prior to functional imaging, an anatomical MRI was obtained using an MP-RAGE imaging sequence (TR = 2530 ms, TI = 1100 ms, TE = 3.44 ms). Functional MRI was performed using the Functional Biomedical Informatics Research Network protocol (fBIRN, www.birncommunity.org) (Greve et al., 2011) such that TR = 2 s, flip angle = 77°, TE = 30 ms, number of slices = 30, echo spacing = 500 ms, with posterior-anterior phase encoding and a bandwidth of 2290 Hz/pixel. The in-plane resolution was 3.43 mm with a slice thickness of 5 mm with a 1 mm skip and axial slices approximately aligned with the AC-PC line. Each functional run provided an fMRI time course of 150 data points. The scanner trigger pulse was acquired to allow the synchronization of the NIRS and fMRI recordings.
Cardiac and respiration waveforms were measured for all subjects using a pulse oximeter and either a pressure monitor or stress gauge attached to the chest with an elastic belt. Both acquisitions were sampled at 25Hz.
NIRS pre-processing
The changes in optical density recorded at each of the 28 channels were converted to changes in concentration of HbO and HbR using the modified Beer-Lambert relationship, with a pathlength correction factor of 6 and a partial volume correction factor of 50 (Boas et al., 2004; Huppert et al., 2006). A NIRS channel was excluded from subsequent processing if the baseline detected intensity of either wavelength was below or above the acceptable range of the system (40 to 70 decibels respectively) or if the channel was determined to be excessively noisy on visual inspection. Previous work has shown that NIRS-fMRI correlations are dominated by low frequency components (Greve et al., 2009; Tong and Frederick, 2010). As a result our HbO and HbR waveforms were low-pass filtered at 0.1 Hz (Butterworth, 5th order) before being synchronized to the fMRI trigger pulse and down-sampled to the fMRI sample rate of 0.5 Hz. If no channels were rejected, this process would produce 56 time series (28 for HbO and 28 for HbR), each with 150 data points.
fMRI pre-processing
All fMRI scans were motion corrected to the middle time point using the AFNI 3dvolreg program (Cox and Jesmanowicz, 1999). Non-brain voxels were masked out using the FSL Brain Extraction Tool (Smith, 2002). Anatomical images were analyzed using FreeSurfer (surfer.nmr.mgh.harvard.edu) (Dale et al., 1999; Fischl et al., 1999) to construct models of the cortical surface of each individual. Each fMRI time series was smoothed using a 5 mm full-width at half-maximum Gaussian in the 3D volume. The middle time point of each run was registered to the subject’s anatomical image using the FreeSurfer bbregister program (Greve and Fischl, 2009) which allowed the subject-specific cortical mask to be mapped into the functional image space. All reported results are from cortical gray matter.
RETROICOR processing
Pulsatile and respiration regressors were created using the RETrospective Image CORrection (RETROICOR) approach (Glover et al., 2000). Pulse and respiration waveforms were modeled using a fundamental and harmonic sine and cosine function to produce 4 regressors for pulse and 4 for respiration. These regressor waveforms were down-sampled to the fMRI acquisition rate on a slice-specific basis, so as to compensate for the finite delay between the acquisition of consecutive slices (Jones et al., 2008). The pulse oximetry data of one run in one subject was rejected due to a recording error. In this case only the 4 respiration RETROICOR regressors were used in further processing.
Linear models and Error Variance Reduction
To assess the utility of NIRS regression of physiological noise, we constructed a number of linear models of the resting-state fMRI signal. All regression analyses were performed using FreeSurfer (surfer.nmr.mgh.harvard.edu) and bespoke Matlab (The Mathworks Inc.) software functions. The first linear model consisted of 3 regressors corresponding to 0th, 1st and 2nd order temporal drift polynomials and a further 6 regressors constructed from motion correction parameters. This ‘reference’ model is used as a comparison throughout, and forms the basis of all other models.
A slice-specific RETROICOR model was computed by augmenting the reference model with all 4 pulse and all 4 respiration RETROICOR waveforms for each slice as defined above. The regression was performed on a slice-specific basis and the results averaged across all cortical voxels.
The NIRS models were computed by adding one or more of the down-sampled NIRS signals to the reference model. The NIRS models are separated into ‘HbO models’ (those containing one or more HbO regressors), ‘HbR models’ (those containing one or more HbR regressors) and ‘HbO & HbR models’(those containing both the HbO and HbR regressors from one or more NIRS channels). To provide a simple metric of the efficacy of NIRS modelling of resting-state fMRI time series, we computed the variance of the residual error of each model and compared it to that of the reference model. The larger the reduction in variance the better the model of the fMRI time series (a percentage variance reduction (PVR) of 100 %, indicates that the model is able to remove all the variance in the fMRI time series unaccounted for by the reference model). The PVR achieved by the RETROICOR model was also calculated. A general linear model (GLM) of the fMRI time series (Y ), with design matrix ( X ) has a residual error (ε) given by:
| (1) |
where β is the vector of parameter estimates calculated via:
| (2) |
The residual variance is given by:
| (3) |
where d is an estimate of model’s degrees of freedom and is a function of the number of regressors included in the model. As the model is altered (for example by adding regressors to the design matrix, X), the residual variance (var(ε)) will change depending on how well the model accounts for data. Note that the variance does not necessarily decrease with the addition of regressors to the linear model.
