Abstract
The Campylobacter jejuni JlpA protein is a surface-exposed lipoprotein that was discovered as an adhesin promoting interaction with host epithelium cells, an early critical step in the pathogenesis of C. jejuni disease. Increasing evidence ascertained that JlpA is antigenic, indicating a role of JlpA in immune response during the infectious process. Here, we report the crystal structure of JlpA at 2.7Å resolution, revealing a catcher's mitt shaped unclosed half β-barrel. Although the apparent architecture of JlpA is somewhat reminiscent of other bacterial lipoproteins such as LolB, the topology of JlpA is unique among the bacterial surface proteins reported to date and therefore JlpA represents a novel bacterial cell surface lipoprotein. The concave face of the structure results in an unusually large hydrophobic basin with a localized acidic pocket, suggesting a possibility that JlpA may accommodate multiple ligands. Therefore, the structure provides framework for determining the molecular function of JlpA and new strategies for the rational design of small molecule inhibitors efficiently targeting JlpA.
Keywords: Crystal structure, Bacterial lipoprotein, JlpA, Campylobacter jejuni
1. Introduction
In bacteria, cell surface-exposed lipoproteins are a set of proteins anchored to the outer membrane by virtue of their N-terminal modification with N-acyl-S-diacylglyceryl group. The ability of lipoproteins to decorate bacterial membranes provides for a wide variety of structural and functional roles in host-pathogen interactions, from surface adhesion to translocation of virulence factors into host cells (Tokuda and Matsuyama, 2004). Given their broad distribution among bacteria and their unique structural features, it is not surprising that at least one of the toll-like receptors (TLR2) is designed to detect lipoproteins as an innate immune response to the presence of bacteria (Aliprantis et al., 1999). Of note, surface-exposed lipoproteins that are essential for survival in the host are of considerable therapeutic interest as potential vaccine targets.
Campylobacter jejuni, a Gram-negative microaerophilic spiral ε-proteobacterium, is an important food-borne pathogen and a leading cause of human acute bacterial gastroenteritis worldwide (Poly and Guerry, 2008; Zilbauer et al., 2008). Symptoms of human C. jejuni infection range from mild diarrhea to dysentery characterized by severe abdominal pain, fever, vomiting, and inflammation. Of note, C. jejuni gastroenteritis has been linked to Guillian-Barré Syndrome (GBS), a devastating post-infection autoimmune neuropathy. The development of GBS is associated with lipooligosaccharides that mimic human gangliosides (Nachamkin et al., 2002). Despite the high occurrence of human disease and knowledge of multiple genome sequences, the pathogenesis of C. jejuni is still poorly understood. Moreover, considering the continuous problem of colonization of livestock combined with antibiotic resistance, it is imperative to further insights into how C. jejuni causes the observed clinical spectrum.
Adherence of bacteria to host epithelial cells is a crucial step in a bacterial infection. C. jejuni initially colonizes the small bowel and then moves to the colon, the target organ of disease. After successful colonization of the mucus lining of the gut, C. jejuni can adhere to epithelial cells (Poly and Guerry, 2008). Adherence of C. jejuni is a multifactorial process in which multiple binding factors may be required to bind to their specific receptors to achieve an efficient interaction with host cells. A number of C. jejuni proteins, such as PEB1, MOMP, CadF, JlpA, and CapA, have been reported to bind cultured epithelial cells. Disruption of the peb1A gene encoding PEB1 showed reduced C. jejuni adherence to human HeLa cells by 50 to 100 fold (Pei et al., 1998). The outer membrane protein MOMP encoded by the porA gene was found to bind to INT 407 cells (Moser et al., 1997). CadF appears to bind to cell matrix protein fibronectin, as a cadF mutant showed a 50% reduction in adhesion to human INT 407 cells compared to a wild-type C. jejuni isolate (Monteville et al., 2003). CapA, a putative autotransporter, has been reported to play a role in adherence and invasion of Caco-2 cells in vitro (Ashgar et al., 2007). However, the relative contribution as well as significance of these factors to disease remains uncertain, given the absence of small animal models of diarrhea for C. jejuni.
