Abstract
The microenviroment contributes to directing mammary epithelial cell (MEC) development and the progression of breast cancer. Three-dimensional culture models have been used to support formation of structures that display varying degrees of disorganization that parallel the degree of cancer. Synthetic hydrogels were employed to investigate the mechanisms by which specific adhesion signals in the microenvironment directed development. Polyethylene glycol-based hydrogels supported 3D growth of MECs and directed formation of a range of phenotypes that were functions of genotype, and identity and concentration of adhesion peptides RGD and YIGSR. Non-cancerous and cancerous MECs responded differentially to the same adhesion cues and produced variable structural organizations. An analysis of dynamic signaling pathways revealed differential activities of transcription factors within the MAPK and JAK/STAT pathways in response to genotype and adhesion. These results directly implicate adhesion in cancer development and demonstrate that AP1, CREB, STAT1, and STAT3 all contribute to the genotype dependence of cellular response to adhesion peptides. The tools presented in this work could be applied to other systems and connect extracellular cues with intracellular signaling to molecularly dissect tissue development and further biomaterials development.
1. Introduction
Tissue development occurs through a complex spatial and temporal organization of cells and extracellular matrix (ECM) proteins, and the formation and maintenance of specific cellular architectures are essential to normal function and homeostasis. The organizations of tissues are drastically altered in the progressions of cancers, with the degree of disorder paralleling the progression of disease development [1, 2]. These altered phenotypes result from differential patterns of cell growth, cell-cell communication, and ECM remodeling that manifest when cells acquire cancerous mutations and differentially express genes. As cancer progresses, cells influence the microenvironment around them by secreting ECM proteins and matrix-digesting proteases [3, 4]. Tissue density can also be affected [5] as rearrangements of ECM proteins alter mechanical forces within tissues [6]. The impact of cells on the microenvironment during cancer is multifaceted and interconnected.
Whereas cells can transform microenvironments as cancer progresses, signals within the microenvironment can reciprocate and transform cells. As such, cancerous growth can be amplified by establishing feedback loops between the microenvironment and mutated cells or differentially signaling non-mutated cells [6, 7]. Signals transmitted by the microenvironment stem from ECM proteins, which have biochemical and mechanical contributions, and soluble molecules, such as growth factors, which are secreted by cells [8]. Adhesion signals play important roles in signaling cells during cancer progression, as the identity and presentation of adhesion sequences can differentially affect growth [9, 10]. As cells degrade matrices through proteolysis, soluble fragments of ECM proteins are generated and ligate cells, which can induce cancer-like behaviors such as structural reorganization and migration [11–13] and upregulation of proteases leading to greater matrix degradation [3, 14]. Tumor aggressiveness has also increased in the cases of growing non-invading cancerous cells in the presence of stromal cells derived from cancerous tissue [15] or in matrices of increasing rigidity [16, 17], a characteristic of cancerous tissue [5]. Alternatively, the microenvironment can direct malignant cells to exhibit less disorganized phenotypes. Preconditioning a matrix with embryonic stem cells [18] and blocking ECM protein binding with an antibody against β1-integrin [19] have each diminished cell invasiveness. The microenvironment is thus an important contributor to the phenotypes of tissues and interplays with the genotypes of cells.
The contributions of the microenvironment on tissue phenotype have been investigated using three-dimensional (3D) culture systems, which are able to replicate many of the phenotypes observed in native tissues [20]. 3D matrices have importance in cancer biology since they support development of normal and cancerous cells into tissues of varying disorder and distinctive architectures that are not observed in 2D monolayers [20, 21]. Mutations that are associated with cancerous processes in native tissues manifest into different tissue architectures in 3D systems [22, 23]. Further, gene expression patterns from breast cancer cells cultured in 3D have been correlated with patient survival [24], and cells have also shown hormone and therapeutic responsiveness that is more faithful to in vivo responses [25, 26]. However, to date, 3D studies with cancer cells have been largely performed in natural matrices, which contain numerous signals that complicate the performance of mechanistic studies. Synthetic systems provide an alternative 3D culture system in which the biological cues can be modulated. Systems based on poly(ethylene glycol) (PEG) have become widely used in regenerative medicine for tissue development [27–30] and stem cell differentiation [31–33], but have been used less frequently to study the contributors to abnormal growth, such as signals associated with cancer. A desirable quality about PEG hydrogels is their ability to be controlled for adhesion, degradation, and mechanical properties [34], and present cells with a range of microenvironments to investigate their influences on tissue phenotype.
In this report, we have applied a PEG-based 3D culture platform to investigate the impact of cell adhesion on structural phenotype and signaling pathways during normal and cancerous mammary tissue development. Synthetic hydrogels were initially screened for cell viability and the ability to reproduce structures observed in Basement Membrane Extract (BME) (i.e. Matrigel). Peptide motifs derived from ECM proteins were then used to investigate the influence of adhesion identity and concentration on tissue architectures formed by cells derived from wild-type or transformed non-cancerous breast tissue or poorly-differentiated breast tissue. The influence of adhesion peptides on cell growth was further characterized by analyzing dynamic patterns of signaling pathways. The goal of this work was to identify the mechanisms by which specific adhesion signals can impact tissue development within a minimal context and the differential response due to the genotype.
2. Materials and Methods
2.1 Cell maintenance
MCF-10A (10A) cells and MCF-10A.ErbB2 (10A.ErbB2) cells, 10A cells stably overexpressing ErbB2 receptors using retroviral infection [22], were generously provided by H. Band (U. Nebraska, Omaha, NE) and maintained in DFCI media [35]. MDA-MB-231 (231) cells were generously provided by J. S. Jeruss (Northwestern University, Chicago, IL) and maintained in DMEM/F-12 media. All three cell lines were cultured at 37°C and 5% CO2 in a humidified incubator.
2.2 Synthetic hydrogel materials
Poly(ethylene glycol) tetravinyl sulfone (PEG-VS) was synthesized from 20 kDa 4-arm PEG-OH (Creative PEGworks, Winston Salem, NC) using techniques previously described [27, 30]. Adhesion peptides Ac-GCGYGRGDSPG (RGD) and Ac-GCGYIGSRSPG (YIGSR), nonadhesive scramble peptide Ac-GCGYGRDGSPG (RDG), plasmin-sensitive degradable crosslinking peptide Ac-GCYK↓NRGCYK↓NRCG, and non-degradable D-isomerized crosslinking peptide Ac-GCYDKNDRGCYDKNDRCG were synthesized and purified at the Northwestern University IBNAM Chemistry Core.
2.3 3D BME culture
Working over ice, one part cells in phosphate buffered saline (PBS) was blended with four parts phenol red-free, growth factor reduced BME (Trevigen, Gaithersburg, MD) and 50 µl of solution was deposited into cell culture plate inserts. 10A and 10A.ErbB2 cells were each encapsulated at 2×105 cell/ml and 231 cells were encapsulated at 1×106 cell/ml. Hydrogels were incubated at 37°C for 45 min and the corresponding media was subsequently added above and below the insert. Hydrogels were maintained for 10 d with media changes every other day.
2.4 3D PEG culture
PEG-VS was dissolved in HEPES buffered saline (HBS) (1 M HEPES, 0.1 M NaOH, pH 7.6) such that its final concentration when blended with peptides and cells would be 7.5%. RGD, RDG, or YIGSR were dissolved in HBS and reacted with PEG-VS at 37°C for at least 30 min. Cells in HBS (5×106 cell/ml) and crosslinking peptide in HBS were blended with PEG-VS at a ratio of 1:1:3 cells:crosslinking peptide:PEG-VS/adhesion peptide. Droplets of 5 µl were cast in triplicate on ethanolized Parafilm and incubated at 37°C for 15 min. Hydrogels were then transferred to wells of a 96-well plate and repeatedly washed with the corresponding cell media during the first few hours. Hydrogels were cultured for 10 d with media changes daily for the first three days followed by media changes every other day.