HRF Simulation
To further quantify the utility of NIRS regressors in the removal of physiological noise, and therefore the improved recovery of a functional response, we introduced a series of simulated HRFs into the resting state fMRI time series of each data set. We produced a simple HRF using a single gamma function (Figure 7) with a peak latency of 6 seconds, down-sampled to match TR and scaled to provide a 2 % peak increase in the BOLD signal of each relevant voxel. A ‘stimulus paradigm’ of ten HRFs was introduced into each 300-second run in an event-related fashion. To increase the reliability of our results, 30 different stimulus paradigms were used in which each inter-HRF-interval was a randomly selected period between 2 and 24 seconds. For each stimulus paradigm, subject, run and linear model, the HRF was estimated using a finite impulse response approach. For comparison, this process was also performed for the RETROICOR model. The HRF recovered by each model was compared to the true HRF time series using two metrics: the mean-squared error (MSE) and Pearson’s correlation coefficient (R2). Within the context of the GLM, the MSE is given by:
| (4) |
where the efficiency of the model (ξ) is:
| (5) |
and C is the contrast matrix pertaining to the HRF (Dale, 1999). The value of the MSE achieved by a given model is thus dependent on both the residual variance and the efficiency of that model.
As the correlation between a given NIRS channel and the fMRI signal will vary significantly across the fMRI volume (Tong and Frederick, 2010), the selection of specific regions of interest in which to apply the simulated functional response so as to best test the use of NIRS signals as nuisance regressors is not trivial. To estimate the average effect of NIRS regression, the simulated HRFs were first added to (and recovered from) the BOLD time series of every cortical voxel, without any consideration for the correlation between the NIRS signals and the fMRI volume.
Selected voxels and optimized time-lag
This simulation was repeated for models containing only a single NIRS regressor in groups of 100 voxels selected on the basis of the value of the correlation between the fMRI time series and each NIRS regressor. This process was performed in order to quantify the success of a given NIRS regressor with respect to the value of the baseline correlation between it and the BOLD-fMRI volume. The baseline correlation is denoted to distinguish it from the correlation between synthetic and recovered HRFs (R2). We calculated using the fMRI time series prior to the introduction of the simulated HRF in order to minimize computational expense. Three groups of 100 voxels, with in the range 0–0.1, 0.1–0.2 and 0.2–0.3, were selected for each subject, run and HbO or HbR signal.
The results of Tong and Frederick (2010) suggest that the NIRS-fMRI correlation traverses the cerebral vasculature in a wave-like pattern. In an attempt to take advantage of this phenomenon, the HRF simulation was also performed across the whole cortex using models containing a single NIRS regressor, but with a voxel-specific time-lag introduced between that NIRS regressor and the fMRI time series. Because the NIRS recording time for each run exceeded that of the fMRI (NIRS recording began ~ 30 seconds prior to the fMRI sequence and continued for ~ 30 seconds after) we were able to shift the NIRS signals in time relative to the fMRI recording. We produced 20 different time lags, from − 10 to + 10 seconds in 1 second intervals, relative to the fMRI time series for every HbO and HbR signal. For every cortical voxel, HbO signal, HbR signal and for every run, we were able to compute the time lag which optimized the baseline correlation ( ). A voxel and NIRS-signal specific linear model was then used to recover the simulated HRF at each voxel using this optimum time lag, and the results were averaged across the cortex.
Synthetic NIRS Signals
Near-infrared spectroscopy signals will always contain some noise contribution from non-physiological sources. When the NIRS signals are low-pass filtered, this noise is also low-pass filtered. When such waveforms are used in the general linear model, it is possible that they could provide a good model of resting state fMRI oscillations simply because the constituent noise spans the frequency space below 0.1 Hz. The use of NIRS regressors could therefore produce a significant reduction in fMRI noise that is not related to the physiological measurements of HbO and HbR. To account for this possibility, our calculations of variance reduction and our HRF simulations were all repeated using synthetic NIRS data. These waveforms were obtained by producing two random vectors and passing them through the entire processing stream, including the Beer-Lambert transformation and low-pass filtering. This is effectively equivalent to repeating all our processing under the null hypothesis that there is no relationship between HbO, HbR and the BOLD signal.
Results
The average proportion of the cortex found to be significantly correlated with a given NIRS regressor (using a two-tailed, voxel-wise t-test, p < 0.01) was 17.8 % for HbR signals and 16.8 % for HbO signals, with subject means ranging from 5–22 %. Subject 6 showed very little correlation despite there being no obvious issues with either the NIRS or fMRI recordings. Figure 2 shows an example of a significance map for a single, well-correlated HbO regressor. The average proportion of the cortex significantly correlated with the synthetic NIRS signals was 3.7 %.
Figure 2.
An example of the significance map for a single subject, run and NIRS-HbO regressor. Note that significantly correlated voxels are spread throughout the cortex as well as in deeper regions of the brain.