Another putative adhesin is the JlpA protein, a surface-exposed lipoprotein (Jin et al., 2001; Jin et al., 2003). The jlpA gene encodes JlpA of 372 amino acid residues with a molecular mass of 42.3 kDa. JlpA contains a typical signal peptide and lipoprotein-processing site at the N-terminus. Previous work demonstrated that the JlpA molecule contains lipid as confirmed by incorporation of [3H]-palmitic acid and binds to HEp-2 cells, establishing JlpA as a novel C. jejuni lipoprotein adhesin virulence factor (Jin et al., 2001). A jlpA mutant showed about 19% adherence to human HEp-2 cells relative to the wild-type strain, although a more recent study failed to detect any JlpA-mediated binding to chicken cells in vitro (Jin et al., 2001; Flanagan et al., 2009). Another study reported that JlpA interacts with HEp-2 cell surface heat shock protein 90α (Hsp90α) and initiates signaling pathways leading to activation of NF-κB and p38 MAP kinases, suggesting that JlpA contributes to the inflammatory responses associated with C. jejuni infection (Jin et al., 2003). Moreover, recent work has demonstrated that JlpA is a glycoprotein and is immunogenic during human infection (Scott et al., 2009). Here, we report the crystal structure of JlpA at 2.7Å resolution. The JlpA structure reveals an unclosed half β-barrel fold with a wide hydrophobic concave face, representing a novel bacterial surface lipoprotein.
2. Expression, purification and crystallization of JlpA
The DNA fragment encoding the mature form of JlpA lacking the signal peptide (JlpA18–372) was amplified by PCR using C. jejuni strain 81–176 genomic DNA and was inserted into NdeI / BamHI sites of pET19b (Novagen) to express an N-terminal His-tagged protein. For the phasing purpose, JlpA18–372 variants (JlpAI45M/I160M, JlpAI45M/L284M, JlpAI160M/I284M, and JlpAI45M/I160M/I284M) were produced by using the plasmid pET19b∷JlpA18–372 as template and QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene).
The plasmids containing the genes encoding JlpA18–372 and its variants were expressed in E. coli BL21(DE3) or B384(DE3) using liquid Luria-Bertani or SelenoMet (Molecular Dimensions Ltd.) media supplemented with 100 μg/ml ampicillin. Bacteria were grown to an A600 0.8 at 37°C, and expression was induced with 0.2 mM IPTG at 23°C for 3 h. Cells were harvested by centrifugation and resuspended in buffer-A (20 mM Tris-HCl, pH 8.0, 250 mM NaCl, 5 mM β-mercaptoethanol) containing 10 mM imidazole, 0.1 % Triton X-100 and protease inhibitors (Roche). After sonication and centrifugation, supernatants were subjected to a binding reaction with Ni-NTA agarose (Qiagen) for 30 min. Protein-resin complexes were packed onto a column and proteins were eluted using a step gradient method using 50, 100, 250, and 500 mM imidazole in buffer-A. Subsequently, fractions containing target proteins were purified using a HiTrap-Q column (GE Healthcare) with a linear gradient of 50 mM to 1M NaCl in 20 mM Tris-HCl (pH 8.0). The peak fractions were further purified using a HiPrep 16/60 Sephacryl S-100 HR column (GE Healthcare) equilibrated with buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl. Purified JlpA and its variants were concentrated to ~7 mg/ml for crystallization. Crystals of both native and selenomethionine-derivatized JlpA (SeMet-JlpA) were obtained at 16°C using the hanging drop vapor diffusion method with a reservoir solution containing 32–35% (v/v) 2-methyl-2,4-pentanediol (MPD), 0.03M CaCl2 and 100 mM Na-acetate, pH 5.6 (see Supplementary Information for further details of purification and crystallization).
3. Data collection, structure determination and refinement
Since crystals were extremely unstable, single crystals were swiftly collected from crystallization drops and flash-cooled in liquid nitrogen, without additional cryo-protectant. Data sets were collected at 100K using native crystals and SeMet-JlpA crystals on Advanced Photon Source beamline 19ID, Argonne (Table 1). Data sets were processed with HKL3000 (Minor et al., 2006).
