Abstract
Background
Wiskott-Aldrich Syndrome (WASP) family proteins participate in many cellular processes involving rearrangements of the actin cytoskeleton. To the date, four WASP subfamily members have been described in Drosophila: Wash, WASp, SCAR, and Whamy. Wash, WASp, and SCAR are essential during early Drosophila development where they function in orchestrating cytoplasmic events including membrane-cytoskeleton interactions. A mutant for Whamy has not yet been reported.
Results
We generated monoclonal antibodies that are specific to Drosophila Wash, WASp, SCAR, and Whamy, and use these to describe their spatial and temporal localization patterns. Consistent with the importance of WASP family proteins in flies, we find that Wash, WASp, SCAR, and Whamy are dynamically expressed throughout oogenesis and embryogenesis. For example, we find that Wash accumulates at the oocyte cortex. WASp is highly expressed in the PNS, while SCAR is the most abundantly expressed in the CNS. Whamy exhibits an asymmetric subcellular localization that overlaps with mitochondria and is highly expressed in muscle.
Conclusion
All four WASP family members show specific expression patterns, some of which reflect their previously known roles and others revealing new potential functions. The monoclonal antibodies developed offer valuable new tools to investigate how WASP family proteins regulate actin cytoskeleton dynamics.
Keywords: Wiskott-Aldrich Syndrome, Wash, WASp, SCAR, Whamy, Drosophila, actin nucleation, cytoskeleton, oogenesis, embryogenesis, pole cells, CNS, cellularization, morphogenesis, gastrulation, monoclonal antibody, mitochondria tubulation, golgi
INTRODUCTION
WASP family members participate in cytoskeleton reorganization and signal transduction by acting as effectors of Rho family GTPases and polymerizing actin through the Arp2/3 complex (reviewed in: Millard et al., 2004; Takenawa and Suetsugu, 2007; Campellone and Welch, 2010; Rottner et al., 2010; Firat-Karalar and Welch, 2011). WASP family members have been implicated in the formation of lamellipodia and filopodia which are important for processes including morphogenesis, membrane/vesicle trafficking, angiogenesis, the inflammatory immune response, pathogen infection, and cancer metastasis (Lommel et al., 2001; Snapper et al., 2001; Suetsugu and Takenawa, 2003; Yamazaki et al., 2003; Yan et al., 2003; Yamazaki et al., 2005; Yamaguchi and Condeelis, 2007; Derivery et al., 2009; Duleh and Welch, 2010; Hanisch et al., 2010; Romer et al., 2010). All WASP family members have a conserved C-terminal VCA module important for actin monomer binding, as well as binding the Arp2/3 complex to stimulate de novo actin polymerization leading to branched actin filament structures. Structural divergence of the known WASP family members outside the VCA region leads to variation in their activity and regulation, and was used to subdivide them into subclasses (Bompard and Caron, 2004). Eight WASP family members representing five WASP protein subfamilies have been described to date in mammals: WASP (WASP and N-WASP), SCAR/WAVE (SCAR/WAVE1, WAVE2, and WAVE3), WASH, WHAMM, and JMY (Derry et al., 1994; Miki et al., 1996; Machesky and Insall, 1998; Linardopoulou et al., 2007; Campellone et al., 2008; Zuchero et al., 2009; Veltman and Insall, 2010).
Knockout of either mouse N-WASP or Scar2, both of which are ubiquitously expressed, results in embryonic lethality (Snapper et al., 2001; Yan et al., 2003; Vartiainen and Machesky, 2004). Scar1 is most highly expressed in brain, and Scar1-null mice exhibit defects in brain function (Soderling et al., 2003). Mouse mutants for WASH, WHAMM, and JMY have not yet been reported. However, over-expression and depletion studies in cells have uncovered a role for WHAMM in maintaining Golgi structure and in the regulation of ER to Golgi transport (Campellone et al., 2008). JMY was identified as a transcriptional cofactor that is regulated by the Mdm2 oncoprotein and augments the response of the p53 tumor suppressor protein to DNA damage (Shikama et al., 1999; Coutts et al., 2007; Coutts and La Thangue, 2007). More recently, silencing JMY expression in neuronal cells has been shown to increase neurite outgrowth, suggesting that JMY normally functions to suppress neurite formation (Firat-Karalar et al., 2011). WASH, like N-WASP, has been shown to participate in endocytosis trafficking. In mammalian cells, WASH localizes to early and recycling endosomes, where it plays a role in endosome sorting by facilitating tubule fission via Arp2/3 activation (Derivery et al., 2009; Gomez and Billadeau, 2009), as well as maintaining the shape of those compartments (Duleh and Welch, 2010). In Dictyostelium, WASH mediates F-actin coating of mature lysosomes and is required for the exocytosis of indigestible materials (Carnell et al., 2011). WASH has also been shown to accumulate in fibroblasts at Salmonella entry sites, suggesting that it facilitates pathogen entry in an Arp2/3 dependent manner and independent of membrane ruffling (Hanisch et al., 2010).
Drosophila wasp, scar, and washout (wash) loss of function mutants exhibit impairments in cell fate decisions and cell morphology during oogenesis and embryonic development (Ben-Yaacov et al., 2001; Zallen et al., 2002; Schenck et al., 2004; Linardopoulou et al., 2007; Richardson et al., 2007; Berger et al., 2008; Gildor et al., 2009; Liu et al., 2009; Sens et al., 2010; Mukherjee et al., 2011). Wash is an essential gene that is required during pupal development (Linardopoulou et al., 2007) and functions downstream of Rho GTPase to regulate actin and microtubules dynamics during Drosophila oogenesis (Liu et al., 2009). WASp is required for the establishment of lineage and cell fate in the developing central nervous system (Ben-Yaacov et al., 2001), microvilli formation (Zelhof and Hardy, 2004), and bristle development (Bogdan et al., 2004). SCAR is known as the major regulator of actin dependent processes mediated by Arp2/3, including axon development, egg chamber structure, and adult eye morphology (Zallen et al., 2002; reviewed in: Vartiainen and Machesky, 2004). Both WASp and SCAR have roles in myoblast fusion, where they appear to be essential during the formation of muscles in both embryos and adults (Richardson et al., 2007; Berger et al., 2008; Gildor et al., 2009; Sens et al., 2010; Mukherjee et al., 2011). No mutants have yet been reported for Drosophila Whamy. Consistent with WASP family proteins carrying out their effects through activation of the Arp2/3 complex, Drosophila Arp2/3 mutants have similar defects in cytoplasmic organization and cytoskeleton dynamics in many morphogenetic events taking place during oogenesis and early embryogenesis (Hudson and Cooley, 2002; Stevenson et al., 2002; Vartiainen and Machesky, 2004).
Here we present the spatial and temporal localization of the different WASP family proteins during Drosophila development to gain a better understanding of their roles in the wide spectrum of morphogenetic processes in which they function. Our study shows how the expression and localization of WASP family proteins mirrors their known roles and reveals potentially new interesting functions for this protein family.