2.5 Evaluation of cell viability
After 10 d of culture in PEG, cell viability was assessed by two methods in parallel: live/dead staining or alamarBlue. For live/dead staining, hydrogels were stained with Calcein AM and Ethidium Homodimer (Invitrogen, Carlsbad, CA) for 50 min. Fluorescence images with a 5× objective were taken using an epifluorescence microscope (Leica, Bannockburn, IL). Alternatively, viability was assessed using alamarBlue (Invitrogen) following manufacturers instructions. In short, media was replaced with media containing 10% (v/v) alamarBlue solution and hydrogels were incubated at 37°C for 4 h. Media was subsequently transferred to a 96-well plate and fluorescence was measured at 565/595 using a Synergy 4 microplate reader (Biotek, Winooski, VT).
2.6 Immunfluorescence confocal micrscopy
BME and PEG hydrogels were both processed the same way using an adaptation of fixing and staining techniques for overlay BME culture [23]. Cells were fixed for 30 min with 4% paraformaldehyde, washed with PBS containing 100 mM glycine, permeabilized with 0.5% Triton-X in PBS for 5 min, washed with immunofluorescence buffer (IF) [23], blocked with 2% bovine serum albumin for 1 h, stained with anti-laminin V-AlexaFluor 488 conjugated protein (1:200 dilution; Millipore, Billerica, MA) overnight, washed with IF, counterstained with TOPRO-3 (5 µM, Invitrogen) for 10 min, and washed with PBS. BME hydrogels within culture inserts were transferred to a 100 mm culture dish after cutting the membrane of the insert. PEG hydrogels were transferred directly to a dish using forceps. Hydrogels were immersed in PBS and a 40× immersion lens was used to capture images using an upright Leica confocal microscope.
2.7 Structure assessment
Images for all conditions from at least three independent experiments were used for analysis. To determine sizes of structures, phase images of hydrogels after 10 d of culture were taken using a phase microscope (Leica). ImageJ was used to correlate pixel size with physical diameters of structures and the raw diameters were normalized by the average diameters for blank hydrogel conditions. Images taken using confocal microscopy were used to semiquantitatively analyze samples according to the following: for nuclear staining, structures were classified as (I) immature with few cells, (II) spherical with a filled lumen, (III) spherical with a hollow lumen, (IV) non-spherical but organized, or (V) non-spherical and disorganized; for laminin V staining, the distribution was classified as (I) immature and mostly cytoplasmic, (II) mixed cytoplasmic with some basally localized, or (III) mostly basally localized. Total populations of each category were manually counted.
2.8 Lentivirus constructs and production
Transcription factor (TF) reporter constructs containing enhancer elements for AP1, CREB, STAT1, or STAT3, along with a negative control construct without enhancer element (TA) (Panomics, Madison, WI), were transferred to an HIV-1 backbone vector (VSV-G). Lentiviruses were produced and concentrated as described previously [36].
2.9 Signaling pathway profile analysis
10A, 10A.ErbB2, or 231 cells were transduced in parallel with a lentivirus TF-specific reporter gene (AP1, CREB, STAT1, STAT3, TA, or PBS negative control; 1e4 physical particles/cell) by centrifugation at 800 g and 32°C for 45 min and seeded into wells of a polystyrene plate. After 3 d of culture, cells were split and seeded into PEG hydrogels containing 1 mM RDG, RGD, or YISGR in triplicate as described in a previous section. On days 1, 3, 6, and 10 d, 1 mM d-luciferin (Caliper) was added to wells and light production was quantified with bioluminescence imaging as described previously [37, 38]. Light fluxes were transformed such that the values at 1 d of imaging were the same across all replicates and fluxes at each time for each TF were normalized by the average TA value for each adhesion condition. Cell types were measured in independent experiments, each carried out in three independent experiments.
2.10 Statistical analysis
All statistical analyses were performed using the open software R [39]. For cell viability and size data, a student’s t-test with false discovery rate (FDR) adjustment for multiple comparisons was used (α=0.05). For categorical data, a χ2 test was performed to determine if there were significant differences in the populations (α=0.05). Correspondence analysis was performed using the ‘ca’ package developed for R [40]. With this approach, data were mapped onto single points in Euclidian space. The linear distance between points of the same variable correlated with similarity (closer points were more similar) and the angular distance from the origin between points of different variables corresponded to the strength of associations (points with similar angular distances were better associated). For analysis of signaling pathway data, only data above background determined by a student’s t-test (α=0.01) were employed. A multiple regression model was used on log2-transformed data and p-values were corrected by FDR adjustment. Models for each TF were as follows:
where TFi is the normalized TF activity. Cell and Peptide are qualitative variables that are represented by the combination of two binary variables. For this system, the reference condition is 10A cells and blank hydrogels. Time is the temporal series at which TF activity was measured, 1, 3, 6 and 10 d. Final models were obtained by stepwise removal of terms that had a p-value>0.01, starting with interaction terms that had the greatest p-values. Post-hoc multiple testing was used to calculate significant differences between time, peptide condition, and cell type by FDR (α=0.05).
3. Results
3.1 3D phenotypes of MECs in natural system
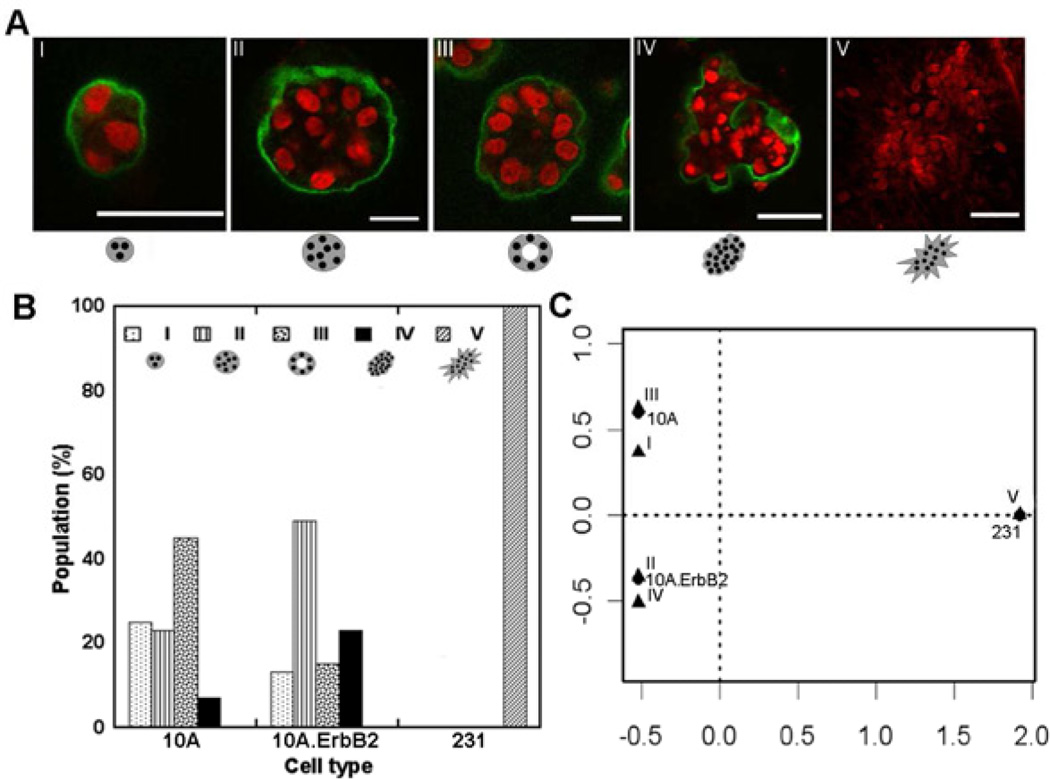
BME is a natural matrix capable of supporting growth of both non-cancerous and cancerous MECs into distinct phenotypes. 10A, 10A.ErbB2, or 231 cells, representing three different stages of breast cancer, were cultured. 10A cells are a MEC line derived from non-cancerous tissue [14, 41]. 10A.ErbB2 cells are an ErbB2 (HER2/Neu) over-expressing cell line that represented a premalignant transformation commonly observed in breast cancer [22]. 231 cells were derived from an invasive cancer and represent a poorly-differentiated stage that has acquired numerous cancerous mutations [14]. The observed structures produced by these cell lines were classified into five categories of organizations illustrated in Figure 1A: (I) immature with few cells, (II) spherical with a filled lumen, (III) spherical with a hollow lumen, (IV) non-spherical but organized, or (V) non-spherical and disorganized. Structure classes I–IV were observed for 10A and 10A.ErbB2 cells, yet the relative distribution of each structure varied (Fig. 1B). The majority of 10A cells formed class III and majority of 10A.ErbB2 cells formed class II. Class V was not observed for 10A nor 10A.ErbB2 cells, and was the exclusive structure for 231 cells (Fig. 1B).