Whole cortex variance reduction
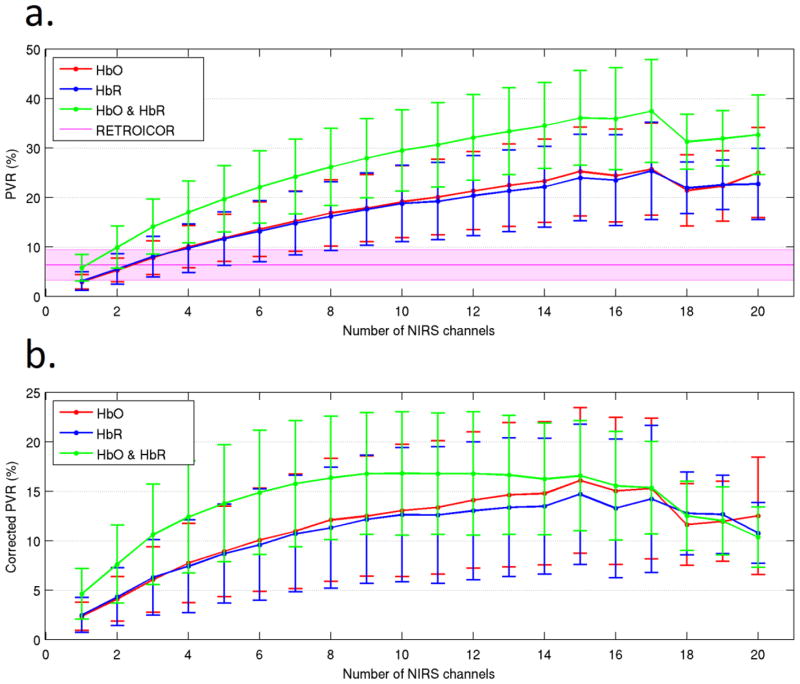
The percentage variance reduction (PVR) was computed for the HbO, HbR, HbO & HbR, RETROICOR and synthetic NIRS models. The results of this process are shown in Figure 3a as a function of the number of NIRS channels (n) in each linear model.
Figure 3.
The percentage variance reduction obtained by the HbO, HbR and HbO & HbR models as a function of the number of NIRS channels incorporated into each model. Values of PVR are obtained in comparison to the reference model. The corresponding results for the pulse and respiration RETROICOR model are also included. Figure 3b shows the corrected PVR values, which were obtained by subtracting the PVR achieved using synthetic NIRS regressors from that of the real NIRS regressors. These corrected results represent the extent to which the variance of the BOLD-fMRI signal can be modeled by the physiological content of the NIRS data. In each figure the error bars (and shaded region for the RETROICOR model) represent the standard deviation across all runs.
Each data point in Figure 3a corresponds to the mean PVR achieved over the whole cortex and averaged over all runs and either every possible combination of n channels or 28 random combinations of n channels, whichever is smaller. For example, the HbO data point corresponding to 1 NIRS channel is the average PVR obtained across every possible model containing a single HbO regressor (if no channels were excluded, there would be 28 possible models), for each subject and run. For the HbO data point corresponding to 2 NIRS channels, 28 pairs of non-excluded HbO regressors were randomly selected, and the average PVR achieved by those pairs of regressors is presented. The upper limit on the number of random combinations was set at 28, as it was computationally prohibitive to use every possible model in most cases (for example, 28 choose 2 = 378 possible models). This process was performed in order to minimize the sensitivity of the results to channel selection. This same process was applied to obtain the HbR model PVR results in Figure 3. For the HbO & HbR models, 1 NIRS channel corresponds to 2 regressors (the HbO and HbR regressors corresponding to each single NIRS channel), 2 NIRS channels corresponds to 4 regressors and so on. This randomized selection approach was also applied for the whole-cortex HRF simulation results below.
As shown in Figure 3a, the PVR increases with the addition of NIRS regressors for all models until 15 NIRS channels are applied, at which point the PVR is equal to 36 % for the HbO & HbR model. The PVR is significantly larger than zero for all models (one-tail t-test, p < 0.01). The addition of the slice-specific respiration and pulse RETROICOR regressors to the reference model also produces a significant reduction in variance, averaging 6.4 %. The PVR achieved by the HbO & HbR model becomes significantly larger than that of the RETROICOR model when 2 NIRS channels are applied (one-tail, paired t-test, p < 0.01). It is worth noting that the use of 8 synthetic NIRS regressors (the same number of regressors used in the RETROICOR model) produces an average PVR of 4.7 %.
Figure 3b shows the corrected PVR, achieved by subtraction of the variance reduction calculated using the synthetic NIRS waveforms. This provides a lower-bound estimate of the average PVR attributable to each NIRS model. A peak value of 17.0 % is achieved for the HbO & HbR model incorporating 9 NIRS channels. The PVR remains significantly larger than zero for all models after subtraction of the synthetic NIRS results (one-tail t-test, p < 0.01).
Whole cortex HRF simulation
Figure 4a shows the average mean squared error (MSE) between the synthetic and recovered HRFs for HbO, HbR, HbO & HbR, RETROICOR and the synthetic HbO & HbR models (synthetic HbO and synthetic HbR models were also calculated but for simplicity are not displayed). These data are normalized to the value obtained using the reference model (i.e. that without NIRS regressors) and are shown as a function of the number of NIRS channels applied (n). For each NIRS model, either all possible combinations, or 28 combinations of n NIRS channels were randomly selected and the results provided are an average of all combinations, as described above for figure 3. The MSE decreases with the addition of NIRS regressors for HbO, HbR and HbO & HbR models. This decrease reaches significance (one-tail paired t-test, p < 0.01) with the use of a single NIRS channel for the HbO & HbR model and with the use of 2 NIRS channels for both the HbO and HbR models. The minimum, equal to reduction of 21 %, is found for the HbO & HbR model when 10 NIRS channels are applied.
Figure 4.