RESULTS AND DISCUSSION
Generation and validation of Wash, WASp, SCAR, and Whamy monoclonal antibodies
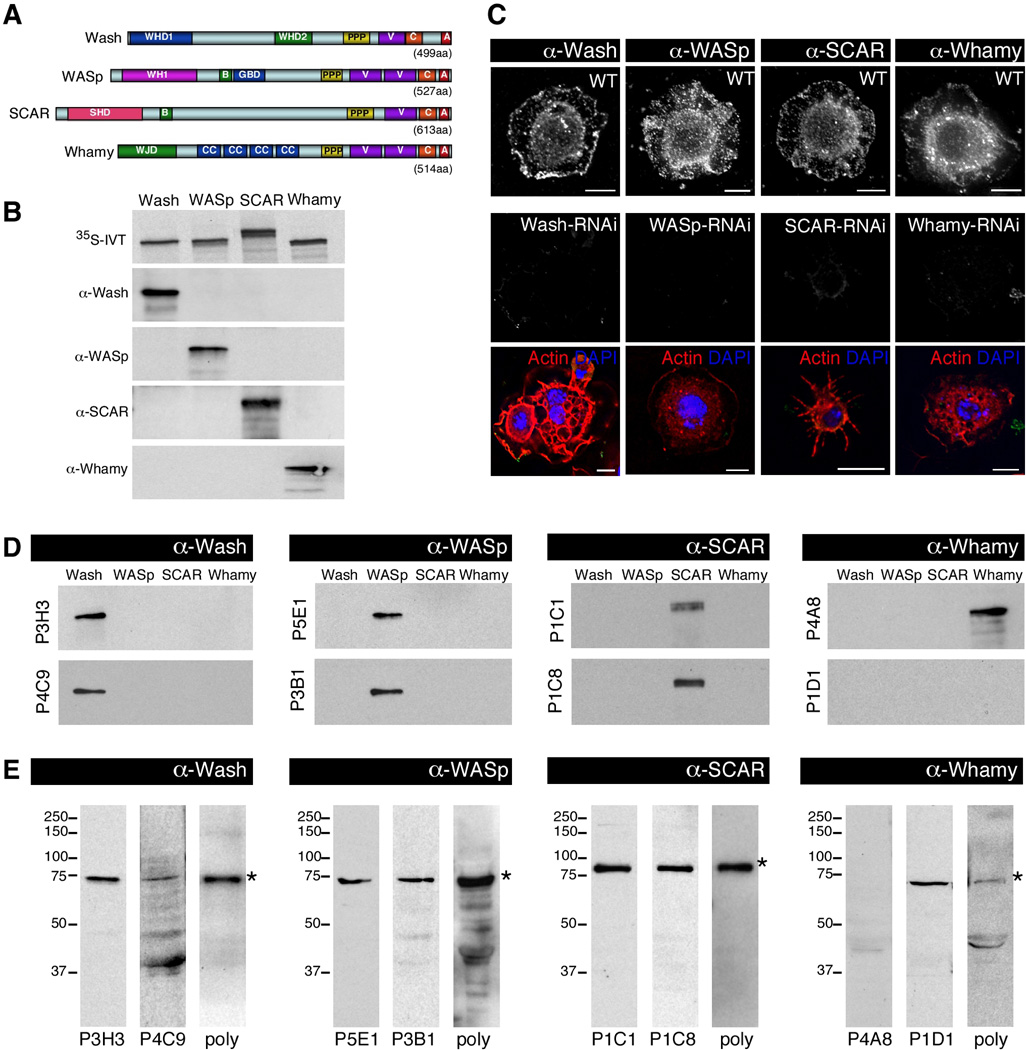
Four subfamilies of WASP family proteins have been identified in Drosophila to date each represented by the single gene: Wash (CG13176), WASp (CG1520), SCAR (CG4636), and Whamy (CG12946) (Fig. 1A) (Ben-Yaacov et al., 2001; Zallen et al., 2002; Linardopoulou et al., 2007; Liu et al., 2009). To examine their spatial and temporal expression patterns we generated mouse polyclonal antibodies against full-length Drosophila Wash, WASp, SCAR or Whamy proteins. Western blot analysis of in vitro translated (IVT) full-length WASP family proteins showed that each of these polyclonal antisera specifically recognized the protein against which it was generated (Fig. 1B). The same full-length WASP family protein constructs were translated in the presence of 35S-Methionine to show the relative levels of the translated proteins (Fig. 1B, upper panel). Since mutants that remove WASP family proteins do not yet exist for Whamy and only reduce protein levels for Wash, Scar and WASp, we confirmed the specificity of these polyclonal antisera by staining wildtype S2 cells and S2 cells treated with RNAi specific for each of the WASP family members (Fig. 1C). Each of the polyclonal antibodies recognized their respective proteins in wildtype cells and this recognition was greatly reduced or eliminated in the corresponding RNAi treated cells (Fig. 1C).
Fig. 1.
Generation and validation of Wash, WASp, SCAR, and Whamy monoclonal antibodies. A: Diagrams of Drosophila WASP family proteins indicating their characteristic domains. WHD1: Wash homology domain 1; WHD2: Wash homology domain 2; PPP: proline rich domain; VCA: actin and Arp2/3 binding; WH1: WASp homology 1; B: basic domain; GBD: GTPase binding domain; CC: coiled coil; SHD: SCAR homology domain; WJD: WHAMM and JMY homology domain. Amino acids are indicated (aa). B-C: Specificity of the mouse polyclonal antibodies generated against WASP family proteins by western blot analysis and cell staining. (B) Autoradiograph of 35S-labeled in vitro translated (IVT) full-length WASP family proteins showing the relative levels of the translated proteins (upper panel). Western blots of IVT WASP family proteins probed with mouse polyclonal antibodies generated against Wash, WASp, SCAR, and Whamy as indicated. (C) Wildtype (top row) and WASP family RNAi treated (middle and bottom rows) Drosophila S2 cells stained with the indicated WASP family mouse polyclonal antisera (middle row) or with phalloidin (to visualize actin) and DAPI (to visualize nuclei; bottom row). Scale bars: 10µm. D-E: The specificity of the monoclonal antibodies used in this study was validated by western blot analysis. (D) Western blots of in vitro translated WAS family proteins probed with monoclonal antibodies generated against Wash (P3H3 and P4C9), WASp (P5E1 and P3B1), SCAR (P1C1 and P1C8) and Whamy (P4A8 and P1D1). (E) Western blots of Drosophila 0–2hours embryo whole cell extract performed with WASP family monoclonal antibodies or with WASP family polyclonal (poly) antibodies as indicated. Endogenous WASP family protein is indicated with an asterisk.
We then generated monoclonal hybridoma lines derived from the mice corresponding to the WASP family polyclonals described above. The specificity of the monoclonal lines was validated by western blot analysis of IVT and endogenous proteins (Fig. 1D–E). We examined two different monoclonal lines generated for each of the WASP family proteins: P3H3/P4C9, P5E1/P3B1, P1C1/P1C8, P4A8/P1D1, that respectively recognize Wash, WASp, SCAR and Whamy (Fig. 1D-E). Each monoclonal line specifically recognized the in vitro translated WASP family member against which it was generated, except one Whamy monoclonal line. Whamy (P1D1) does not recognize IVT proteins using western blot analysis (Fig. 1D), but does recognize endogenous Whamy protein (Fig. 1E). Each monoclonal line, as well as the polyclonal sera, specifically recognized the endogenous WASP family protein against which it was generated (marked by an asterisk) in whole cell embryo extracts, except one Whamy monoclonal line. Whamy (P4A8), does not recognize endogenous Whamy protein using western blot analysis (Fig. 1E), but does recognize IVT Whamy protein (Fig. 1D).
As all of the monoclonal lines exhibited specificity for the WASP family member against which it was generated, for the remainder of this study we used the following lines: P3H3 for Wash, P5E1 for WASp, P1C1 for SCAR, and an equal mixture of P1D1 and P4A8 for Whamy, unless otherwise indicated. All eight monoclonal lines have been sent to the Developmental Studies Hybridoma Bank (University of Iowa; http://dshb.biology.uiowa.edu/) for distribution.