Figure 1. 3D phenotypes of MECs in BME.
Cells cultured in BME displayed different organizational phenotypes, shown by confocal microscopy (A; red-TOPRO3 nuclear stain, green-laminin V; scale bars represent 50 µm). Organizations of cells were categorized into five classifications as described in the text with examples shown in A. Confocal images of structures for 10A, 10A.ErbB2, and 231 cells were categorized and differences existed between the cell populations (p < 2e-16) (B), with correspondence analysis revealing the most compelling associations (C).
As the data were categorical and semi-quantitative, a correspondence analysis was used to identify trends in the data. 10A cells were most associated with class III, 10A.ErbB2 cells were most associated with class II and closely associated with class IV, and 231 cells were most associated with class V (Fig. 1C). These observations were consistent with previous observations in BME [14, 22]. Deposition of a basement membrane, as exemplified by basal laminin V localization, was also considered to characterize structures. However, no differences were observed between laminin V localization by 10A and 10A.ErbB2 cells (data not shown) and 231 cells did not express laminin V.
3.2 Establishment of synthetic system for 3D culture of MECs
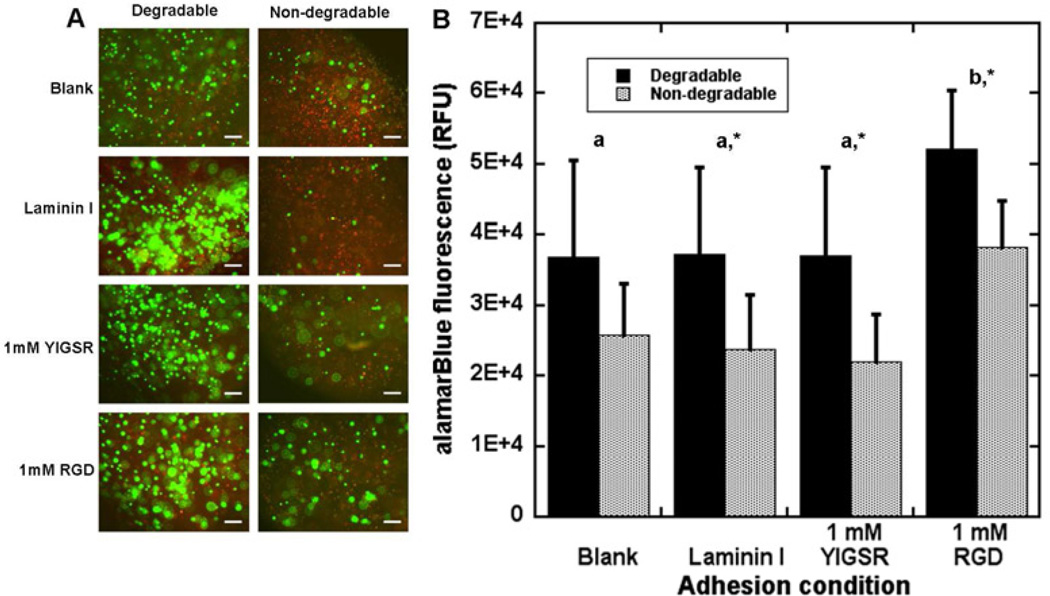
PEG conditions were subsequently investigated to identify environments that support MEC survival. The initial screening studies employed 10A cells, and investigated hydrogels with varying matrix degradation and peptides or proteins that support cell adhesion. Plasmin-mediated degradation of BME by MECs has previously been observed [42] and this mechanism was incorporated into the synthetic framework using crosslinking peptides containing a sequence that was responsive to plasmin (YKNR) [30, 43]. Degradable and non-degradable (D-isomer) crosslinking peptides were investigated, with adhesion supported by blending laminin I (0.1 mg/ml) into the hydrogel precursor solution or modifying PEG units with adhesion peptides (YIGSR or RGD; 1 mM each). For all adhesion conditions, the degradable crosslinker led to greater cell viability (Fig. 2A and B), with significant increases for all conditions except hydrogels lacking adhesive support (Fig. 2B). Within degradable hydrogels, cells in the presence of RGD were also more viable than in the other adhesion conditions. These studies demonstrated that degradation improved viability and subsequent studies only considered degradable hydrogels.
Figure 2. Screening PEG conditions for 10A cell survival.
PEG hydrogels cross-linked with a plasmin-sensitive peptide (degradable) or a D-isomerized peptide (non-degradable) were either left blank, blended with laminin I, or functionalized with 1 mM YIGSR or RGD and used to culture 10A cells. After 10 d of culture, degradable hydrogels resulted in greater viability as seen with live/dead staining (A; green-Calcein AM, red-ethidium homodimer; scale bars represent 200 µm) or an alamarBlue assay (B). Values represent average values +/− s.d. from samples in triplicate in three independent experiments. Letters represent significantly different levels for degradable conditions and ‘*’ represents a significant difference between degradable and non-degradable hydrogels for the specific adhesion condition (p<0.05).
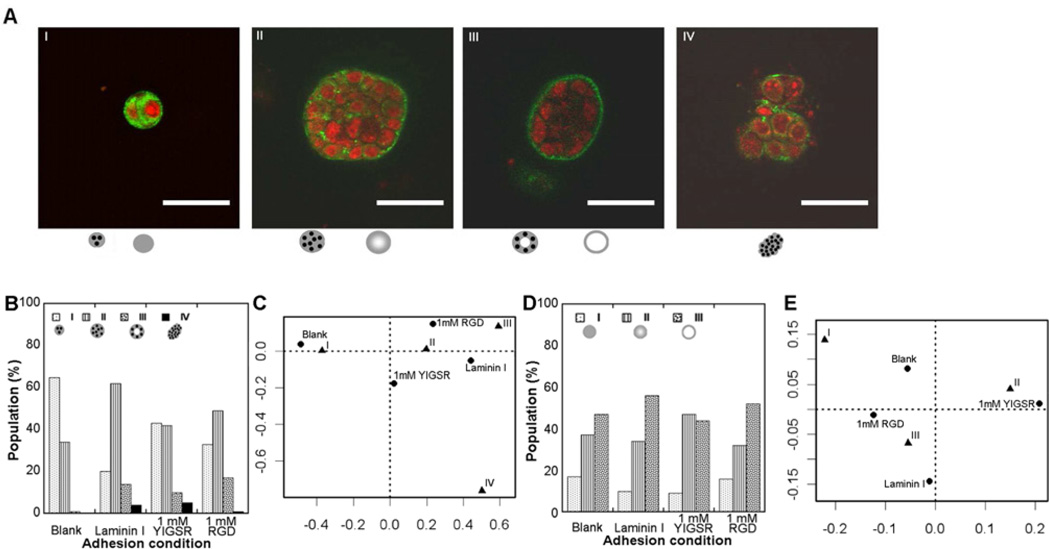
Degradable PEG hydrogels supported the formation of cellular structures I – IV by 10A cells (Fig 3A), consistent with the structures formed in BME (Fig. 1A). The relative distribution of these structures among the four categories depended on the identity of the adhesion molecule (Fig. 3B), with blank hydrogels most associated with class I, laminin I most associated with class II, YIGSR associated nearly equally with classes I and II, and RGD associated with classes II and III (Fig. 3C).
Figure 3. Screening PEG conditions for 10A cell phenotype.
10A cells growing within PEG hydrogels displayed the first four cell organization classifications seen within BME as shown by confocal microscopy (A; red-TOPRO3 nuclear stain, green-laminin V; scale bars represent 50 µm). Structures varied in character with laminin V distribution and three classes of structures are shown (A I–III). Confocal images were categorized according to cell organizations and differences between populations grown with different adhesion conditions were seen (p=2e-6) (B) with associations seen with categorical analysis (C). Images were also categorized according to laminin V distribution and showed variations among populations (D) yet there were no significant differences with adhesion condition (p=0.35).