The result of the HRF simulation applied across all cortical voxels for HbO, HbR, HbO & HbR and synthetic NIRS models as a function of the number of NIRS channels applied. The corresponding results for the pulse and respiration RETROICOR model are also included. Figure 4a shows the mean-squared error (MSE) between the synthetic and recovered HRFs, normalized to the value obtained by the reference model. For clarity, the normalized MSE for up to ten NIRS channels are shown on a larger scale on the inserted axes. Figure 4b shows Pearson’s correlation coefficient (R2) between the synthetic and recovered HRFs. The mean changes in MSE and R2 for the RETROICOR model compared to the reference model are negligibly. In each figure the error bars (and shaded region for the RETROICOR model) represent the standard deviation across all runs.
The slice-specific RETROICOR results are highly variable but on average the RETROICOR model fails to reduce the MSE between the synthetic and recovered HRFs, and results in an average increase in MSE of 0.1 % (standard deviation of 14 %) compared to the reference model.
Figure 4b shows the corresponding results for the correlation coefficient (R2) between the synthetic and recovered HRFs. Again, the data are normalized to the reference model. The value of R2 between the synthetic and recovered HRFs follows a similar pattern to that observed in the MSE results; an improvement is observed with the number of NIRS channels applied for all models up to 10 NIRS channels, at which point the HbO & HbR model produced an increase in R2 of 2.0 %. This increase in R2 above the reference model achieves significance for all NIRS models when a single NIRS channel is applied (one-tail paired t-test, p < 0.01). The RETROICOR model also fails, on average, to produce an improvement in R2, producing a mean decrease of 0.04% (standard deviation 0.4 %).
The equivalent results calculated using the synthetic NIRS data show an approximately exponential increase in MSE and decrease in R2 as synthetic regressors are added. This detrimental effect reaches significance for both MSE and R2 metrics for all synthetic NIRS models using a single NIRS channel.
Selected voxels and optimized time-lag
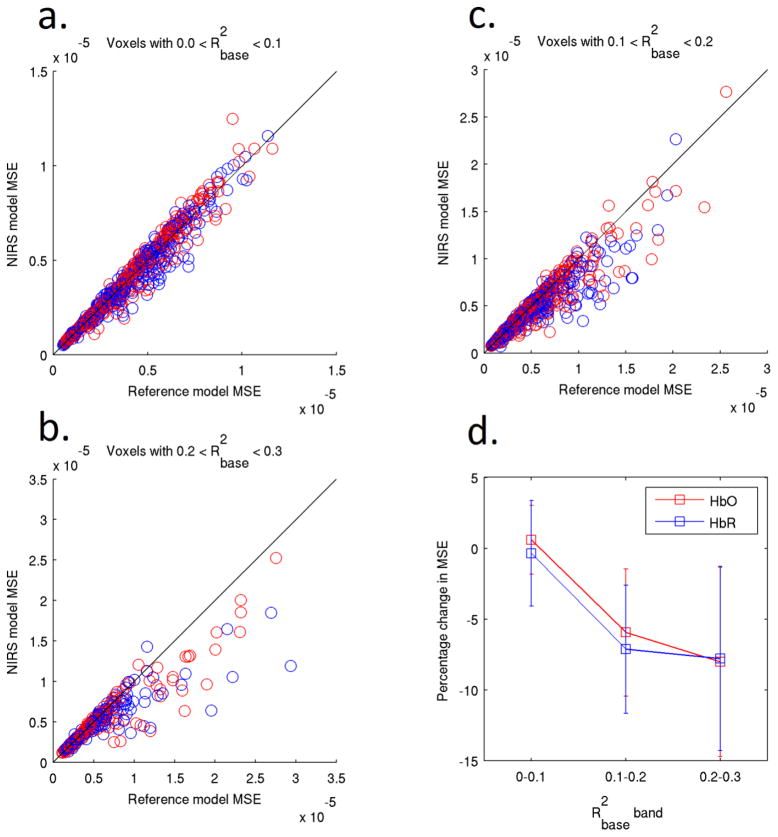
Figures 5a–5c show scatter plots of the MSE for single-regressor HbO and HbR models plotted against the corresponding MSE of the reference model. Data points on the 45° line correspond to instances where the NIRS regressor model has had no effect on the MSE. These simulations were performed in 100 voxels specific to each subject, run and HbO or HbR regressor such that lies between 0–0.1, 0.1–0.2 and 0.2–0.3 for figures 5a–5c respectively. This process was not repeated for the synthetic NIRS signals because their correlation with the fMRI data was insufficient.
Figure 5.
The single-regressor NIRS model MSE plotted against the reference model MSE for 100 voxels selected using the value of the baseline NIRS-fMRI correlation for each voxel. The data is separated into three bands on the basis of . The average reduction in MSE for all data points is shown in figure 5d. The MSE between the synthetic and recovered HRFs decreases as a function of . The error bars in figure 5d show the standard deviation across all data points of figures 5a–5c.