WASP family expression during Drosophila oogenesis
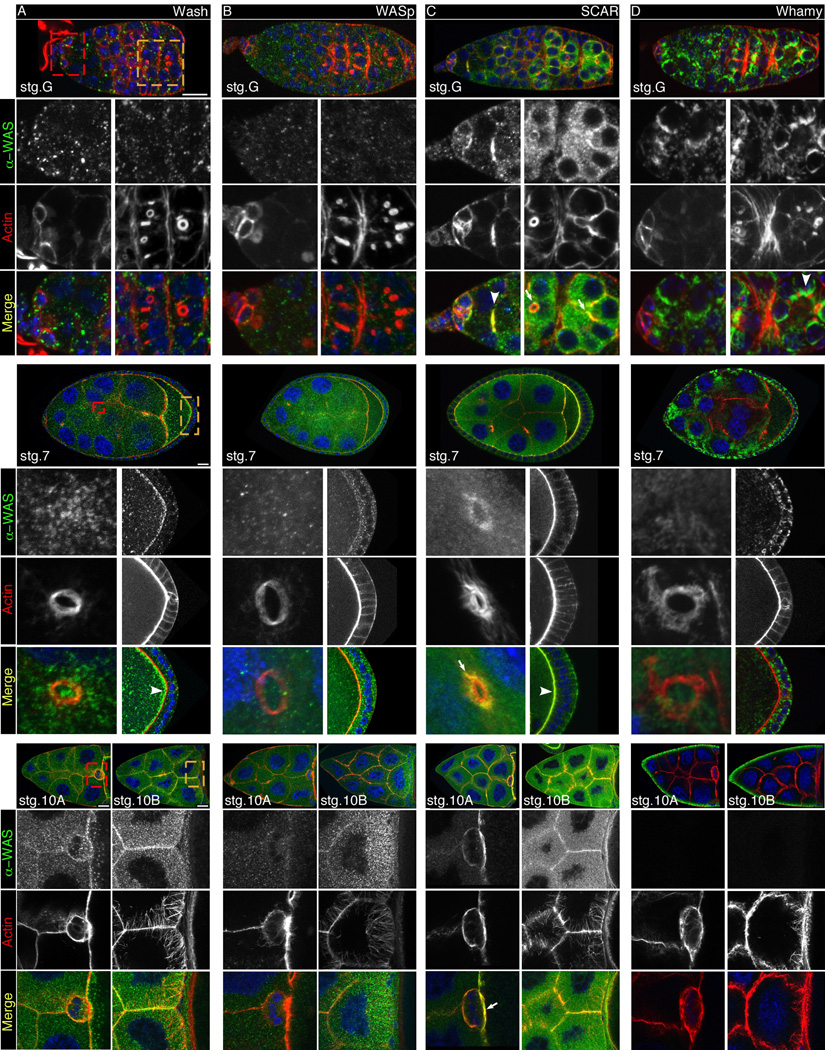
Drosophila oogenesis, a complex developmental process involving the coordinated differentiation of germ line and somatic cells, requires many intricate and dynamic cytoskeletal rearrangements (Fig. 2A) (reviewed in: Horne-Badovinac and Bilder, 2005; Bastock and St Johnston, 2008; He et al., 2011). The germline stem cells (GSCs), located in the germarium, divide asymmetrically renewing the stem cell and giving rise to a differentiated cystoblast. The cystoblast then undergoes four rounds of incomplete divisions to produce a cyst of 16 cells that are interconnected by actin-rich ring canals. One cell within a cyst will become the oocyte, while the remaining 15 develop into oocyte growth supporting nurse cells. Cysts are then surrounded by somatically derived follicle cells to produce egg chambers. These completed egg chambers then “bud off” from the germarium through the formation of somatic cell stalks, thereby forming an assembly line of successively older individual egg chambers. Border cells, a small population of follicle cells originating at the anterior pole of the egg chamber, delaminate from their neighbors and crawl between the nurse cells to position themselves at the anterior border of the oocyte and subsequently form the micropyle. During late oogenesis, the nurse cells dump their contents rapidly into the oocyte, a process that is dependent on contraction of the actin cytoskeleton. Finally, the somatic follicle cells overlying the oocyte secrete first a vitelline membrane and then an eggshell including a micropyle, the structure through which sperm enter the egg, and dorsal appendages, specialized structures that facilitate gas exchange in the developing embryo. The Arp2/3 complex is required during oogenesis for ring canal expansion (Hudson and Cooley, 2002; Zallen et al., 2002). GFP fusions to the Arpc1 and Arp3 subunits of the Arp2/3 complex were present throughout the cytoplasm of germ cells, but enriched at ring canals where they co-localized with actin (Hudson and Cooley, 2002). Consistent with Arp2/3 expression, all four Drosophila WASP family proteins exhibit dynamic spatial and temporal patterns of expression during oogenesis, suggesting multiple roles for each of these proteins.
Fig. 2.
WASP family protein expression during Drosophila oogenesis. A: Schematic of Drosophila oogenesis. ooc: oocyte; NC: nurse cells; SC: germline stem cell; FSC: follicle cell stem cell; FC: follicle cell; cysts: developing cysts; BC: border cells; mp: micropyle; DA: dorsal appendages. B-E: Expression patterns of Wash (B), WASp (C), SCAR (D) and Whamy (E) throughout oogenesis. The germarium (G) and individual egg chambers are shown from different stages (stg.2–stg.14) of oogenesis as indicated. (B) Wash becomes enriched at the oocyte cortex at stg.7 (arrows in stg.7, stg.9), at the nurse cell cortex at stg.10A-10B (arrows), and is highly expressed in dorsal appendages (stg.13–14) (arrows). (C) During stg.6–7 WASp becomes enriched in the posterior follicle cells overlying the oocyte (arrows in stg.6). WASp is also highly expressed in the border cells (arrow in stg.10A and stg.11), nurse cell cortices (arrow in stg.10B), and the lumen of the dorsal appendages (stg.13–14) (arrow in stg.13). (D) SCAR expression can be detected in the germarium, where it accumulates highlighting developing cysts (arrows in stg.G). SCAR also accumulates at the periphery of the oocyte (arrows in stg.6–7) and remains at the oocyte cortex through stg.10A. Beginning at stage 10B SCAR exhibits cortical accumulation within the nurse cells (arrow in stg.11). SCAR is highly expressed in dorsal appendages (stg.12–14) (arrows in stg.14). (E) Whamy exhibits asymmetric localization in nurse cells from stg.G-9, where it accumulates on the apical (outermost) side of the cell (arrows in stg.4 and stg.8). This asymmetric distribution is also observed in the follicle cells in stages 7–8 (arrow). Similar to Wash and SCAR, Whamy is enriched at the dorsal appendages (stg.14). Scale bars: stg.G-stg.6, 10µm; stg.7–stg.9, 20µm; stg.10–14, 100µm. Egg chamber stage (stg.) is indicated and in all panels single slice (2µm thickness) confocal micrographs are shown.
Wash is expressed in all germline and somatic cells throughout oogenesis (Fig. 2B). Puncta of Wash expression can be seen in addition to its uniform expression (Fig. 2B, stg.G-6; and Fig. 4A). Wash does not co-localize with actin-rich structures such as ring canals or nurse cell actin bundles, and is not enriched in GSCs or border cells (Fig. 4A). Cytoskeletal elements must be temporally and spatially coordinated for cells to carry out complex functions, including the cytoplasmic movements required for dispersing or localizing intracellular components. We have shown that Wash is essential for the timing of one such movement, ooplasmic streaming, a process necessary for the proper localization of cytoplasmic components from the nurse cells into the oocyte (Liu et al., 2009). Consistent with its role in stabilizing the actin and microtubule cytoskeleton during ooplasmic streaming, Wash is enriched at the oocyte cortex at stages 7–10, prior to the onset of ooplasmic streaming (Fig. 2B, stg.7–10) and co-localizes with F-actin (Fig. 4A, stg.7). Females with reduced wash expression lay smaller eggs with fused or absent dorsal appendages (Linardopoulou et al., 2007). Correspondingly, Wash expression is enriched in the dorsal appendages and micropyle (Fig. 2B, stg.13–14 and Fig. 3F).
Fig. 4.
WASP family protein co-localization with F-actin during Drosophila oogenesis. A-D: Confocal micrograph projections showing WASP family proteins (green or grayscale) co-localizing with F-actin (red or grayscale) and co-stained with DAPI (blue) as indicated. Wash (A), WASp (B), SCAR (C) and Whamy (D) expression patterns in the germarium (upper panels) and individual egg chambers from stages 7 (middle panels) or stages 10A and 10B (lower panels) showing co-localization with actin rich structures. Higher magnification views of germarium region 1 (yellow dashed box) and regions 2b-3 (red dashed box), stage 7 ring canals (red dashed box) and oocyte cortex (yellow dashed box), stage 10A border cells (red dashed box) and nurse cell actin bundles in stage 10B (yellow dashed box). Scale bars: Germarium: 10µm, stages 7, 10A and 10B: 100µm.
Fig. 3.
WASP family proteins show distinct patterns of expression in follicle cells, as well as in the oocyte. A-Aiii: Surface Z-projection showing the asymmetric localization of Whamy in follicles cells (α-Whamy, green; DAPI, blue). (Ai-Aiii) Higher magnification views of regions indicated in A. B-E”: Surface Z-projections of egg chambers stained with α-Whamy (green or grayscale) and alpha-tubulin to visualize microtubules (MTs) (B-B”, red or grayscale), the golgi protein Lava lamp (C-D”, red or grayscale), or the mitochondria marker MitoTracker (E-E”, red or grayscale). DAPI (blue) is included to visualize nuclei in D”. F-I: Surface Z-projections of stage 8, 10, 12 and 14 egg chambers stained with α-Wash (F), α-WASp (G), α-SCAR (H), or α-Whamy (I). Scale bars: A-Aiii, B-B”, E-E”: 10µm; C-D”: 100µm, F-I, 20µm.