The distribution of laminin V within the structures was more heterogeneous within PEG relative to the observations within BME, which was exclusively localized to basement membranes of structures in BME (Fig. 3A). The laminin V distribution was classified as (I) immature and mostly cytoplasmic, (II) mixed cytoplasmic with some basally localized, or (III) mostly basally localized. Differences in laminin V distribution within each adhesion condition were observed (Fig. 3D), yet no significant differences in the populations were observed between conditions.
3.3 Influence of adhesion on MEC structure phenotype
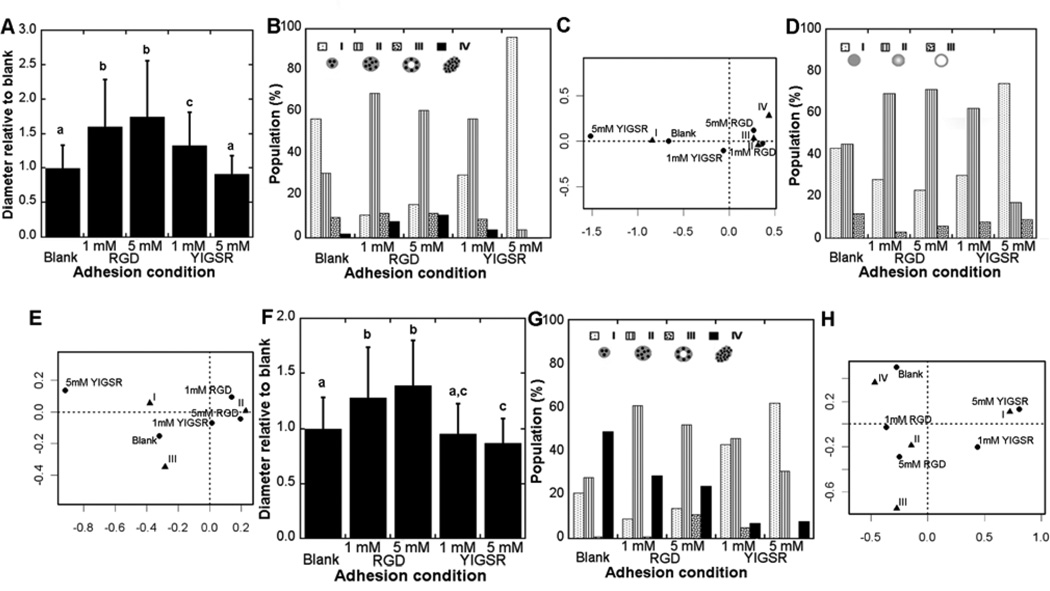
Using degradable PEG hydrogels, the influence of adhesion identity and concentration on MEC phenotype was investigated. RGD and YIGSR both supported the formation of structures by 10A cells in the previously described screening studies and were employed in subsequent studies as they provide for more consistent and reproducible system relative to full length proteins. Both adhesion peptides were incorporated into hydrogels with increasing concentrations (1 or 5 mM), along with a scrambled control peptide (RDG) to ensure the same extent of PEG modification, which provides for consistent crosslinking across conditions.
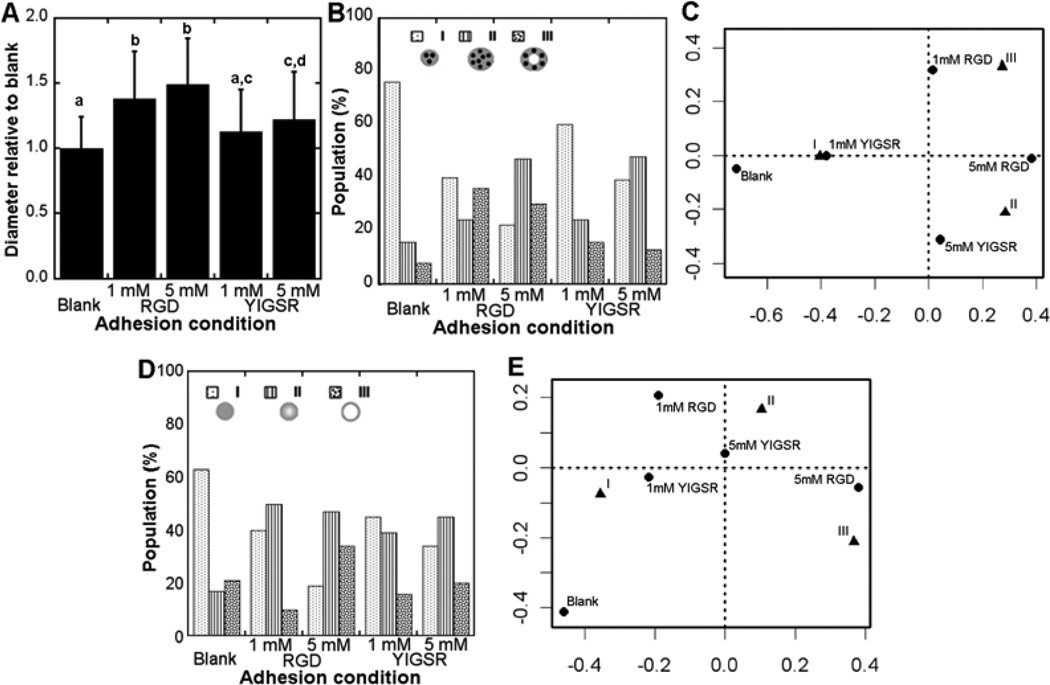
The 10A cells produced phenotypes I – IV, yet the distribution of phenotypes varied with peptide identity and concentration. Relative to blank hydrogels, incorporation of RGD at either concentration resulted in significant increases in the diameter of the structure (Fig. 4A). Incorporation of YIGSR also resulted in larger structures, with the effect significant only at the greatest concentration (Fig. 4A). Within blank hydrogels, class I was the greatest percentage of structures. The presence of RGD and YIGSR shifted the distribution toward classes II and III, with the 5 mM peptide concentration having the greatest percentage of classes II and III (Fig. 4B and C). RGD modification of hydrogels was more closely associated with class III, the most abundant phenotype observed in BME, relative to YIGSR modification (Fig. 4C). The laminin V distribution was primarily class I in blank hydrogels, and shifted toward increased class II and III in the presence of peptides (Fig. 4D and E). A class III laminin V distribution, indicating a localized basement membrane as seen most often in BME, corresponded most closely with hydrogels containing 5 mM RGD (Fig. 4E). Notably, the scrambled peptide was employed to maintain the same theoretical extent of PEG modification and thereby obtain similar crosslinking, which was not done in the screening experiments (Fig. 3). Greater populations of immature structures and cytoplasm localized laminin V were observed using scrambled peptides, though the trends between adhesion conditions were similar.
Figure 4. Influence of adhesion on 10A cell phenotype.
10A cells in PEG hydrogels produced a variety of phenotypes that depended on adhesion identity and concentration. Sizes of structures were measured and normalized to blank hydrogels for a given experiment (A), with mean +/− s.d. values taken from three independent experiments. Letters represent significance levels (p<0.05). Confocal images for structures were classified as described in Fig. 3 for cell organizations and laminin V distributions. Cell organization had differences between adhesion conditions (p=5e-8) (B) with associations revealed by correspondence analysis (C). Laminin V distribution varied with adhesion condition (p=7e-5) (D) with associations revealed by correspondence analysis (E).
10A.ErbB2 cells, which overexpress the single oncogene ErbB2, and 231 cells, which are derived from an advanced stage of cancer, also had phenotypes that were a function of adhesion peptide identity and concentration. The diameter of 10A.ErbB2 structures was increased in the presence of RGD (both 1 and 5 mM); however, for YIGSR, larger structures were only observed at the low concentration (1 mM) (Fig. 5A). An increase in the percentage of class II structures was observed in the presence of RGD peptides, with a shift to class I structures in the presence of 5 mM YIGSR (Fig. 5B and C). Development of class IV structures was associated with increasing RGD peptide concentration (Fig. 5C), though the percentage of the population with this morphology was relatively small. Laminin V distribution shifted toward an increase in class II for RGD peptides, with increased class I in the presence of YIGSR (Fig. 5D and E). For 231 cells, RGD peptides produced structures with increased diameter at both concentrations (1 and 5 mM), and 5 mM YIGSR produced structures that were statistically smaller than what was observed in blank hydrogels (Fig. 5F). A large population of 231 structures was of class IV in blank PEG, and the presence of RGD lead to increased percentages of class II and III (Fig. 4G and H). Structures were largely class I, with no class III structures observed, as YIGSR concentration increased (Fig. 4G). As in BME, 231 cells did not express laminin V (data not shown) and this classification was not used. Taken together, 231 cells exhibited a more aggressive phenotype (Class V) than 10A.ErbB2 cells in blank hydrogels, but RGD and YIGSR produced similar effects on structure phenotype – more organization with RGD and less maturity with YIGSR.