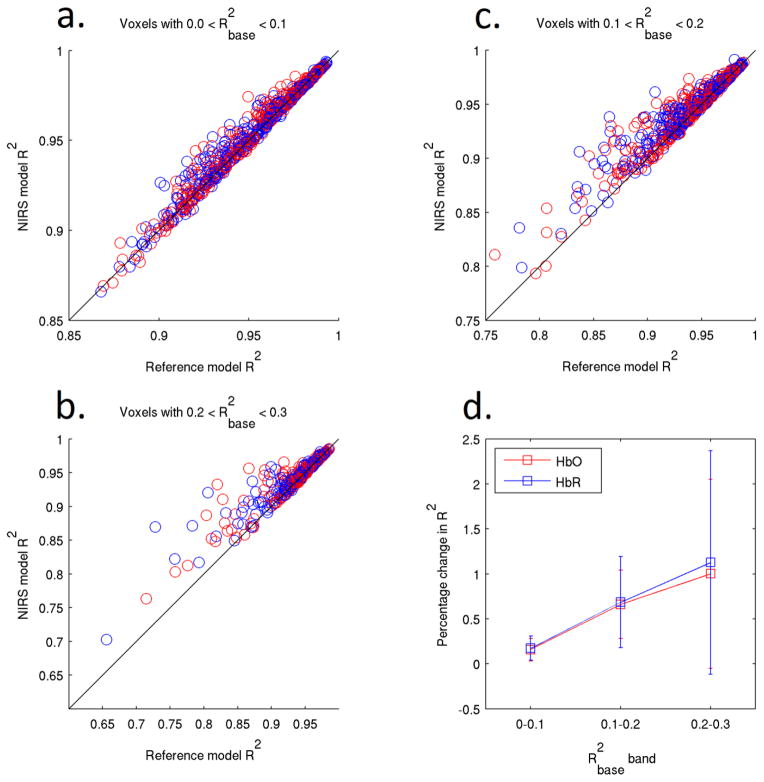
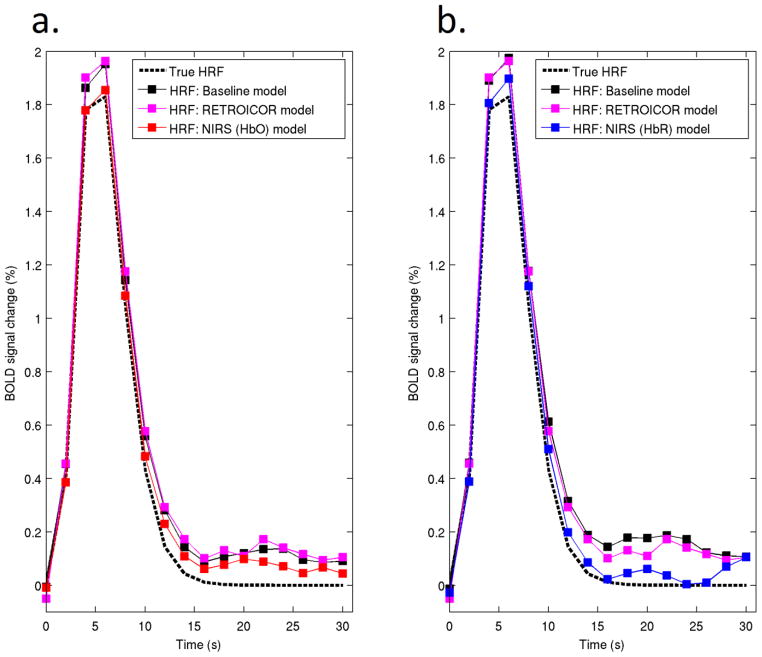
For voxels in the 0–0.1 baseline correlation band (Figure 5a), NIRS regression provides no significant change in MSE for either the HbO or HbR models when compared to the baseline model (two-tailed t-test, p = 0.53). Both the 0.1–0.2 and 0.2–0.3 baseline correlation bands (Figures 5b and 5c) show a significant decrease in MSE for both HbO and HbR models (p < 0.01). Figure 5d shows the mean percentage change in MSE for each of these three bands. Figures 6a–6d are equivalent to Figures 5a–5d but present the value of the correlation coefficient (R2) between the synthetic and recovered HRF instead of the MSE. Figure 6d shows that the mean value of R2 increases as a function of the baseline correlation . Figure 7 provides one example of the effect of HbO (Fig. 7a) and HbR (Fig. 7b) regressors in well-correlated voxels ( ), for a single run and subject.
Figure 6.
The single-regressor NIRS model R2 plotted against the reference model R2 for 100 voxels selected using the value of the baseline NIRS-fMRI correlation for each voxel. The data is separated into three bands on the basis of . The average increase in R2 for all data points is shown in figure 6d. The value of R2 between the synthetic and recovered HRFs increases as a function of . The error bars in figure 6d show the standard deviation across all data points of figures 6a–6c.
Figure 7.
An example of the HRFs recovered using the reference model, pulse and respiration RETROICOR model and by single-regressor HbO (figure 7a) and HbR (figure 7b) models for a single subject, run and NIRS channel for voxels well correlated each NIRS regressor ( ). The true, single-gamma HRF is also shown in both figures.
Figure 8 shows the results of the HRF simulation where the optimum time lag has been introduced between each NIRS HbO or HbR signal and the fMRI time series of each voxel. Figures 8a and 8b respectively show the MSE and R2 between the synthetic and recovered HRFs, normalized to the values obtained by the reference model. In each case the result of the comparable zero-time lag models (equivalent to the first data points of figures 4a and 4b) are provided for comparison. The use of the optimum time lag causes a significant reduction in MSE (one-tailed paired t-test, p < 0.01) for all models compared to the zero-time lag equivalent, including the synthetic NIRS model. The reduction in MSE obtained for the optimum time-lag HbO & HbR model is 13.6 % compared to 6.3 % for zero time lag. Correspondingly, there is an increase in R2 when the optimum time lag regressors are applied, but this does not quite reach significance for the HbO (p = 0.011), HbR (p = 0.014) or HbO & HbR (p = 0.10) models. The improvement is very significant for the synthetic NIRS data (p < 10−6). For both metrics, the use of optimum time lag produces a greater improvement for HbO regressors than for HbR regressors.