WASp is expressed at all stages of oogenesis, with specific accumulation in the posterior follicle cells at stage 6 (Fig. 2C, stg.6), the time at which Gurken (epidermal growth factor ligand) signal is being expressed to establish the perpendicular axes of the oocyte (reviewed in: Riechmann and Ephrussi, 2001). Interestingly, the enrichment of WASp in the posterior follicle cells is diminished by stage 7 (Fig. 2C, stg.6–7 and Fig. 4B), suggesting that WASp could be acting as an oocyte organization factor, affecting signaling from the posterior follicle cells to the oocyte. WASp does not co-localize with actin-rich structures and is not enriched in GSCs or border cells (Fig. 4B). Later in oogenesis WASp is enriched in nurse cell cortices (Fig. 2C, stg.10B and Fig. 3G, stg.10), where the nurse cells come into contact with the oocyte (Fig. 2C, stg.11; Fig. 4B, stg.10A) and the micropyle (Fig. 2C, stg.13).
Unlike other members of the WASP family, SCAR is highly expressed during oogenesis and co-localizes with actin-rich structures (Fig. 2D; and Fig. 4C). In the germarium, SCAR is initially enriched at the border of GSCs and cap cells, and again at the border of GSCs and cystoblasts (Fig 4C, stg.G). SCAR then accumulates within the cytoplasm of the developing cysts and maintains a higher level of expression in the germline compared to the surrounding somatic follicle cells (Fig. 2D, stg.G-10A; Fig 4C, stg.G), and is enriched in the developing ring canals where it co-localizes with actin (Fig 4C, stg.G,7). As oogenesis progresses, SCAR becomes highly enriched at the oocyte cortex (Fig. 2D, stg.6–10A; Fig. 4C, stg.7) and nurse cell cortices (Fig. 2D, stg.9–12; Fig. 3H, stg.10; and Fig. 4C, stg.10A–10B). Like Wash and WASp, SCAR is also expressed in the dorsal appendages (Fig. 2D, stg.12–14 and Fig. 3H, stg.12–14).
Whamy exhibits an unusual asymmetric subcellular localization as it accumulates on the apical (outermost) side of each nurse cell and somatic follicle cells (Fig. 2E, stg.G-10A; Fig. 4D). Whamy does not co-localize with actin-rich structures (Fig. 4D), but can be seen to thread its way through ring canals (Fig. 4D, stg.G). In z-projections of the egg chamber epithelia (Fig. 3A-Aiii and Fig. 3I, stg.8), Whamy is highly enriched in a “crescent shaped” pattern surrounding the nuclei of certain follicles cells, suggesting a possible role for Whamy in the polarization of the developing egg chamber. This asymmetric localization varies in different regions of the egg chamber. In the more anterior region of the egg chamber Whamy is homogenously distributed within the cytoplasm of the cells (Fig. 3Aii), whereas in the middle of the egg chamber it appears to surround the nuclei forming a crescent shape (Fig. 3Aiii). The transition in Whamy subcellular localization from one region to another is quite sharp (Fig. 3Ai).
While studies in cells have uncovered a role for WHAMM in maintaining Golgi structure and in the regulation of ER to Golgi transport (Campellone et al., 2008), in vivo functions for fly Whamy have not yet been reported. However, Whamy’s asymmetric localization is intriguing in the light of a recent study showing that Drosophila egg chambers undergo circumferential rotation around their long axes during stages 7–9, a process that is proposed to involve polarized cell behaviors (Haigo and Bilder, 2011). Therefore, we co-stained ovaries with Whamy and alpha-tubulin to visualize microtubules, the golgi protein Lava lamp (Sisson et al., 2000), or the mitochondrial marker MitoTracker (Fig. 3B-E”). We do not observe co-localization of Whamy with microtubules (Fig. 3B-B”) or golgi (Fig. 3C-C”) in stages 7–9 egg chambers when Whamy is asymmetrically enriched. However, Whamy and Lava lamp staining overlap in stage 10 egg chambers where Whamy and golgi exhibit perinuclear accumulation (Fig. 3D-D”). Most strikingly, Whamy expression pattern is identical to that exhibited by mitochondria, as MitoTracker co-localizes with all aspects of Whamy subcellular localization (Fig. 3E-E”). Mitochondria dynamics and morphology are important for many cellular functions including development, aging and cell death, and when defective have been linked to human diseases, such as inherited neuropathies (Okamoto and Shaw, 2005). The co-localization of Whamy with mitochondria is interesting in light of a possible role for Whamy in mitochondria tubulation, which could be important for energy metabolism or cell shape changes necessary for proper egg chamber rotation.
WASP family proteins are expressed throughout Drosophila embryogenesis
The formation of tissues and organs during embryogenesis involves a series of exquisite morphogenetic processes executed through precisely orchestrated tissue contractions, foldings and migrations that are highly dependent on dynamic coordination of the actin and microtubule cytoskeleton (reviewed in: Sweeton et al., 1991; Harden, 2002; VanHook and Letsou, 2008; Harris et al., 2009). Following the formation of a hollow ball of cells (called a cellular blastoderm), the posterior end of the Drosophila embryo migrates up and over the top (dorsal) surface in a process called germband extension, which is responsible for delivery of specific cell types to the interior of the embryo. After this is accomplished, the reverse process (called germband retraction) occurs leaving a large open hole on the dorsal surface of the embryo. Dorsal closure then takes place to close the hole by zipping up the surrounding epidermis over the extraembryonic amnioserosa cells, which overlays the dorsal hole. The zippering phase occurs through complex actin cable formation and filopodial dynamics at the leading edge of the epithelium. Towards the end of dorsal closure the embryo also undergoes another morphogenetic process in which it internalizes its head structures, known as head involution. All four Drosophila WASP family proteins exhibit dynamic spatial and temporal patterns of expression during embryogenesis consistent with their involvement in numerous developmental processes.
Wash expression throughout Drosophila embryogenesis
Wash is an essential gene in Drosophila (Linardopoulou et al., 2007; Liu et al., 2009). Homozygous zygotically mutant flies die at the transition from the 3rd larval instar to pre-pupae, where they display an elongated body phenotype and spiracle-eversion defects (Linardopoulou et al., 2007). Wash is expressed uniformly in the cytoplasm of all cells throughout embryogenesis, as well as exhibiting spatially and temporally specific enrichments above these uniform levels in different cells and/or regions (Fig. 5). During syncytial stages 1–2 (Fig. 5A-B”), Wash localizes to the cytoplasm surrounding migrating nuclei (energids). Wash is also enriched in cells undergoing shape changes; during gastrulation Wash accumulates in cells forming the cephalic and ventral furrows (Fig. 5F-H”). Rho family small GTPases have been shown to play an important role in these processes by regulating the actin cytoskeleton in cells undergoing shape changes. One possible function for Wash in this context could be linking the actin cytoskeleton with signals received from Rho GTPase (Leptin, 2005; Liu et al., 2009). In later embryonic stages, Wash is highly expressed in posterior spiracles whose proper formation also requires regulation by Rho GTPases (Fig. 5N-N”) (Hu and Castelli-Gair, 1999; Simoes et al., 2006). We do not observe specific accumulation of Wash in the developing nervous system or musculature. Interestingly, Wash is enriched in the posterior midgut and malpighian tubules during late embryogenesis (Fig. 5P-R”).
Fig. 5.