Table 1.
Statistics from crystallographic analysis.
| Data Collection | Native | Se-Met | ||
|---|---|---|---|---|
| peak | inflection | remote | ||
| Wavelength (A) | 0.97943 | 0.97940 | 0.97954 | 0.97172 |
| Space group | P41212 | P41212 | ||
| Cell parameters (Å) | a = b = 111.63, c = 170.40 | a = b = 111.56, c = 171.51 | ||
| Resolution (Å) | 20.00–2.70 (2.80–2.70) | 50.00–2.95 (3.00–2.95) | 50.00–2.95 (3.00–2.95) | 50.00–3.20 (3.26–3.20) |
| Reflections (Total / Unique) | 147966 / 27364 | 205330 / 22977 | 179940 / 22384 | 145684 / 17951 |
| Completeness (%)a | 90.5 (76.0) | 97.5 (84.2) | 94.6 (72.9) | 96.4 (80.7) |
| Rmerge (%)a,c | 6.6 (33.9) | 7.9 (36.4) | 7.7 (37.6) | 7.6 (33.8) |
| Mean <I/σ(I)>a | 22.8 (2.2) | 28.6 (2.4) | 25.0 (2.1) | 22.3 (2.5) |
| Redundancya | 5.5 (2.8) | 9.0 (5.1) | 8.1 (4.1) | 8.2 (4.6) |
| Phasing | ||||
| Overall FOM after SHARP phasingb (centric/acentric) | 0.19728 / 0.31024 | |||
| Structure Refinement | ||||
| Resolution (Å) | 2.70 | |||
| No. reflections (working/test) | 25985 / 1371 | |||
| R(%)a, d/Rfreee(%) | 24.2 / 27.2 | |||
| Total number of atoms | 5510 | |||
| Protein molecules | 2 | |||
| Average B factor (Protein Å2) rms deviation | 80.3 | |||
| Bonds(Å) | 0.019 | |||
| Angles (°) | 1.67 | |||
| Ramachandran plot (%) | ||||
| Most favored regions | 88.8 | |||
| Additional allowed regions | 10.9 | |||
| Generously allowed regions | 0.3 | |||
The values in parentheses correspond to statistics for the highest resolution shell.
The figure of merit .
, where Ii is the intensity of an observation, 〈I〉 is the mean value for that reflection, and the summations are over all equivalents.
, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated with 5% of the data excluded from the refinement.
In initial experiments we performed multiple-wavelength anomalous dispersion (MAD) phasing (Hendrickson et al., 1985) using a SeMet-JlpA crystal that diffracted to 3.3Å resolution. However, the electron density map prevailed with β strands, which appeared to form a large β-sheet, and building a structure model was not possible. In fact, JlpA (strain 81–176) contains 5 Met sites (excluding the first Met site), but three of them were clustered at the C-terminal region of JlpA, suggesting uncertain occupancy of `Se' sites for these Met residues in MAD phasing. To improve phasing, JlpA variants containing 2 to 3 additional Met sites were produced, as described above. Among these, one variant, JlpAI45M/I160M, yielded crystals. Along with improved cryo-crystallography, the structure was finally solved by MAD phasing from a SeMet-JlpAI45M/I160M crystal using autoSHARP (Vonrhein et al., 2006). After solvent flattening, a starting model was generated by RESOLVE (Terwilliger, 2000). When one of two molecules in the asymmetric unit was manually traced ~95% by COOT (Emsley and Cowtan, 2004), the structure model was used as a template for molecular replacement with program MOLREP within the CCP4 suite (CCP4, 1994) against native crystal datasets.
Further rounds of model building and refinement were performed by using COOT, CNS 1.21 (Brunger, 2007), PHENIX (Terwilliger et al., 2008) and REFMAC5 (Murshudov et al., 1999). Final refinement cycles were carried out by REFMAC5 incorporating TLS corrections (Winn et al., 2003). Structural validation was performed with the Protein Data Bank validation server (http://deposit.pdb.org/validate/). Crystallographic parameters are listed in Table 1.