Figure 5. Influence of adhesion on cancerous MEC phenotypes.
10A.ErbB2 and 231 cells in PEG hydrogels both produced a variety of phenotypes that depended on adhesion identity and concentration. Sizes of 10A.ErbB2 structures were measured and normalized to blank hydrogels for a given experiment (A), with mean +/− s.d. values taken from three independent experiments. Conditions with different letters were significantly different (p<0.05). Confocal images for structures were classified as described in Fig. 3 for cell organizations and laminin V distributions. 10A.ErbB2 cell organization showed differences between adhesion conditions (p=2e-15) (B) with associations revealed by correspondence analysis (C). Laminin V distribution within 10A.ErbB2 structures varied with adhesion condition (p=8e-5) (D) with associations revealed by correspondence analysis (E). Sizes of 231 structures were measured and normalized to blank hydrogels for a given experiment (F). 231 cell organization had differences between adhesion conditions (p<2e-16) (G) with associations revealed by correspondence analysis (H).
3.4 Signaling pathway analysis within PEG hydrogels
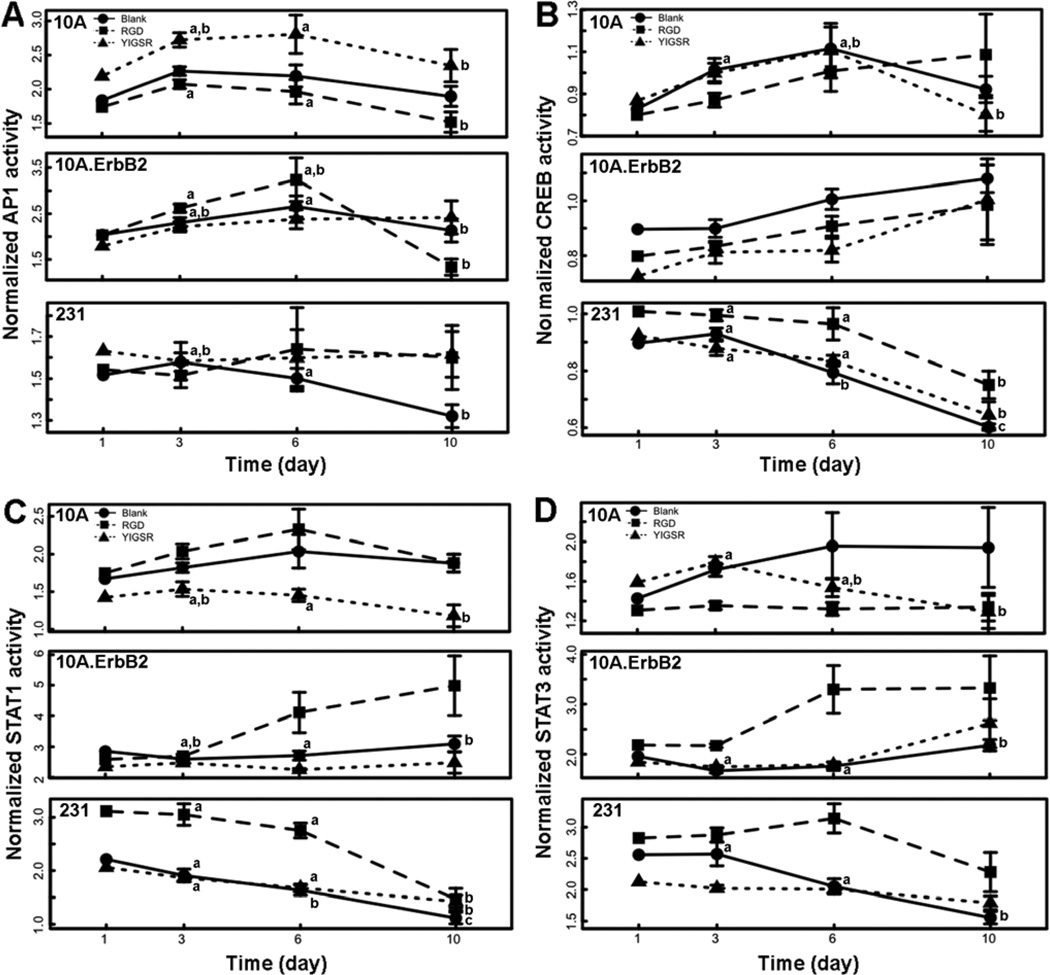
In order to investigate mechanisms underlying the influences of adhesion peptides on cell growth and how signaling responses vary between cell types, a cell-based signaling assay was used to quantify dynamic TF activities. Lentiviral delivery of reporter genes was used to quantify signals throughout the 10 d culture in the MAPK (AP1 and CREB) and JAK/STAT (STAT1 and STAT3) pathways, both of which transduce adhesion signals from the microenvironment [44, 45].
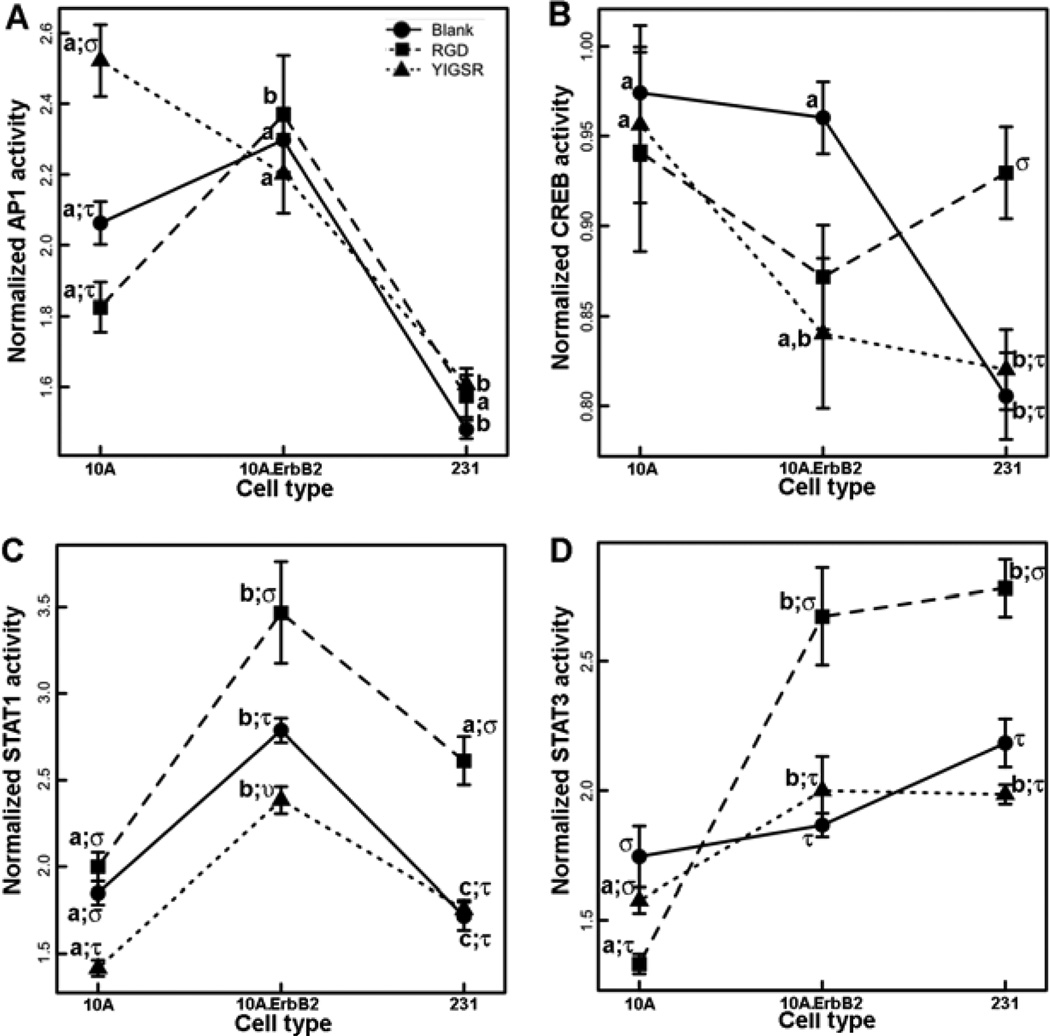
The activities of TFs were measured in order to capture dynamic behaviors of signaling pathways (Figure 6), and a multiple regression model was used to test the significance of cell type, peptide identity, time, and their interactions on pathway activities (Table 1, S1). AP1 and CREB activities were nonlinear over time (Table 1, S1) while STAT1 and STAT3 had more linear temporal behaviors. Relative to 10A cells, STAT1 and STAT3 activity profiles generally increased for 10A.ErbB2 cells and decreased for 231 cells (Fig. 6). CREB activity decreased in 231 cells as well (Fig. 6B). Also, temporal activities of all TFs were interdependent on both cell type and peptide identity, represented by significant interaction terms of the regression models in Table 1, indicating the responses to adhesion signals were reliant upon genotype. 10A cells showed increased AP1 activity upon stimulation with YIGSR, an effect not observed in the cancerous cells (Fig. 7A). 231 cells had increased CREB activity upon stimulation with RGD, an effect not observed in the other cell lines (Fig. 7B). Within both cancerous cell lines, RGD stimulated STAT1 and STAT3 activities (Fig. 7C and D). While YIGSR decreased STAT1 activity in 10A and 10A.ErbB2 cells, there was not a significant effect in 231 cells (Fig. 7C).