Wash expression throughout Drosophila embryogenesis. A-R: Confocal micrograph projections showing Wash expression in successively older wildtype embryos (α-Wash (P3H3); grayscale or green; DAPI, blue). Embryonic stage (stg) is indicated. (A’-R”) Higher magnification views of region from A-R, respectively, without (A’-R’) and with (A”-R”) DAPI staining to visualize nuclei. Wash is expressed during embryogenesis throughout the whole embryo. Note increased Wash accumulation in energids at stages 1–2 (arrows in A). Wash exhibits dynamic subcellular localization in mitotic domains (arrows in F’). During gastrulation (stg.6–8), Wash is enriched in cephalic furrow (arrows in F), ventral furrow (arrows in G), and the posterior midgut invagination (arrow in H). In later staged embryos, Wash accumulates in spiracles (stg.13; arrow in N’) and malpighian tubules (stg.15–17; arrows in Q). Lateral views are shown throughout with the exception of G-G” (ventral view). Scale bars: A-R, 50µm; A’-R”, 20µm.
WASp expression throughout Drosophila embryogenesis
In Drosophila, WASp is essential during processes involving asymmetric cell divisions mediated by Notch signaling pathway and neurogenesis. wasp mutant flies fail to eclose from the pupal case (or die shortly after if they do eclose), and they lack neurosensory sensory bristles (Ben-Yaacov et al., 2001; Tal et al., 2002). WASp is also known to be involved in myoblast fusion, a process necessary for the proper formation of somatic musculature in Drosophila ((Richardson et al., 2007; Schafer et al., 2007; Berger et al., 2008; Gildor et al., 2009; Sens et al., 2010; Mukherjee et al., 2011).
Similar to Wash, WASp is expressed uniformly in the cytoplasm of all cells throughout embryogenesis, as well as exhibiting spatially and temporally specific enrichments different cells and/or regions (Fig. 6). During gastrulation, WASp is highly expressed in the ventral furrow (Fig. 6G-G”), as well as in the dorsal transverse folds and posterior midgut invagination (Fig. 6I-I”). WASp is also enriched in the stomach, posterior midgut, and malpighian tubules (Fig. 6PR”). When using the polyclonal antibody against WASp, we observed an accumulation of protein in the ventral epidermal denticles. Although we did not recapitulate this staining using monoclonal antibody P5E1, this expression can be observed with P3B1, the other WASp monoclonal antibody (Fig. 6S-T”). Despite the requirement of WASp in neurogenesis and myoblast fusion, we do not observe specific accumulation of WASp in developing neural or muscle precursors.
Fig. 6.
WASp expression throughout Drosophila embryogenesis. A-R: Confocal micrograph projections showing WASp expression in successively older wildtype embryos (α-WASp (P5E1); grayscale or green; DAPI, blue). Embryonic stage (stg) is indicated. (A’-R”) Higher magnification views of region from A-R, respectively, without (A’-R’) and with (A”-R”) DAPI staining to visualize nuclei. WASp is expressed during embryogenesis throughout the whole embryo. Note the enrichment of WASp in ventral furrow (arrows in G) and the posterior midgut invagination (arrows in F and I). Beginning with germband retraction (stg.12) WASp accumulates in the amnioserosa (arrows in N) and remains until dorsal closure is finished (stg.15). In later staged embryos, WASp is enriched in the midgut (arrow in Q), malpighian tubules (arrows in R), and ventral denticles (arrows in S). Lateral views of embryos are shown in all cases, with the exception of G-G” (ventral view). (S-T) WASp polyclonal antibody (S; 1:100) and a second WASp monoclonal antibody (T; WASp (P3B1); 1:10) were used to show WASp accumulation in the ventral denticles. Scale bars: A-T, 50µm; A’-R”, 20µm; S’-T”, 10µm.
SCAR expression throughout Drosophila embryogenesis
SCAR is expressed in all stages of embryogenesis. In the early stages SCAR is thought to be an important regulator of the cortical actin cytoskeleton, either activating the Arp2/3 complex to polymerize actin or organizing the actin cytoskeleton during furrow formation (Zallen et al., 2002).
Similar to Wash and WASp, SCAR is expressed uniformly in the cytoplasm of all cells throughout embryogenesis, as well as exhibiting spatially and temporally specific enrichments in different cells and/or regions (Fig. 7). At stage 2 of embryogenesis SCAR accumulates at the actin caps overlying each nuclei (Fig. 7A-B”) and in pseudocleavage furrows (Fig. 7C-C”, also see Fig. 9D). During gastrulation, SCAR accumulates in the cells undergoing apical constriction necessary for their invagination on the ventral surface (Fig. 7F-F”), as well as in the cephalic furrow and posterior midgut invagination (Fig. 7G-H”). SCAR undergoes dynamic subcellular changes during mitosis as reflected by its expression in mitotic domains (Fig. 7I-I”).
Fig. 7.
SCAR expression throughout Drosophila embryogenesis. A-R: Confocal micrograph projections showing SCAR expression in successively older wildtype embryos (α-SCAR (P1C1); grayscale or green; DAPI, blue). Embryonic stage (stg) is indicated. (A’-R”) Higher magnification views of region from A-R, respectively, without (A’-R’) and with (A”-R”) DAPI staining to visualize nuclei. SCAR is expressed during embryogenesis throughout the whole embryo. SCAR accumulates in energids at stages 1–2 (arrows in B’) and in pseudocleavage furrows at stg.3 (arrows in C’). During gastrulation (stg.6–8), SCAR is enriched in cephalic furrow (arrows in G), ventral furrow (arrows in F’), and the posterior midgut invagination (arrow in H’). Scar exhibits altered subcellular localization in mitotic domains (arrows in I’). In later staged embryos, SCAR is enriched in the central nervous system (stg.13–17; arrows in O, Q) and brain (stg.17; arrow in R). Lateral views are shown throughout with the exception of F-F” (ventral view), G-G” (dorsal view), and Q-Q” (ventral view). Scale bars: A-R, 50µm; A’-R”, 20µm.
Fig. 9.
WASP family expression during cellularization. Expression pattern of WASP family proteins (green or grayscale) in confocal cross-sections of nuclear cycle (nc) 8–14B embryos expressing an actin reporter (sGMCA; red) and co-stained with DAPI (blue). A: Schematic representation of embryonic nuclear divisions (nc8–14B) and pole cell formation. B-E: Wash (B), WASp (C), SCAR (D), or Whamy (E) expression throughout the cellularization process. Note that SCAR is expressed in actin caps (arrows in nc10–11), in newly formed furrows (arrow in nc14A–14B), and at the cell cortex in forming pole cells (Pole Cells). All images are the same magnification. Scale bar in B (nc8): 10µm.
During germband extension, SCAR accumulates in hemocytes (the Drosophila equivalent of macrophages) migrating from the head to the tail (Fig. 7 M-M”), consistent with a proposed role for role in phagocytosis (Pearson et al., 2003). SCAR is required for proper neural development and has been shown to accumulate in cells of the developing central nervous system (CNS) (Zallen et al., 2002). Consistent with this, our SCAR monoclonal similarly shows accumulation of SCAR in neurons in the developing nerve cord (Fig. 7O-Q”). Interestingly, in later embryos we also observe a high accumulation of SCAR in the neurons in the brain (Fig. 7R-R”). Despite a role for SCAR in myogenesis, we do not observe a specific accumulation of SCAR in muscle precursors.
Whamy expression throughout Drosophila embryogenesis
Whamy expression pattern during early through mid-embryogenesis is similar to the other members of the WASP family. Whamy is highly expressed in energids (Fig. 8A-C”) and in cells undergoing shape changes during gastrulation (Fig. 8F-H”), but does not co-localize with actin-rich structures (Fig. 9E). At stages 13–14 Whamy accumulates in the hindgut and clypeolabrum, and remains highly expressed in these tissues during later stages (Fig. 8N-O”). Interestingly, Whamy localizes to midgut cells in the same asymmetric subcellular localization pattern that we observed in ovaries (Fig. 8, P-P”). In the later stages of embryogenesis, Whamy is enriched in the central nervous system (Fig. 8Q-Q”) and somatic musculature (Fig. 8S-S”). No mutant analyses for Whamy have yet been reported.
Fig. 8.