4. Molecular architecture of JlpA
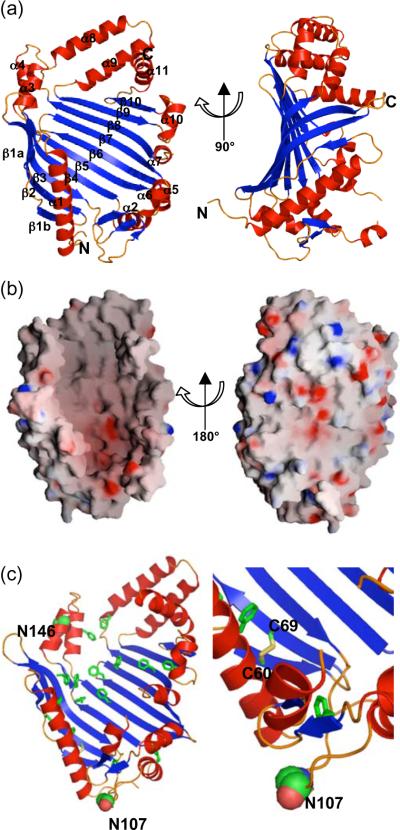
JlpA adopts a single domain structure with an α/β-fold composed of curved 10-stranded antiparallel β-sheet, which forms a large central β sheet, and 11 α-helices with 3 small strands, which encircle the central β sheet (Fig. 1a). The large β-sheet has the strand order β1 to β10. The JlpA structure has a `catcher's mitt' shape with overall dimension of ~80 × 50 × 25 Å (Fig. 1b). One remarkable feature of the JlpA structure is the partition of the molecular surface: while the concave side forms a wide hydrophobic basin with localized acidic pockets, the convex face reveals a polar surface throughout (Fig. 1b). While the floor of the concave face made by the major β-sheet is rather flat, all α-helices together with loop regions and three minor β-strands are arranged to create the basin walls. The hydrophobic basin is rich in Phe residues (Fig. 1c). The overall basin results in a surface area of ~4041 Å2 and a volume of ~13,197 Å3. Two internal Cys residues of JlpA, Cys-60 of β1 and Cys-69 of β2, form a disulfide bond in the crystal structure (Fig. 1c).
Figure 1.
Structure of JlpA. (a) Cartoon representation of two views of JlpA. The secondary structural elements are labeled. “N” and “C” indicate the N-terminus and C-terminus of JlpA. (b) Surface electrostatic potential of JlpA. The surface was contoured and displayed using the program GRASP (Nicholls, 1991). Color-coding is according to charge, with blue for the most positive regions (25 kbT), red for the most negative regions (−25 kbT), and linear interpolation in between. (c) Phe residues (in stick representation) lining of the hydrophobic basin. Two Asn sites of N-glycosylation, N107 and N146, are shown in sphere representation. The disulfide bond Cys60–Cys69 is indicated in stick model with a close-up view (right panel).
Two protomers in the asymmetric unit are very similar and superimposed with an RMSD of 0.3 Å (Fig. 2a and b). Yet, localized conformation differences between two protomers are observed for several regions that correspond to highly flexible loops and/or peptide segments with missing electron density residues (Fig. 2b). Interestingly, these regions are all mapped on top of the basin walls, including the connecting loop between α1 and β1 (47QDSGI51), a large segment between β2 and β3 (76TLAKDNNDEYQELF89), the C-terminal end of α3 and the loop between α3 and α4 (144DINASLFQQDP154), a region including α6 and α7 (233INELLNMVNYEQASDFS249), connecting loop between α8 and α9, helix α10, and the C-terminal helix α11 (Fig. 3a and b). The crystallographic interface between two protomers in the asymmetric unit is formed by the N-terminal region and a portion of β1 and β2, resulting in a very small molecular contact surface (Fig. 2a). Interestingly, cysteine residues Cys-18 (the lipidation site of JlpA in vivo) of two protomers make an inter chain disulfide bond (Fig. 2a). Considering the little molecular contact of the interface, this disulfide bond appeared to be important for crystallization and crystal packing and irrelevant to the JlpA function. Taken together, the functional assembly of JlpA is a monomer both in solution and crystals.