Figure 8.
The values of the MSE and R2 between the synthetic and recovered HRFs for the zero time-lag and optimum time-lag, single-channel HbO, HbR, HbO & HbR and synthetic NIRS models, normalized to the equivalent values of the reference model. A significant (two tailed paired t-test, p < 0.01) improvement is apparent for both metrics and all models, though the improvement is more distinct for HbO models than for HbR models. In each case the error bars represent the standard deviation across all runs.
Discussion
Our results confirm that low-frequency oscillations present in near-infrared spectroscopy data correlate significantly with a large proportion of BOLD-fMRI voxels, spread throughout the cortex. At frequencies below 0.1 Hz, NIRS signals are subject to a range of physiological oscillations. These include the spontaneous changes in vascular dilation referred to as vaso-motion (Gustafsson, 1993) and the global oscillations in arterial blood pressure known as Mayer waves (Julien, 2006). As the effects of vaso-motion tend to be location-specific, the systemic oscillations caused by Mayer waves affecting both the scalp-dominated resting-state NIRS signal and BOLD-fMRI voxels across the brain seem the most likely source of this correlation.
Whole-cortex PVR and HRF simulation
The utility of NIRS signals in the regression of physiological noise can be established in a variety of ways. A simple assessment of the efficacy of a given linear model is the variance of that model’s residual error. Our results indicate that the inclusion of NIRS signals in the model can reduce this variance by as much as 36 %. To achieve this, a large number of NIRS channels (> 15) are required. NIRS regression produces an average PVR of 6.0 % for a single NIRS channel, comparable to that achieved by the slice-specific RETROICOR approach (6.4 %), (Figure 3a).
As is clear from Figure 3, a significant proportion of the variance reduction achieved by NIRS regression is not due to the physiological information present in the NIRS data, but can equally be achieved by the regression of random vectors occupying the same frequency space. However, even when this effect is taken into account, an average PVR of 17 % can still be achieved by the HbO & HbR model (Figure 3b). These corrected PVR results suggest that applying 10 NIRS channels provides the best model of physiological noise in the BOLD signal. The value of 17 % represents a lower-bound estimate on the average PVR which can be obtained by a model containing 10 or more NIRS channels.
The results of our explicit simulation of an event-related BOLD-fMRI experiment confirm that NIRS can be used to improve the recovery of the HRF time course. There is a significant improvement in both the mean-squared error (MSE) and the correlation coefficient (R2) between the synthetic and recovered HRFs when NIRS regressors are applied. Note that both the HbO and HbR models provide an improvement in MSE and R2 and that the HbO & HbR model is consistently better than either single-chromophore model. This may suggest that the HbO and HbR signals provide independent contributions to the model of resting-state fMRI fluctuations, but could also be the result of spatial averaging if the HbO and HbR signals are correlated with different regions of the cortex.
Figure 4 shows that recovery of the HRF improves even if regressors are applied in a ‘blind’ fashion; across the whole cortex and without consideration for the baseline NIRS-fMRI correlation. The minimum MSE and maximum R2 are achieved across the whole cortex when approximately 10 NIRS channels are included in the linear model. The MSE does not monotonically decrease (as the PVR increases in Figure 3a) because the MSE is dependent on both variance and efficiency. Although adding NIRS regressors decreases the residual variance, it can also decrease the model’s efficiency. As the number of NIRS channels increases beyond 10, the decrease in efficiency overcomes the decrease in residual variance, and the MSE begins to increase.
As stated previously, we believe that the majority of the low-frequency contribution to the resting state NIRS signal (and therefore to the NIRS-fMRI correlation) originates in the superficial layers of the head. As a result, the acquisition of NIRS signals suitable for regression could be performed using channels which only sample superficial tissues. Because of the improved signal-to-noise ratio associated with short separation channels, superficial NIRS acquisition is often significantly easier than standard NIRS.
Obtaining multi-channel, superficial NIRS during fMRI experiments is therefore entirely feasible, and may be particularly useful in experiments which require precise recovery of the HRF time course and where the number of stimuli or the scanning duration are limited. Examples of such cases include the study of the hemodynamic response to inter-ictal epileptic activity with fMRI (Bénar et al., 2002) or the computation of the cerebral metabolic rate of oxygen from combined NIRS-fMRI (Huppert et al., 2009). It is important to note that the HRF simulation data presented here are the average results obtained from 30 pseudo-random event-related paradigm designs. It was necessary to use a large number of different designs in order to produce a robust result, which suggests that the efficacy of NIRS regression may be sensitive to experimental design. An investigation of the utility of NIRS in the regression of fMRI noise for different experimental paradigms (including block designs) will therefore be necessary to establish which fMRI experiments will benefit from simultaneous NIRS recording.