Whamy expression throughout Drosophila embryogenesis. A-T: Confocal micrograph projections showing Whamy expression in successively older wildtype embryos (α-Whamy (P1D1/P4A8 mix); grayscale or green; DAPI, blue). Embryonic stage (stg) is indicated. (A’-T”) Higher magnification views of region from A-T, respectively, without (A’-T’) and with (A”-T”) DAPI staining to visualize nuclei. Whamy is expressed during embryogenesis throughout the whole embryo. Note increased Whamy accumulation in energids at stages 2–3 (arrows in B). During gastrulation (stg.6–8), Whamy is enriched in cephalic furrow (arrows in F), ventral furrow (arrows in G), dorsal transverse fold (arrow in H), and the posterior midgut invagination (arrow in H’). In later staged embryos, Whamy is enriched in the hindgut (stg.13–14; arrow in N), midgut (stg.15, dashed box in P represents higher magnification views in P’ and P”), and muscle (arrows in S). Lateral views are shown throughout with the exception of G-G”(ventral view) and Q-Q” (ventral view). Scale bars: A-T, 50µm; A’-M”, 20µm; N’-T”, 10µm.
WASP family expression during cellularization
The initial stages of Drosophila embryonic development occur in a syncytium, in which nuclei divide within a common cytoplasm (Fig. 9A) (reviewed in: Mazumdar and Mazumdar, 2002). As the somatic nuclei are dividing synchronously for thirteen replicative cycles, they migrate to the embryo surface forming the syncytial blastoderm. The embryos then undergo a special cytokinesis, known as ‘cellularization’, during which there is simultaneous cell formation over the whole surface of the embryo. The cleavage furrows that form to separate the blastoderm nuclei during cellularization accumulate high levels of F-actin and myosin-II and require coordinated growth and extension of plasma membrane. The Arp3 subunit of the Arp2/3 complex has been shown previously to localize to actin-rich structures in early embryos: the margins of expanding actin caps, to mature pseudocleavage furrows during syncytial blastoderm divisions, and to somatic cleavage furrows in post-cellularization embryo divisions (Stevenson et al., 2002).
To visualize the spatial and temporal localization of WASP family proteins during the early nuclear divisions, we examined nuclear cycle 8–14B embryos stained with Wash, WASp, SCAR, and Whamy monoclonal antibodies (Fig. 9B-E). Wash, SCAR, and Whamy proteins are expressed throughout the cytoplasm during blastoderm stages and they also exhibit dynamic enrichments above this uniform expression (Fig. 9B, D-E). WASp, on the other hand, is expressed in a uniformly punctate pattern and at very low levels (Fig. 9C). Wash is initially distributed throughout the apical and basal cytoplasm (Fig. 9B, nc11–13), then it accumulates in the basal cytoplasm of the blastoderm embryo (Fig. 9B, nc14A–14B). SCAR has been shown previously to co-localize with F-actin-rich structures in the blastoderm embryo (Zallen et al., 2002) and most closely resembles the expression patterns observed for Arp2/3 complex components in the early embryo. SCAR is enriched in the actin caps overlying each nucleus once they reach the periphery (Fig. 9D, nc10) and then is associated with the invaginating pseudocleavage (Fig. 9D, nc12–14A) and cleavage furrows (Fig. 9D, nc14B). Whamy is initially expressed in the basal cytoplasm at high levels (Fig. 9E, nc8–13), then it accumulates in the apical cytoplasm upon cellularization (Fig. 9E, nc14A–14B).
The first syncytial nuclei to reach the periphery are in the posterior of the embryo. These nuclei, along with their specialized posterior pole cytoplasm, form cells to become the Drosophila germline precursors cells or pole cells. While pole cells are usually considered transcriptionally quiescent, we find that all four WASP family proteins are uniformly expressed in pole cells (Fig. 9B-E, Pole Cells). In addition, SCAR is highly enriched in the furrows as pole cells bud from the embryo and subsequently divide (Fig. 9D, Pole Cells).
WASP family proteins exhibit dynamic nuclear localization
In addition to their cytoplasmic roles, the Arp2/3 complex and WASP family proteins have been implicated in nuclear processes. Before Wash was appreciated as a WASP family protein, it was identified in a chromatin-remodeling complex along with ISWI and TRF2 (Hochheimer et al., 2002). The Arp2/3 complex and the WASP family proteins N-Wasp and JMY have been observed in the nucleus and implicated in transcriptional regulation, at least in part, through regulating nuclear actin polymerization (Suetsugu and Takenawa, 2003; Jin et al., 2004; Wu et al., 2006; Coutts et al., 2007; Coutts and La Thangue, 2007; Yoo et al., 2007). JMY was also recently shown to nucleate linear actin filaments within the nucleus (Zuchero et al., 2009), but the significance of this activity to its transcriptional activation properties is not yet known.
Interestingly, Drosophila Wash, WASp, SCAR and Whamy exhibit dynamic subcellular localization changes during mitosis, including localization within the nucleus (Fig. 10A-D”). Consistent with this, the WASP family monoclonal antibodies recognize their respective WASP family protein in Drosophila 0–12 hour embryo nuclear extracts (Fig. 10E). Following cellularization, at least 25 mitotic domains, which are clusters of cells undergoing locally synchronous mitosis, partition the embryo blastoderm surface (Foe, 1989). Cells within a particular mitotic domain share morphological and cell fate characteristics (Foe, 1989; Arora and Nusslein-Volhard, 1992). Despite all four WASP family proteins localizing to the nucleus, they have different temporal subcellular localization patterns, perhaps suggesting specific nuclear roles for each family member. For example, Whamy is nuclear in cells surrounding the mitotic domain and it becomes perinuclear in mitotic cells (Fig.10D-D”), where it could play a role in the trafficking of molecules within dividing cells. Another striking example of WASP family protein nuclear localization is in the developing CNS. We find that WASp expression is enriched in the nuclei of neuroblasts during the onset of embryonic CNS development (Fig. 10F). Consistent with this nuclear WASp accumulation, wasp mutant analysis has suggested a role for WASp in cell fate decisions during CNS development (Ben-Yaacov et al., 2001).
Fig. 10.
WASP family proteins exhibit dynamic subcellular localization during gastrulation and CNS development. A-D””: Single section (1µm thickness) confocal micrographs of stg.7 wildtype embryos showing the expression pattern of Wash (A-A”’), WASp (B-B”’), SCAR (C-C”’), or Whamy (D-D”’) in mitotic domains, with (A-D, A’”-D’”) and without (A’-D”) DAPI staining to visualize nuclei (a-WASP family member, green or grayscale; DAPI, blue). Note that all WASP family proteins exhibit dynamic subcellular localization with exclusive cytoplasmic (cf. arrowheads in C”) or both nuclear and cytoplasmic (cf. arrows in C”) localization. (A”-D””) Higher magnification views of the regions outlined with dashed boxes in A’-D’, respectively. Scale bars: A-D’, 20µm; A”-D”’, 7µm. E: Western blots of Drosophila 0–12hour embryo nuclear extracts performed with WASP family monoclonal antibodies as indicated. F-F’”: WASp accumulates in the nuclei of CNS precursors. Ventral surface projections of early stg.9 wildtype embryos stained with WASp (green or gray). Neuroblasts are identified by Hunchback (Hb) nuclear staining (red; F, F”). (F”-F”) Higher magnification views of the region outlined by dashed box in F, with (F”) and without (F’”) α-Hb co-staining. Scale bars: F-F’, 20µm; F”-F”’, 10µm.
Concluding remarks
WASP family proteins have been proposed to play key roles in the intricate orchestration of cellular processes essential for morphogenetic events through their tight spatial and temporal coordination of actin dynamics with other cellular machineries (reviewed in: Takenawa and Suetsugu, 2007; Rottner et al., 2010). Consistent with every cell and virtually every cellular process requiring some form of actin regulation, we find that Drosophila WASP family proteins are expressed in all cells throughout development. In addition to their ubiquitous expression, all WASP family members show enrichment in spatially and temporally specific patterns at both the cell and subcellular levels. These enrichments can be overlapping among WASP family members (i.e., accumulation at the oocyte cortex for Wash and SCAR or accumulation in the forming ventral furrow for all WASP family members) or unique to particular WASP family members (i.e., the posterior follicle cell localization of WASp or the asymmetric expression of Whamy in follicle cells). In general, these enrichments mirror times and places in which that particular WASP family member has been shown to function. Together, these observations suggest that the functions of these proteins are not redundant to each other. For example, WASp expression is enriched in the nervous system and wasp mutant flies exhibit neural defects. As not all areas of enriched WASP family protein expression have yet been associated with a phenotype, it will be interesting to determine if the expression patterns are predictive of protein function. The monoclonal antibodies generated will provide valuable tools for future studies to unravel the functions of these essential and highly conserved actin regulatory proteins.