Figure 6. Dynamic activity of signaling pathways.
Signaling pathways within living cells were quantified within PEG hydrogels either left blank (circles) or containing RGD (squares) or YIGSR (triangles) peptides by assessing activity of AP1 (A), CREB (B), STAT1 (C), or STAT3 (D) over time. The means ± s.e. from triplicate hydrogels from three independent experiments are plotted. Significant differences over time within each peptide condition are represented by letters (a, b, c) (p<0.05).
Table 1.
Significant results of multiple regression models for signaling pathway analysis.
| TF | Variable | Significant Interactions |
|---|---|---|
| AP1 | Cell Type (S) Peptide (S) Time (S) Time2 (S) |
10A.ErbB2:RGD 231:RGD YIGSR:Time |
| CRE | Cell Type (S) Peptide (S) Time (S) Time2 (S) |
10A.ErbB2:YIGSR 231:RGD 231:Time |
| STAT1 | Cell Type (S) Peptide (S) Time (S) Time2 (NS) |
231:RGD 231:YIGSR 10A.ErbB2:Time 231:Time |
| STAT3 | Cell Type (S) Peptide (S) Time (S) Time2 (NS) |
10A.ErbB2:RGD 231:RGD 10A.ErbB2:Time 231:Time |
NS-Not Significant; S-Significant (p<0.01)
Figure 7. Cell type and peptide identity influence signaling pathway activity.
Normalized TF activities were averaged across all time points to identify only the influences of cell type and peptide identify (blank – circles; RGD – squares; YIGSR – triangles) on AP1 (A), CREB (B), STAT1 (C), or STAT3 (D) activities. The means ± s.e. from triplicate hydrogels and three independent experiments are plotted. Significant differences between cell types within each peptide condition are represented by letters (a, b, c) and significant differences between peptide condition within each cell type are represented by symbols (stu) (p<0.05). For example, AP1 activity within 10A cells significantly increased with YIGSR (represented by different symbols) and was significantly greater in 10A cells than 231 cells in the presence of YIGSR (represented by different letters).
4. Discussion
As numerous factors can contribute to tissue development and cancer progression, a minimal synthetic system was employed that reproduced the 3D phenotypes of normal and cancerous mammary tissue, and allowed for mechanistic studies of the microenvironmental contribution. BME is widely used to study 3D growth of cancer but is composed of dozens of ECM molecules and soluble factors without an exact composition and varies with lot [46]. Matrices composed solely of pure ECM components, including collagen and laminin, are better defined, but biochemical and mechanical signals are interdependent. Synthetic systems, alternatively, contain minimal biochemical signals and are more easily tunable to investigate the effects of specific microenvironmental contributions [34]. These matrices are composed of a backbone that is responsible for creating the 3D structure and can be decorated with biological stimuli that allow for adhesion and degradation, in turn directing cellular processes as tissues develop. The observations of this study more definitively pinpoint adhesion as a major contributor to tissue phenotype without the overlapping signals of other factors in the microenvironment.
Signaling pathways within biological systems are dynamic [47, 48], and thus an important parameter in our signaling pathway analysis was the ability to capture the temporal behavior of the TFs during development of structures. The TFs investigated exhibited different dynamic profiles that varied with the cellular context. While STAT1 and STAT3 activities had general increases over time for 10A.ErbB2 cells, these same TFs decreased over time within 231 cells (Fig. 6). By using bioluminescence imaging to track reporter gene activity, the dynamic profiles of TF activities could be quantified non-invasively and directly within hydrogels as cells developed into structures. While tissue growth is the culmination of many pathway events and individual pathway activities can only partially explain phenotypic changes, cell-based signaling pathway analyses are an enabling approach for studying the dynamic mechanisms and a library of TF reporters could be used to monitor more widespread intracellular events [37, 38]. Furthermore, this technology may allow phenotype to be connected with alterations in genotype, microenvironment, or both in combination.
10A cells developed into structures within PEG hydrogels that resembled those observed in BME and adhesion significantly influenced structural phenotypes and signaling pathway activities. Adhesive signals from the microenvironment originate from receptors on the surfaces of cells that bind ECM proteins and transduce a variety of signals that direct processes throughout tissue growth [49]. RGD is a sequence present in multiple ECM proteins, including fibronectin, vitronectin, and laminin, that has been shown to ligate a variety of integrin receptors [50] YIGSR is derived from laminin and binds a non-integrin 67 kDa receptor [51]. Both peptides supported growth of 10A cells into multi-cellular spherical structures, with some exhibiting hollow lumens indicative of a greater level of organization resulting from apoptosis of the interior cells and senescence [41] (Fig. 4). RGD more reliably produced developed structures, but the number of well-developed structures increased with concentrations of both adhesion peptides, indicating that increasing the receptor ligation advanced the development of 10A cells. RGD has been seen to promote growth and organization in other tissues, including other epithelial cells and stem cells [52]. RGD significantly lowered the activity of STAT3, consistent with the reported down-regulation of active STAT3 upon integrin engagement within a 3D microenvironment [53]. YIGSR increased AP1 activity and decreased STAT1 activity (Fig. 6C, Fig. 7C). The full laminin protein has been shown to induce AP1 activity in a cell-type dependant manner, though the observations were not specifically attributed to YIGSR [54]. Ligation of the 67 kDa receptor has been reported to lead to de-phosphorylation of p38 [51], which in turn can affect STAT1 activity [55, 56], possibly explaining the decrease.
Relative to 10A cells, 10A.ErbB2 and 231 cells developed structures with varied levels of organization and differentially responded to adhesion peptides, reflecting an altered interpretation of signals from the microenvironment. The 10A.ErbB2 and 231 cells displayed distinct phenotypes both within BME and PEG hydrogels. Whereas 10A and 10A.ErbB2 cells primarily formed structures consisting of few cells (class I) within blank PEG hydrogels, 231 cells readily grew into non-spherical structures that resembled the architectures of early cancers observed by others in BME [21]. The ability to grow without support from adhesion may be due to the increased rate of cell cycle progression and the unresponsiveness to anti-growth cues from the microenvironment that are characteristic of cancerous cells [57] and is consistent with 231 cells being derived from an advanced stage of cancer. Further, relative to 10A cells, 10A.ErbB2 and 231 cells differentially responded to adhesion peptides to impact phenotype. Whereas 10A structure size increased in the presence of both RGD and YIGSR (Fig. 4), 10A.ErbB2 and 231 structures were both smaller and less developed in the presence of YIGSR (Fig 5). Ardini et al. demonstrated a mechanism of 231 cell mobility in laminin that required 67 kDa receptor binding and subsequent degradation by cathepsin B [58]. Expression of the 67 kDa receptor has been correlated with cancer malignancy [59, 60] and cells derived from an advanced stage of cancer may have a greater reliance on cathepsin mediated proteolysis for invasion. Hence, whereas 10A cells could develop into advanced structures in the presence of YIGSR, cancerous cells were less able to invade the plasmin-sensitive hydrogels.