Figure 2.
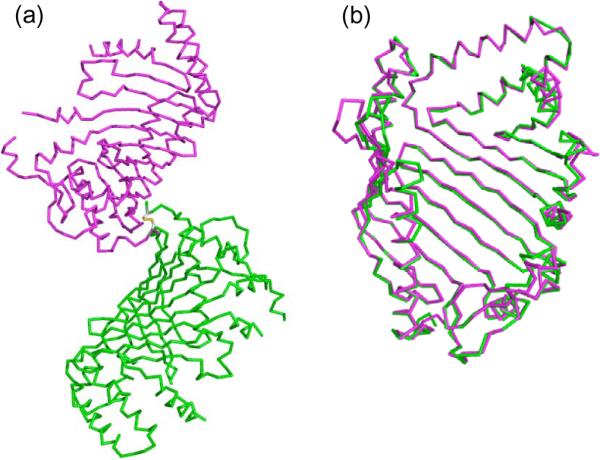
Two protomers in the asymmetric unit. (a) The crystal packing of the two JlpA molecules (protomer A, magenta; and protomer B, green). The inter chain disulfide bond Cys18-Cys18 is indicated in stick model. (b) Superposition of two protomers in the asymmetric unit.
Figure 3.
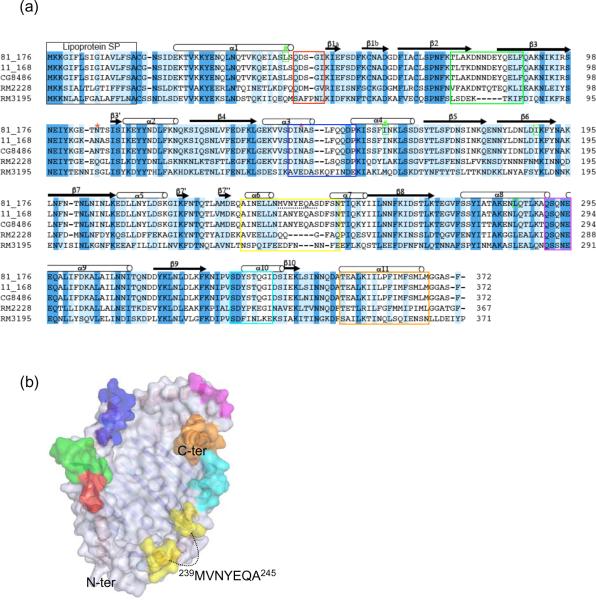
Sequence alignment of JlpA homologs. (a) Sequence alignment of JlpA proteins from 4 representative strains of Campylobacter spp. Four positions (Leu or Ile) selected for `Met' mutation are indicated with green boxes. Two Met introduced sites yielding crystals are highlighted with `#'. The secondary structural elements are shown above the sequences. (b) Location of variable regions on the molecular surface of JlpA. The color-coding corresponds to the boxed peptide regions in red, green, yellow, magenta, and orange in A. The dashed line indicates 239MVNYEQAS245, a segment disordered in the crystal structure.
5. Structural conservation across the JlpA family and antigenicity
Alignment of JlpA from multiple strains of C. jejuni and other Campylobacter species illustrates remarkable sequence conservation [~97% identity within C. jejuni isolates (GenBank accession numbers: YP_001000663, YP_002344378, and AAG29817), ~50% identity between C. jejuni and C. coli (GenBank: EAL56375) JlpAs, and ~36% identity between C. jejuni and C. upsaliensis (GenBank: EAL52450) JlpAs], suggesting that they share the same overall structure. Especially, the two internal cysteines Cys-60 and Cys-69 forming a sulfide bond are strictly conserved. In considering that a number of secreted and membrane proteins in both bacteria and eukaryotes contain disulfide bonds, the disulfide bond between Cys-60 and Cys-69 may play an important role in the folding and stability of JlpA in the course of secretion to the cell surface.