EXPERIMENTAL PROCEDURES
Fly strains and genetics
Wildtype flies (Oregon R) or transgenic flies expressing an actin reporter (sGMCA) (Kiehart et al., 2000) were cultured on yeast-cornmeal-molasses-malt extract media and maintained at 25°C.
Antibody generation
Polyclonal antibodies for Wash, WASp, SCAR, and Whamy were generated by immunizing BALB/c BYJ Rb (8.12) 5BNR/J mice (Jackson Labs) with the full-length WAS protein fused to GST. The monoclonal lines were generated in the FHCRC Antibody Development Shared Resource Facility as described (Wayner et al., 1989; Harmon et al., 1995) and have been sent to the Developmental Studies Hybridoma Bank at the University of Iowa for distribution (http://dshb.biology.uiowa.edu/).
Drosophila S2 cell maintenance and immunostaining
Drosophila S2 cells were maintained at 25°C in Schneider’s Drosophila Medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum, 25mM glutamine, 50 units penicillin and 50µg streptomycin (Invitrogen, Carlsbad, CA). For staining, cells (1×106) were plated on Concanavalin A (ConA; Sigma) coated coverslips and allowed to spread for 1 hour at 25°C. Cells were fixed for 15 minutes using 4% formaldehyde in phosphate-buffered saline (1× PBS). Cells were washed three times for 15 minutes each with PbT (1× PBS, 0.1% Tween, 0.1% BSA, 0.05% Azide), then blocked for 2 hours using 5% normal goat serum in PbT. Each of the WASP family polyclonal antibodies was used at 1:100 dilution in PbT. To visualize microtubules, α-alpha-tubulin monoclonal antibody DM1A (Sigma, St Louis, MO) was used at 1:2000. Cells were incubated with the primary antibodies for 1 hour at room temperature. After this incubation coverslips were carefully washed three times with PbT. Secondary antibodies (Alexa conjugates; Invitrogen, Carlsbad, CA) diluted 1:2000 in PbT were added and slides were incubated for 1 hour at room temperature. For actin visualization 3 units of Alexa Fluor 568 phalloidin (Invitrogen, Carlsbad, CA) were included with the secondary antibodies. Cells were carefully washed with PbT three times and then incubated with DAPI (1µg/ml) for 15 minutes. Coverslips were then washed two times with PbT and mounted on slides in SlowFade Gold with DAPI media (Invitrogen, Carlsbad, CA). Stained cells were visualized using a DeltaVision RT microscope with a 100× Oil objective. The following filters were used: DAPI 360/40 457/40, FITC 490/20 526/38, RD-TR-Cy3 555/28 617/63. Post-acquisition images were de-convolved using SoftWoRX software, processed using ImageJ and assembled using Canvas software.
Immunostaining of Drosophila ovaries
Female flies were fattened for 2 days on yeast-cornmeal-molasses-malt extract media with supplemental dried active baker's yeast at 25°C. Ovaries were dissected into Drosophila Ringer's buffer at room temperature and fixed using 1× PBS 6% formaldehyde/heptane for 10 minutes. After three washes with PTW (1× PBS, 0.1% Tween-20), ovaries were permeabilized in 1× PBS plus 1% Triton X-100 for 2 hours at room temperature, then blocked using PAT (1× PBS, 0.1% Tween-20, 1% bovine serum albumin (BSA), 0.05% Azide) for 2 hours at 4°C. Antibodies were added at the following concentrations: α-Wash (P3H3) 1:20, α-WASp (P5E1) 1:10, α-SCAR (P1C1) 1:50, α-Whamy (P1D1/P4A8) 1:20, α-Lava Lamp 1:5000 (Sisson et al., 2000), α-alpha-Tubulin 1:200 (Abcam Inc., Cambridge, MA); and ovaries were incubated overnight at 4°C. Primary antibody was then removed and ovaries were washed three times with XNS (1× PBS, 0.1% Tween-20, 0.1% BSA, 2% normal goat serum) for 30 min each. Alexa conjugates secondary antibodies (Invitrogen, Carlsbad, CA) diluted in PbT (1:2000) were then added and ovaries were incubated overnight at 4°C. Ovaries were washed 8 times with PTW at room temperature and then incubated for 1 hour with DAPI (1µg/ml, to visualize DNA) and/or phalloidin (2units/ml, to visualize actin). After three additional washes with PTW, ovaries were mounted on slides in SlowFade Gold with DAPI media (Invitrogen, Carlsbad, CA) and visualized using a Zeiss confocal microscope as described below. For co-localization with MitoTracker, ovaries were dissected at room temperature in Schneider’s media and incubated for 15 min with 500nM MitoTracker Orange CMTMRos (Invitrogen, Carlsbad, CA), then briefly rinsed with Schneider’s media. Ovaries were then fixed, blocked and stained as described above.
Immunostaining of Drosophila embryos
Female flies were fattened for 2 days on yeast-cornmeal-molasses-malt extract media with supplemental dried active baker's yeast at 25°C. Embryos were collected and aged at 25°C or room temperature. Embryos were then dechorionated with bleach, fixed with 4% formaldehyde in a 1×PBS/heptane bi-layer for 20 minutes, and devitellinized in methanol. After three washes with PTW (1×PBS /0.1% Tween-20), embryos were blocked using PAT (1×PBS, 0.1% Tween-20, 1% BSA, 0.05% Azide) for 2 hours at 4°C. Antibodies were added at the following concentrations: α-Wash (P3H3) 1:20; α-WASp (P5E1) 1:10; α-SCAR (P1C1) 1:50; α-Whamy (P1D1/P4A8) 1:20; and the embryos were incubated overnight (~12 hours) at 4°C. Primary antibody was then removed and embryos were washed three times with XNS (1× PBS, 0.1% Tween-20, 2% normal goat serum) for 30 min each. Secondary antibodies (1:1000–2000; Alexa conjugates; Invitrogen, Carlsbad, CA) in PbT (1× PBS, 0.1% Tween-20, 0.1% BSA, 0.05% Azide) were then added as appropriate and incubated overnight (~12 hours). Embryos were washed 4–8 times with PTW. Embryos were then incubated for 15 minutes with DAPI (1µg/ml, to visualize DNA). After 3–6 additional washes with PTW, embryos were mounted on slides in SlowFade Gold with DAPI media (Invitrogen, Carlsbad, CA). Embryos were visualized using a Zeiss confocal microscope or a Nikon/Perkin-Elmer spinning disk microscope as described below.
Microscopy
Stained embryos and ovaries were visualized using a Zeiss LSM-510META confocal microscope with excitation at 488 nm or 543 nm and emission collection using a BP-505–550, BP-560–615 or LP-560 filter. For DAPI visualization we used a two-photon 780 nm laser line and a BP415–450 filter. Images were obtained using a Plan-Apochromat 20 times/0.75 dry objective. The embryo images in Figure 9 were obtained on a Nikon Eclipse Ti stand (Nikon Instruments, Melville, NY, USA), with 60×/1.4NA objective lens and controlled by Volocity software (v.5.3.0, Perkin Elmer, Waltham, MA, USA). Images were acquired with 491nm, 561nm, and 405nm lasers, with a Yokogawa CSU-X1 confocal spinning disc head equipped with a Hamamatsu C9100-13 EMCCD camera (Perkin-Elmer). The following settings were used: exposure time, 200–300 milliseconds; camera sensitivity, 150; gain, 1. The images correspond to 27µm/0.25µm steps. Post-acquisition images were processed in ImageJ or Volocity and assembled using Canvas software.
In vitro translation of WAS family proteins
WAS proteins were in vitro translated using TNT Quick Coupled Transcription/Translation Systems (Promega) following the manufacturer’s protocol. The following constructs were used: pCITE-Wash, pCITE-WASp, pCITE-SCAR, and pCITE-Whamy. Full length WASP family genes were amplified and cloned into pCITE-4b (Novagen/Merck, Rockland, MA) vector using specific restriction sites: Wash (amino acids 1–500) SalI–NotI, WASp (amino acids 1–527) SalI–NotI, SCAR (amino acids 1–613) SacI–NotI and Whamy (amino acids 1–515) BamHI–NotI.