The differential responses of cells to adhesion peptides were further reflected in signaling pathways and illustrated the interplay of genotype and adhesion. 10A.ErbB2 cells had greater AP1 activity than 10A cells upon RGD stimulation (Fig. 7A), and increased expression of α integrin subunits have been associated with cells overexpressing ErbB2 [61], including units that bind RGD. This observation is consistent with RGD contributing to the MAPK pathway. In addition, RGD stimulated increases in STAT1 in both 10A.ErbB2 and 231 cells but not 10A cells (Fig. 7C). Engagement of integrins leads to activation of FAK, which in turn can activate STAT1 [62]. FAK is overexpressed in many cancers [63] and can act in combination with ErbB2 [64]. These changes may explain the increased sensitivity of STAT1 within cancerous cell lines to RGD and illustrate mechanisms by which genetic changes alter signaling networks. Also, whereas YIGSR decreased STAT1 activity in 10A cells, the effect was less pronounced in 10A.ErbB2 cells and not observed in 231 cells (Fig. 7C). The data suggest there were alterations within cancerous cell networks that compensated for the effects of YIGSR on STAT1. While the trends in signaling pathways as functions of peptide were largely similar between the ErbB2 mutant and 231 cancerous cell line, one important distinction was the increase of CREB in response to RGD present only for 231 cells (Fig. 7B). This observation reflects the altered signaling in 231 cells, such as enhanced levels of phosphorylated ERK [65] that in turn can activate CREB [66]. No direct associations between TF activity and phenotype were made due to the large number of signaling pathways concurrently directing growth, but cell arrays with a larger number of constructs may reveal the mechanism by which intracellular networks are influenced by adhesion.
While adhesion and genotype were both shown to contribute to phenotype, other factors should be considered, including the influence of other adhesion signals, growth factors, degradation, stromal cells, and mechanics. Each of these factors act in combination, as cells interpret many signals simultaneously and one factor can influence the contribution of another. ECMs contain many proteins and cells receive combinations of adhesion signals, which can interplay with each other to direct different cell processes. Other peptides and molecules can be considered with the PEG system. Soluble molecules, such as growth factors, are sequestered within native ECMs or secreted by other cells in the microenvironment and can alter cellular behaviors. These molecules can be added to the culturing media or tethered directly to PEG [67]. Cells use other mechanisms of degradation beyond plasmin, often utilizing matrix metalloproteases (MMPs), which have been seen to be upregulated in cancerous tissues [3]. Crosslinking peptides specific for MMPs have already been utilized in other PEG systems [27]. Neighboring cells secrete factors that alter the growth of tissues, and co-culture models have been used in other natural systems. Mechanical forces have been seen to progress cancerous processes [16, 68], and altered mechanics have been investigated with other synthetic systems. Each factor can be individually, or in combination, be investigated using the PEG-based system to identify how signals within the microenvironment contribute to phenotype and progress cancer.
5. Conclusions
In summary, synthetic hydrogels and signaling pathway analysis can be employed to investigate active molecular processes as tissues develop, including the progression of cancer. Signals resulting from both adhesion and genotype influence tissue phenotype and intracellular signaling profiles and PEG-based hydrogels allow specific microenvironmental signals to be independently investigated. Adhesion peptides directed variable phenotypes by normal and cancer MECs and the cellular response to adhesion cues was a function of genotype, which was evident at the levels of both structure organization and signaling pathway activity. Synthetic systems do not yet capture the inherent complexity of native tissues but instead represent a bottom-up approach for identifying the essential features of specific contributors in the cellular microenvironment. In addition to investigating cancerous processes, the tools presented in this study can be applied to other tissues and should be beneficial for designing biomaterials that produce a desired biological response.
Supplementary Material
Acknowledgements
Viability measurements were made at the Northwestern University High Throughput Analysis Laboratory, confocal microscopy was performed at the Northwestern University Biological Imaging Facility, and bioluminescence imaging was performed at the Northwestern University Center for Advanced Molecular Imaging. We thank M. Dimri and the labs of H. Band and V. Band (University of Nebraska) for assistance with BME culture, A. Garcia (Georgia Institute of Technology) and J. Shepard for helpful discussions, and S. Seidlits for synthesis of PEG-VS. This work was funded by the National Institutes of Health (NIH; R21CA125285, UL1DE019587, PL1EB008542, which supports the Biomaterials core of the Oncofertility Consortium Roadmap Grant) and the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust,. MSW and BPB were both supported by an NIH training grant (T32GM008449) and AS was supported by a Baxter Early Career Development Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 2.Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, et al. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339:1277–1284. doi: 10.1056/NEJM199810293391803. [DOI] [PubMed] [Google Scholar]
- 3.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lochter A, Bissell M. Involvement of extracellular matrix constituents in breast cancer. Semin Cancer Biol. 1995;6:165–173. doi: 10.1006/scbi.1995.0017. [DOI] [PubMed] [Google Scholar]
- 5.Sinkus R, Tanter M, Xydeas T, Catheline S. Viscoelastic shear properties of in vivo breast lesions measured by MR elastography. MRI. 2005;23:159–165. doi: 10.1016/j.mri.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 6.Paszek M, Weaver V. The tension mounts: mechanics meets morphogenesis and malignancy. J Mammary Gland Biol Neoplasia. 2004;9:325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- 7.Marx J. All in the Stroma: Cancer's Cosa Nostra. Science. 2008;320:38–41. doi: 10.1126/science.320.5872.38. [DOI] [PubMed] [Google Scholar]
- 8.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García A. Get a grip: integrins in cell–biomaterial interactions. Biomaterials. 2005;26:7525–7529. doi: 10.1016/j.biomaterials.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 11.Garamszegi N, Garamszegi SP, Shehadeh LA, Scully SP. Extracellular matrix-induced gene expression in human breast cancer cells. Mol Cancer Res. 2009;7:319–329. doi: 10.1158/1541-7786.MCR-08-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koshikawa N, Schenk S, Moeckel G, Sharabi A, Miyazaki K, Gardner H, et al. Proteolytic processing of laminin-5 by MT1-MMP in tissues and its effects on epithelial cell morphology. FASEB J. 2004;18:364–366. doi: 10.1096/fj.03-0584fje. [DOI] [PubMed] [Google Scholar]
- 13.Steadman R, Irwin MH, St John PL, Blackburn WD, Heck LW, Abrahamson DR. Laminin cleavage by activated human neutrophils yields proteolytic fragments with selective migratory properties. J Leukoc Biol. 1993;53:354–365. doi: 10.1002/jlb.53.4.354. [DOI] [PubMed] [Google Scholar]
- 14.Kenny HA, Kaur S, Coussens LM, Lengyel E. The initial steps of ovarian cancer cell metastasis are mediated by MMP-2 cleavage of vitronectin and fibronectin. J Clin Invest. 2008;118:1367–1379. doi: 10.1172/JCI33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. The origin of the myofibroblasts in breast cancer. Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J Clin Invest. 1995;95:859–873. doi: 10.1172/JCI117736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Postovit L, Seftor E, Seftor R, Hendrix M. A 3-D Model to study the epigenetic effects induced by the microenvironment of human embryonic stem cells. Stem Cells. 2006;24:501–505. doi: 10.1634/stemcells.2005-0459. [DOI] [PubMed] [Google Scholar]