Among the JlpA sequences from available genomes, JlpA from C. upsaliensis RM3195 (Fouts et al., 2005) represents the least conserved sequence (Fig. 3a). Interestingly, the regions corresponding to variable conformations between two protomers in the crystal structure are particularly divergent in C. upsaliensis RM3195 (Fig. 3b). While the connecting loop between β2 and β3 (70DNNDE84) is absent in RM3195, the region corresponding to loop between α3 and α4 has a short insertion. The connecting loop between α6 and α7 (239MVNYEQA245) is highly dissimilar in RM3195. The antigenic properties of different portions of the JlpA primary structure have not been studied. Therefore, the JlpA structure is very useful to map and probe peptide regions involved in immune responses (Fig. 3b). Indeed, the structure reveals potential epitopes that correspond to several protruding flexible surface loops or peptide segments. These regions in JlpA are invariant among C. jejuni strains and therefore represent obvious targets for use in diagnostic tests and vaccination experiments against C. jejuni.
6. Structural context for JlpA N-linked glycosylation
The C. jejuni genome encodes the PglB oligosaccharide transferase that is responsible for transfer of the heptasaccharide to periplasmic and surface exposed proteins containing an appropriate sequon (D/E-x-N-x-S/T) (Kowarik et al., 2006). JlpA contains two highly conserved N-glycosylation sites, Asn-107 within the sequon 105ETNTS109 and Asn-146 with the sequon 144DINAS148 (Scott et al., 2009). The structure reveals that Asn-107 is in a large loop between β3 and β3' and exposed to solvent (Fig. 1c and 3a), suggesting that the heptasaccharide could be attached to Asn-107 without significant rearrangement of the local secondary structures. On the other hand, Asn-146 occupies the last turn of α helix α3 such that the side chain is partially exposed to solvent. JlpA exists as 3 discrete forms (unglycosylated, singly glycosylated at Asn-146, and doubly glycosylated at Asn-107/Asn-146), suggesting that glycosylation first takes place at Asn-146 and the Asn-107 site cannot be glycosylated without prior glycosylation at Asn-146 (Scott et al., 2009). In fact, the Asn-107 site is close to the N-terminus, implying that this sequon is likely membrane proximal, whereas the Asn-146 is away from the N-terminus (Fig. 1c). Thus, the Asn-146 sequon would be much more accessible to PglB than the Asn-107 sequon, giving an explanation for the preferential glycosylation at the Asn-146 site. It is not known whether or not common structural contexts or requirements exist for bacterial N-linked glycosylation, since only a few structures of C. jejuni glycoproteins have been experimentally determined. Thus, the JlpA sequons described here contribute to valuable information for furthering the mechanism of bacterial N-glycosylation and the design of glycosylation sites into other acceptor proteins in order to generate novel glycoconjugates.