Lysate generation and Western blotting
WASP family antibody specificity was assessed on in vitro translated proteins or protein extracts that were resolved via SDS-PAGE and examined by western blot analysis. Wildtype Drosophila whole-cell (from 0–2 hour embryos) and nuclear (from 0–12 hour embryos) extracts were gifts from Toshi Tsukiyama (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). 15µg of total protein were loaded in each lane and probed with the antibodies at the following concentrations: α-Wash (P3H3; P4C9) 1:10, α-WASp (P5E1; P3B1) 1:5, α-SCAR (P1C1; P1C8) 1:10 and α-Whamy (P1D1/P4A8) 1:10. All polyclonal antibodies were used at 1:200 dilution.
Bullet points.
-
–
Developmental expression patterns of four Drosophila WASP family proteins provided
-
–
Developmental expression patterns of WASP family proteins throughout oogenesis
-
–
Developmental expression patterns of WASP family proteins throughout embryogenesis
-
–
WASP family members exhibit both overlapping and discrete expression patterns
-
–
WASP family expression reflects known functions and reveals potential new functions
ACKNOWLEDGEMENTS
We thank Jeffrey Verboon, James Watts, and Parkhurst lab members for their interest, advice and comments on the manuscript. We are very grateful to Liz Wayner in our Antibody Development Facility for generating the monoclonal lines used in this study, Toshi Tsukiyama for embryo extracts, the Developmental Studies Hybridoma Bank for distributing them, J.C. Sisson for the Lava lamp antibody, and the M.J. Murdoch Charitable Trust for the spinning disk microscope used for imaging. This work was supported by CSIC Mobility Fellowship PA1003163 to AERN and by NIH grant GM097083 to SMP.
Grant Sponsor: NIH; Grant Number: GM097083; Grant Sponsor: CSIC; Grant Number: PA1003163
REFERENCES
- Arora K, Nusslein-Volhard C. Altered mitotic domains reveal fate map changes in Drosophila embryos mutant for zygotic dorsoventral patterning genes. Development. 1992;114:1003–1024. doi: 10.1242/dev.114.4.1003. [DOI] [PubMed] [Google Scholar]
- Bastock R, St Johnston D. Drosophila oogenesis. Curr Biol. 2008;18:R1082–R1087. doi: 10.1016/j.cub.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Ben-Yaacov S, Le Borgne R, Abramson I, Schweisguth F, Schejter ED. Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J Cell Biol. 2001;152:1–13. doi: 10.1083/jcb.152.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Schafer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klambt C, Renkawitz-Pohl R, Onel SF. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci. 2008;121:1303–1313. doi: 10.1242/jcs.022269. [DOI] [PubMed] [Google Scholar]
- Bogdan S, Grewe O, Strunk M, Mertens A, Klambt C. Sra-1 interacts with Kette and Wasp and is required for neuronal and bristle development in Drosophila. Development. 2004;131:3981–3989. doi: 10.1242/dev.01274. [DOI] [PubMed] [Google Scholar]
- Bompard G, Caron E. Regulation of WASP/WAVE proteins: making a long story short. J Cell Biol. 2004;166:957–962. doi: 10.1083/jcb.200403127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol. 2011;193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2007;8:84–90. doi: 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB. Mdm2 widens its repertoire. Cell Cycle. 2007;6:827–829. doi: 10.4161/cc.6.7.4086. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Hsiue PP, Welch MD. The actin nucleation factor JMY is a negative regulator of neuritogenesis. Mol Biol Cell. 2011;22:4563–4574. doi: 10.1091/mbc.E11-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009;10:1043–1050. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 2011;331:1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch J, Ehinger J, Ladwein M, Rohde M, Derivery E, Bosse T, Steffen A, Bumann D, Misselwitz B, Hardt WD, Gautreau A, Stradal TE, Rottner K. Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell Microbiol. 2010;12:84–98. doi: 10.1111/j.1462-5822.2009.01380.x. [DOI] [PubMed] [Google Scholar]
- Harden N. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation. 2002;70:181–203. doi: 10.1046/j.1432-0436.2002.700408.x. [DOI] [PubMed] [Google Scholar]
- Harmon CB, Zelickson BD, Roenigk RK, Wayner EA, Hoffstrom B, Pittelkow MR, Brodland DG. Dermabrasive scar revision. Immunohistochemical and ultrastructural evaluation. Dermatol Surg. 1995;21:503–508. [PubMed] [Google Scholar]
- Harris TJ, Sawyer JK, Peifer M. How the cytoskeleton helps build the embryonic body plan: models of morphogenesis from Drosophila. Curr Top Dev Biol. 2009;89:55–85. doi: 10.1016/S0070-2153(09)89003-0. [DOI] [PubMed] [Google Scholar]
- He L, Wang X, Montell DJ. Shining light on Drosophila oogenesis: live imaging of egg development. Curr Opin Genet Dev. 2011;21:612–619. doi: 10.1016/j.gde.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Hu N, Castelli-Gair J. Study of the posterior spiracles of Drosophila as a model to understand the genetic and cellular mechanisms controlling morphogenesis. Dev Biol. 1999;214:197–210. doi: 10.1006/dbio.1999.9391. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol. 2002;156:677–687. doi: 10.1083/jcb.200109065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, Gandellini F, Stewart DM, Zhu Q, Nelson DL, Notarangelo LD, Ochs HD. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–4019. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. Gastrulation movements: the logic and the nuts and bolts. Dev Cell. 2005;8:305–320. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Mazumdar A, Mazumdar M. How one becomes many: blastoderm cellularization in Drosophila melanogaster. Bioessays. 2002;24:1012–1022. doi: 10.1002/bies.10184. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Gildor B, Shilo BZ, VijayRaghavan K, Schejter ED. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 2011;138:2347–2357. doi: 10.1242/dev.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Pearson AM, Baksa K, Ramet M, Protas M, McKee M, Brown D, Ezekowitz RA. Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect. 2003;5:815–824. doi: 10.1016/s1286-4579(03)00157-6. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev. 2001;11:374–383. doi: 10.1016/s0959-437x(00)00207-0. [DOI] [PubMed] [Google Scholar]
- Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, Johannes L. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schenck A, Qurashi A, Carrera P, Bardoni B, Diebold C, Schejter E, Mandel JL, Giangrande A. WAVE/SCAR, a multifunctional complex coordinating different aspects of neuronal connectivity. Dev Biol. 2004;274:260–270. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- Simoes S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombria JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Field C, Ventura R, Royou A, Sullivan W. Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol. 2000;151:905–918. doi: 10.1083/jcb.151.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Takeshima F, Anton I, Liu CH, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, Klein C, Davidson L, Bronson R, Mulligan RC, Southwick F, Geha R, Goldberg MB, Rosen FS, Hartwig JH, Alt FW. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci U S A. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE. Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol. 2002;12:705–711. doi: 10.1016/s0960-9822(02)00807-2. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Takenawa T. Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J Biol Chem. 2003;278:42515–42523. doi: 10.1074/jbc.M302177200. [DOI] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–789. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tal T, Vaizel-Ohayon D, Schejter ED. Conserved interactions with cytoskeletal but not signaling elements are an essential aspect of Drosophila WASp function. Dev Biol. 2002;243:260–271. doi: 10.1006/dbio.2002.0571. [DOI] [PubMed] [Google Scholar]
- VanHook A, Letsou A. Head involution in Drosophila: genetic and morphogenetic connections to dorsal closure. Dev Dyn. 2008;237:28–38. doi: 10.1002/dvdy.21405. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Machesky LM. The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol. 2004;16:174–181. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–2893. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayner EA, Garcia-Pardo A, Humphries MJ, McDonald JA, Carter WG. Identification and characterization of the T lymphocyte adhesion receptor for an alternative cell attachment domain (CS-1) in plasma fibronectin. J Cell Biol. 1989;109:1321–1330. doi: 10.1083/jcb.109.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8:756–763. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, Fujiwara Y, Bronson R, Snapper SB, Kirschner MW, Geha R, Rosen FS, Alt FW. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW. WASp is required for the correct temporal morphogenesis of rhabdomere microvilli. J Cell Biol. 2004;164:417–426. doi: 10.1083/jcb.200307048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]