Selected voxels and optimized time lag
The results summarized in Figures 5 and 6 provide a measure of the importance of the baseline correlation ( ) to the success of each NIRS model. Although it is difficult to obtain the statistical power required to provide exact quantification of this relationship (due to variation in BOLD noise in different groups of voxels for example), the use of a single NIRS regressor in voxels where will, on average, produce a significant improvement in HRF recovery.
It is also important to note that even in cases of low baseline correlation (Figure 5a and 6a), the use of a single NIRS regressor does not, on average, worsen the recovery of the HRF. This is consistent with Figure 4, which shows that a significant improvement is observed even when NIRS regression is applied across the whole cortex, which must include regions and channels which exhibit a poor NIRS-fMRI correlation. This does not appear to be the case for the synthetic NIRS signals, as the whole-cortex simulation shows that HRF recovery worsens significantly with the use of a single synthetic regressor.
The results of employing the optimum time lag between each NIRS and fMRI time series show a significant decrease in MSE and a modest increase in R2 for HbO, HbR and HbO & HbR models. A comparable improvement in HRF recovery is seen for the synthetic NIRS regressors (Figure 8). An improvement in HRF recovery is clearly expected because by selecting the optimum time-lag we are maximizing the value of the baseline correlation between each NIRS signal and the fMRI data. The fact that the synthetic NIRS regressors (even when optimized for time lag) do not provide the reduction in MSE achieved by the real NIRS signals is further evidence that low-frequency physiological oscillations are common to both fMRI and NIRS recordings. These results are also consistent with the hypothesis that the NIRS-fMRI correlation follows a low-frequency wave pattern throughout the cerebral vasculature, but the improvement this affords to HRF recovery is relatively small; comparable to the addition of a second NIRS channel.
The results of our simulations in pre-selected voxels, and the results of our time-lag optimization are both reliant on a calculation of the baseline correlation ( ) between each NIRS signal and the fMRI data prior to the introduction of the simulated HRF series. The improvement in the recovery of the HRF associated with a higher baseline correlation is therefore to be expected. However, the introduction of the HRF series into the fMRI data produced a negligible change in ; 0.0058 for HbO and 0.0059 for HbR signals. It is therefore reasonable to suggest that NIRS regression could be optimized in a real fMRI study by maximizing , as calculated using the fMRI data during the stimulation period, with little risk of removing the HRF itself. This would obviously require that the NIRS channels are arranged so as not to sample the activated regions of the cortex. Alternatively, could be calculated for a period of NIRS-fMRI data where no stimulation occurs (i.e. during a rest period), but this assumes that the scale and spatial distribution of the NIRS-fMRI correlation will remain constant in time, the veracity of which has yet to be determined.
Conclusions
We have demonstrated that NIRS recordings can be employed to reduce the variance of the residual error in a general linear model of the resting-state BOLD-fMRI. Our results show that the use of a single NIRS channel will reduce variance in the residual error by an average of 6.0 %, comparable to that achieved by RETROICOR methods. The use of 10 NIRS channels can reduce variance by as much as 36 % on average across the whole cortex, though the same number of low-pass filtered white noise regressors can achieve a PVR of 19 %. We have also explicitly shown that the use of even a single NIRS channel can significantly improve the recovery of a simulated hemodynamic response function from BOLD-fMRI data, compared to standard fMRI and RETROICOR approaches. The MSE between the recovered and synthetic HRFs can be reduced by as much as 21 % when 10 NIRS channels are applied. While the exact physiological origins and the inter-subject (and intra-subject) variability of NIRS-fMRI correlations must be examined further, it is clear that they have an important application in the enhancement of fMRI experiments.
Acknowledgments
This work was supported by NIH grants P41-RR14075 and R01- EB006385. L Gagnon is supported by the Fond Quebecois sur la Nature et les Technologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bénar CG, Gross DW, Wang Y, Petre V, Pike B, Dubeau F, Gotman J. The BOLD Response to Interictal Epileptiform Discharges. NeuroImage. 2002;17:1182–1192. doi: 10.1006/nimg.2002.1164. [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Duyn JH. Modulation of spontaneous fMRI activity in human visual cortex by behavioral state. NeuroImage. 2009;45:160–168. doi: 10.1016/j.neuroimage.2008.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: The temporal dynamics of fMRI signal fluctuations related to changes in respiration. NeuroImage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Boas DA, Dale AM, Franceschini MA. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. NeuroImage. 2004;23:S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Burock MA, Dale AM. Estimation and detection of event-related fMRI signals with temporally correlated noise: a statistically efficient and unbiased approach. Hum Brain Mapp. 2000;11:249–260. doi: 10.1002/1097-0193(200012)11:4<249::AID-HBM20>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magnetic Resonance in Medicine. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: The cardiac response function. NeuroImage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. NeuroImage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn Reson Med. 1999;42:1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Dale AM. Optimal experimental design for event-related fMRI. Hum Brain Mapp. 1999;8:109–114. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HBM7>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis: I. Segmentation and Surface Reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Diamond SG, Huppert TJ, Kolehmainen V, Franceschini MA, Kaipio JP, Arridge SR, Boas DA. Dynamic physiological modeling for functional diffuse optical tomography. Neuroimage. 2006;30:88–101. doi: 10.1016/j.neuroimage.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical Surface-Based Analysis: II: Inflation, Flattening, and a Surface-Based Coordinate System. NeuroImage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Franceschini MA, Boas DA. Noninvasive measurement of neuronal activity with near-infrared optical imaging. NeuroImage. 2004;21:372–386. doi: 10.1016/j.neuroimage.2003.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini MA, Joseph DK, Huppert TJ, Diamond SG, Boas DA. Diffuse optical imaging of the whole head. J Biomed Opt. 2006;11:054007. doi: 10.1117/1.2363365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fMRI: the Balloon model, Volterra kernels, and other hemodynamics. Neuroimage. 2000;12:466–77. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Zarahn E, Josephs O, Henson RNA, Dale AM. Stochastic Designs in Event-Related fMRI. NeuroImage. 1999;10:607–619. doi: 10.1006/nimg.1999.0498. [DOI] [PubMed] [Google Scholar]
- Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Phys Med Biol. 2005;50:R1–43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magnetic Resonance in Medicine. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Goldenholz D, Kaskhedikar G, Polimeni JR. BOLD Physiological Noise Reduction using Spatio-Spectral-Temporal Correlations with NIRS ISMRM, Hawaii. Vol. 2009. ISMRM; Hawaii: 2009. [Google Scholar]
- Greve DN, Mueller BA, Liu T, Turner JA, Voyvodic J, Yetter E, Diaz M, McCarthy G, Wallace S, Roach BJ, Ford JM, Mathalon DH, Calhoun VD, Wible CG, Brown GG, Potkin SG, Glover G. A novel method for quantifying scanner instability in fMRI. Magnetic Resonance in Medicine. 2011;65:1053–1061. doi: 10.1002/mrm.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson H. Vasomotion and underlying mechanisms in small arteries. An in vitro study of rat blood vessels. Acta Physiol Scand Suppl. 1993;614:1–44. [PubMed] [Google Scholar]
- Hu X, Le TH, Parrish T, Erhard P. Retrospective estimation and correction of physiological fluctuation in functional MRI. Magnetic Resonance in Medicine. 1995;34:201–212. doi: 10.1002/mrm.1910340211. [DOI] [PubMed] [Google Scholar]
- Huettel S, Song A, Mccarthy G. Functional Magnetic Resonance Imaging. Sinauer Associates; 2004. [Google Scholar]
- Huppert TJ, Allen MS, Diamond SG, Boas DA. Estimating cerebral oxygen metabolism from fMRI with a dynamic multicompartment Windkessel model. Human Brain Mapping. 2009;30:1548–1567. doi: 10.1002/hbm.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ, Hoge RD, Diamond SG, Franceschini MA, Boas DA. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. NeuroImage. 2006;29:368–382. doi: 10.1016/j.neuroimage.2005.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TB, Bandettini PA, Birn RM. Integration of motion correction and physiological noise regression in fMRI. NeuroImage. 2008;42:582–590. doi: 10.1016/j.neuroimage.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien C. The enigma of Mayer waves: Facts and models. Cardiovascular Research. 2006;70:12–21. doi: 10.1016/j.cardiores.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Lina JM, Dehaes M, Matteau-Pelletier C, Lesage F. Complex wavelets applied to diffuse optical spectroscopy for brain activity detection. Opt Express. 2008;16:1029–1050. doi: 10.1364/oe.16.001029. [DOI] [PubMed] [Google Scholar]
- Obrig H, Villringer A. Beyond the Visible - Imaging the Human Brain With Light. J Cereb Blood Flow Metab. 2003;23:1–18. doi: 10.1097/01.WCB.0000043472.45775.29. [DOI] [PubMed] [Google Scholar]
- Prince S, Kolehmainen V, Kaipio JP, Franceschini MA, Boas D, Arridge SR. Time-series estimation of biological factors in optical diffusion tomography. Phys Med Biol. 2003;48:1491–1504. doi: 10.1088/0031-9155/48/11/301. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrink J, Villringer A, Kempf F, Haux D, Boden S, Obrig H. Illuminating the BOLD signal: combined fMRI-fNIRS studies. Magnetic Resonance Imaging. 2006;24:495–505. doi: 10.1016/j.mri.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Strangman G, Culver JP, Thompson JH, Boas DA. A Quantitative Comparison of Simultaneous BOLD fMRI and NIRS Recordings during Functional Brain Activation. NeuroImage. 2002;17:719–731. [PubMed] [Google Scholar]
- Tong Y, Bergethon PR, Frederick BdeB. An improved method for mapping cerebrovascular reserve using concurrent fMRI and near-infrared spectroscopy with Regressor Interpolation at Progressive Time Delays (RIPTiDe) NeuroImage. 2011;56:2047–2057. doi: 10.1016/j.neuroimage.2011.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Frederick BD. Time lag dependent multimodal processing of concurrent fMRI and near-infrared spectroscopy (NIRS) data suggests a global circulatory origin for low-frequency oscillation signals in human brain. Neuroimage. 2010;53:553–564. doi: 10.1016/j.neuroimage.2010.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toronov V, Webb A, Choi JH, Wolf M, Michalos A, Gratton E, Hueber D. Investigation of human brain hemodynamics by simultaneous near-infrared spectroscopy and functional magnetic resonance imaging. Med Phys. 2001;28:521. doi: 10.1118/1.1354627. [DOI] [PubMed] [Google Scholar]
- Villringer A, Planck J, Hock C, Schleinkofer L, Dirnagl U. Near infrared spectroscopy (NIRS): A new tool to study hemodynamic changes during activation of brain function in human adults. Neuroscience Letters. 1993;154:101–104. doi: 10.1016/0304-3940(93)90181-j. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]