- 19.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Kenny P, Lee G, Myers C, Neve R, Semeiks J, Spellman P, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimri M, Naramura M, Duan L, Chen J, Ortega-Cava C, Goswami R, et al. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007;67:4164–4172. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- 23.Muthuswamy S, Li D, Lelievre S, Bissell M, Brugge JS. ErbB 2, but not ErbB 1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fournier MV, Martin KJ, Kenny PA, Xhaja K, Bosch I, Yaswen P, et al. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006;66:7095–7102. doi: 10.1158/0008-5472.CAN-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver VM, Lelièvre S, Lakins JN, Chrenek MA, Jones JCR, Giancotti F, et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhao H, Asztalos S, Chisamore M, Sitabkhan Y, Tonetti DA. Estradiol-induced regression in T47D:A18/PKCalpha tumors requires the estrogen receptor and interaction with the extracellular matrix. Mol Cancer Res. 2009;7:498–510. doi: 10.1158/1541-7786.MCR-08-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4:713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 28.Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. The effects of matrix stiffness and RhoA on the phenotypic plasticity of smooth muscle cells in a 3-D biosynthetic hydrogel system. Biomaterials. 2008;29:2597–2607. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarig-Nadir O, Seliktar D. The role of matrix metalloproteinases in regulating neuronal and nonneuronal cell invasion into PEGylated fibrinogen hydrogels. Biomaterials. 2010;31:6411–6416. doi: 10.1016/j.biomaterials.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 30.Shikanov A, Smith RM, Xu M, Woodruff TK, Shea LD. Hydrogel network design using multifunctional macromers to coordinate tissue maturation in ovarian follicle culture. Biomaterials. 2011;32:2524–2531. doi: 10.1016/j.biomaterials.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraehenbuehl T, Zammaretti P, Vandervlies A, Schoenmakers R, Lutolf M, Jaconi M, et al. Three-dimensional extracellular matrix-directed cardioprogenitor differentiation: Systematic modulation of a synthetic cell-responsive PEG-hydrogel. Biomaterials. 2008;29:2757–2766. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Salinas C, Anseth K. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370–2377. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutolf M, Hubbell J. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 35.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Nat Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin S, Shea LD. Lentivirus immobilization to nanoparticles for enhanced and localized delivery from hydrogels. Mol Ther. 2010;18:700–706. doi: 10.1038/mt.2009.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellis AD, Peňalver-Bernabé B, Weiss MS, Yarrington ME, Barbolina MV, Pannier AK, et al. Cellular arrays for large-scale analysis of transcription factor activity. Biotechnol Bioeng. 2011;108:395–403. doi: 10.1002/bit.22916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss MS, Peñalver Bernabé B, Bellis AD, Broadbelt LJ, Jeruss JS, Shea LD. Dynamic, large-scale profiling of transcription factor activity from live cells in 3D culture. PLoS ONE. 2010;5:e14026. doi: 10.1371/journal.pone.0014026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Team. R: A language and environment for statistical computing. Austria: Vienna; 2008. [Google Scholar]
- 40.Nenadic O, Greenacre M. Corresponence analysis in R, with two- and three-dimensional graphics: the ca package. J Stat Softw. 2007;20:1–13. [Google Scholar]
- 41.Debnath J, Muthuswamy S, Brugge J. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 42.Farina AR, Tiberio A, Tacconelli A, Cappabianca L, Gulino A, Mackay AR. Identification of plasminogen in Matrigel and its activation by reconstitution of this basement membrane extract. BioTechniques. 1996;21:904–909. doi: 10.2144/96215rr03. [DOI] [PubMed] [Google Scholar]
- 43.Jo YS, Rizzi SC, Ehrbar M, Weber FE, Hubbell JA, Lutolf MP. Biomimetic PEG hydrogels crosslinked with minimal plasmin-sensitive tri-amino acid peptides. J Biomed Mater Res. 2010;93:870–877. doi: 10.1002/jbm.a.32580. [DOI] [PubMed] [Google Scholar]
- 44.Aplin AE, Howe AK, Juliano RL. Cell adhesion molecules, signal transduction and cell growth. Curr Opin Cell Biol. 1999;11:737–744. doi: 10.1016/s0955-0674(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 45.Hombría JC-G, Sotillos S. Disclosing JAK/STAT links to cell adhesion and cell polarity. Semin Cell Dev Biol. 2008;19:370–378. doi: 10.1016/j.semcdb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 48.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Maeda T, Sekiguchi K, Sheppard D. Distinct structural requirements for interaction of the integrins alpha5-beta1, alphav-beta5, and alphav-beta6 with the central cell binding domain in fibronectin. Cell Communication and Adhesion. 1996;4:231–250. doi: 10.3109/15419069609010769. [DOI] [PubMed] [Google Scholar]
- 51.Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells. Cancer Lett. 2005;223:1–10. doi: 10.1016/j.canlet.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 52.Chung I, Enemchukwu N, Khaja S, Murthy N, Mantalaris A, Garcia A. Bioadhesive hydrogel microenvironments to modulate epithelial morphogenesis. Biomaterials. 2008;29:2637–2645. doi: 10.1016/j.biomaterials.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee ST, Yun JI, Jo YS, Mochizuki M, van der Viles AJ, Kontos S, et al. Engineering integrin signaling for promoting embryonic stem cell self-renewal in a precisely defined niche. Biomaterials. 2010;31:1219–1226. doi: 10.1016/j.biomaterials.2009.10.054. [DOI] [PubMed] [Google Scholar]
- 54.Kubota S, Tashiro K, Yamada Y. Signaling site of laminin with mitogenic activity. J Biol Chem. 1992;267:4285–4288. [PubMed] [Google Scholar]
- 55.Goh KC, Haque SJ, Williams BR. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsauer K, Sadzak I, Porras A, Pilz A, Nebreda AR, Decker T, et al. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci USA. 2002;99:12859–12864. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 58.Ardini E, Sporchia B, Pollegioni L, Modugno M, Ghirelli C, Castiglioni F, et al. Identification of a novel function for 67-kDa laminin receptor: increase in laminin degradation rate and release of motility fragments. Cancer Res. 2002;62:1321–1325. [PubMed] [Google Scholar]
- 59.Ménard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Research and Treatment. 1998;52:137–145. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- 60.Yamamura K, Kibbey MC, Jun SH, Kleinman HK. Effect of Matrigel and laminin peptide YIGSR on tumor growth and metastasis. Semin Cancer Biol. 1993;4:259–265. [PubMed] [Google Scholar]
- 61.Spangenberg C, Lausch EU, Trost TM, Prawitt D, May A, Keppler R, et al. ERBB2-mediated transcriptional up-regulation of the alpha5beta1 integrin fibronectin receptor promotes tumor cell survival under adverse conditions. Cancer Res. 2006;66:3715–3725. doi: 10.1158/0008-5472.CAN-05-2823. [DOI] [PubMed] [Google Scholar]
- 62.Xie B, Zhao J, Kitagawa M, Durbin J, Madri JA, Guan JL, et al. Focal adhesion kinase activates Stat1 in integrin-mediated cell migration and adhesion. J Biol Chem. 2001;276:19512–19523. doi: 10.1074/jbc.M009063200. [DOI] [PubMed] [Google Scholar]
- 63.Miyazaki T, Kato H, Nakajima M, Sohda M, Fukai Y, Masuda N, et al. FAK overexpression is correlated with tumour invasiveness and lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer. 2003;89:140–145. doi: 10.1038/sj.bjc.6601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benlimame N, He Q, Jie S, Xiao D, Xu YJ, Loignon M, et al. FAK signaling is critical for ErbB-2/ErbB-3 receptor cooperation for oncogenic transformation and invasion. The Journal of Cell Biology. 2005;171:505–516. doi: 10.1083/jcb.200504124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonne GK, Masoumi KC, Lennartsson J, Larsson C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J Biol Chem. 2009;284:33456–33465. doi: 10.1074/jbc.M109.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagstaff SC, Bowler WB, Gallagher JA, Hipskind RA. Extracellular ATP activates multiple signalling pathways and potentiates growth factor-induced c-fos gene expression in MCF-7 breast cancer cells. Carcinogenesis. 2000;21:2175–2181. doi: 10.1093/carcin/21.12.2175. [DOI] [PubMed] [Google Scholar]
- 67.Mann B, Schmedlen R, West J. Tethered-TGF-β increases extracellular matrix production of vascular smooth muscle cells. Biomaterials. 2001 doi: 10.1016/s0142-9612(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 68.Mammoto A, Connor KM, Mammoto T, Yung CW, Huh D, Aderman CM, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.