7. Structural comparison with other proteins
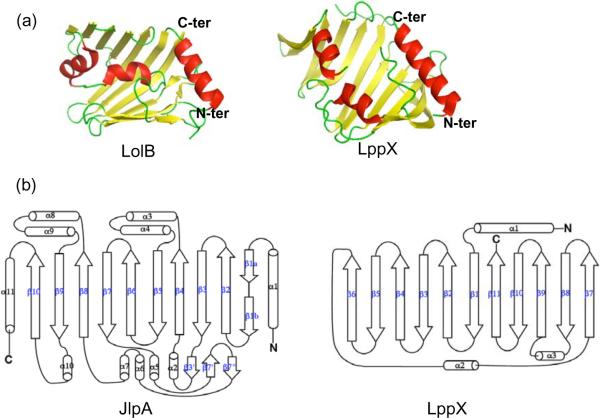
At the primary structure level, JlpA does not have any significant homologs from other organisms. DALI searches for structural homologs of JlpA revealed a few proteins with Z-scores above 7. Yet, all of these proteins were aligned to JlpA with an RMSD over 4. Essentially, all of the top-scored proteins as structural homologs were large bacterial outer membrane proteins such as FauA, FhaC, and OmpG, likely reflecting the β-barrel like topology of JlpA (unclosed half barrel). On the other hand, several bacterial lipoproteins were noteworthy from literature surveys, including LolA, LolB, and LppX (Fig. 4a), albeit DALI searches did not define them as structural homologs. LolA (22.6 kDa) and LolB (23.5 kDa) are lipoprotein localization factors in E. coli (Takeda et al., 2003). LppX (24 kDa) is a lipoprotein of Mycobacterium tuberculosis, required for the translocation of complex lipids such as phthiocerol dimycocerosates (DIM) (Sulzenbacher et al., 2006). The bacterial lipoproteins, LolA, LolB, and LppX, are much smaller than JlpA (42 kDa) and have the β-sheet topology with the strand order β7/ β8/ β9/ β10/ β11/ β1/ β2/ β3/ β4/ β5/ β6, unlike the JlpA topology with the β strand order of 1 to 10 (Fig. 4b). Yet, despite the absence of sequence homology, the apparent structure of JlpA is reminiscent of these bacterial lipoproteins, particularly since LolA, LolB, and LppX also have a hydrophobic cavity in the concave face of the molecule.
Figure 4.
JlpA and LolA/B-like lipoproteins. (a) For clarity, only LolB (PDB: 1IWM) and LppX (PDB: 2BYO) are shown. LolB is an E. coli lipoprotein localization factor and LppX is an M. tuberculosis lipoprotein required for transport of complex lipids to the outer layers of cells. (b) Topology of JlpA (left) and LppX (right). Different orders of β stands are notable.
8. Putative structure-function relationship of JlpA
The predicted lipid-modified N-terminus is located at the one end of the major axis of the central β-sheet (Fig. 1a). The structural features such as protruding flexible surface loops and the position of the N-terminus suggest a topology of JlpA in the outer membrane such that these flexible surface loops are oriented toward the extracellular space. C. jejuni expresses a phase-variable polysaccharide capsule (Bacon et al., 2001) and JlpA would then have not enough capacity to protrude through the bacterial cell surface when the polysaccharide capsule is expressed, since only 5 unstructured residues are available for spacing between the lipidation cysteine residue and helix α1. Likely, JlpA is not extended beyond the polysaccharide layer, which is reminiscent of other bacterial antigenic outer membrane proteins such as the Neisserial surface protein A (NspA). The NspA protein is a homolog of the Opa proteins, which mediate adhesion to host cells, and appears to elicit antibodies. NspA is a β-barrel outer membrane protein having a hydrophobic cleft surface at the extracellular side (Vandeputte-Rutten., 2003). Along these lines, JlpA appears to function as an antigen and an adhesin analogous to antigenic outer membrane proteins, but without being an integral membrane protein.
Adhesive activity of JlpA was previously studied by using HEp-2 cells and a recombinant GST-fusion form of JlpA (Jin et al., 2001). In our ongoing study, we have investigated JlpA binding to the more biologically relevant human intestinal INT407 cells. His-tagged JlpA was fluorescently labeled and added in increasing concentrations to INT407 cells in suspension and binding was monitored by flow cytometry (see Supplementary Information). Unexpectedly, concentrations of JlpA as high as 300 μg/ml failed to reach saturation and 100-fold excess of unlabeled protein was unable to compete for binding with labeled JlpA, while as a control a recombinant form of the B subunit of the heat labile enterotoxin (LTB) of enterotoxigenic E. coli showed saturation in binding under similar conditions (Fig. S1). In view of a large concave surface in the JlpA structure, we speculate that JlpA may contain multiple ligand-binding sites, which may make functional probing by mutagenesis of a single protein ineffective. Further investigations are necessary to decipher the molecular function of the protein.
In summary, the three-dimensional structure of JlpA reveals a unique topology among known bacterial lipoproteins. Nonetheless, the observation that JlpA has an α/β-fold with a large curved β-sheet forming hydrophobic concave surface, characteristics of LolA/LolB proteins and LppX-like proteins, raises a question about a similar role for JlpA as a lipid carrier or translocator of as yet unidentified Campylobacter-specific lipids. Screening of cell extract based ligands would be an interesting approach in the future.
Supplementary Material
Acknowledgements
We thank Stephen J. Savarino for the gift of recombinant LTB and Lanfong Lee for providing wildtype JlpA for assay development. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (19ID) at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported in part by NIH grant AI068943 and Grant E-1616 from the Welch Foundation to H-JY and by the Military Infectious Disease Research Program Work Unit 6000.RAD1.DA3.A0308 to PG. MP is a military service member and PG is a civilian employee of the U. S. government. This work was prepared as part of their official duties. Title 17 USC 105 provides that `Copyright protection under this title is not available for any work of the U. S. government. Title 127 USC 101 defines a US government work as a work prepared by a military service member or employee of the U. S. government as part of that person's official duties.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3UAU.
References
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- Ashgar SS, Oldfield NJ, Wooldridge KG, Jones MA, Irving GJ, et al. CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut. J. Bacteriol. 2007;189:1856–1865. doi: 10.1128/JB.01427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, et al. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 2001;40:769–777. doi: 10.1046/j.1365-2958.2001.02431.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 Suite: Programs for Protein Crystallography”. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 2009;77:2399–2407. doi: 10.1128/IAI.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA, Smith JL, Sheriff S. Direct phase determination based on anomalous scattering. Methods Enzymol. 1985;115:41–55. doi: 10.1016/0076-6879(85)15006-8. [DOI] [PubMed] [Google Scholar]
- Jin S, Joe A, Lynett J, Hani EK, Sherman P, et al. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 2001;39:1225–1236. doi: 10.1111/j.1365-2958.2001.02294.x. [DOI] [PubMed] [Google Scholar]
- Jin S, Song YC, Emili A, Sherman PM, Chan VL. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90alpha and triggers signalling pathways leading to the activation of NF-kappaB and p38 MAP kinase in epithelial cells. Cell Microbiol. 2003;5:165–174. doi: 10.1046/j.1462-5822.2003.00265.x. [DOI] [PubMed] [Google Scholar]
- Kowarik M, Young NM, Numao S, Schulz BL, Hug I, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Monteville MR, Yoon JE, Konkel ME. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology. 2003;149:153–165. doi: 10.1099/mic.0.25820-0. [DOI] [PubMed] [Google Scholar]
- Moser I, Schroeder W, Salnikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol. Lett. 1997;157:233–238. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Liu J, Li M, Ung H, Moran AP, et al. Campylobacter jejuni from patients with Guillain-Barré syndrome preferentially expresses a GD(1a)-like epitope. Infect. Immun. 2002;70:5299–5303. doi: 10.1128/IAI.70.9.5299-5303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Pei Z, Burucoa C, Grignon B, Baqar S, Huang XZ, et al. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 1998;66:938–943. doi: 10.1128/iai.66.3.938-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F, Guerry P. Pathogenesis of Campylobacter. Curr. Opin. Gastroenterol. 2008;24:27–31. doi: 10.1097/MOG.0b013e3282f1dcb1. [DOI] [PubMed] [Google Scholar]
- Scott NE, Bogema DR, Connolly AM, Falconer L, Djordjevic SP, et al. Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 2009;8:4654–4664. doi: 10.1021/pr900544x. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Canaan S, Bordat Y, Neyrolles O, Stadthagen G, et al. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 2006;25:1436–1444. doi: 10.1038/sj.emboj.7601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Miyatake H, Yokota N, Matsuyama S, Tokuda H, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 2003;22:3199–3209. doi: 10.1093/emboj/cdg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P. Crystal structure of Neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J. Biol. Chem. 2003;278:24825–24830. doi: 10.1074/jbc.M302803200. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol. Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Zilbauer M, Dorrell N, Wren BW, Bajaj-Elliott M. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans R. Soc. Trop. Med. Hyg. 2008;102:123–129. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.