Abstract
HMGB1 is a chromatin architectural protein that is released by dead or damaged cells at sites of tissue injury. Extracellular HMGB1 functions as a pro-inflammatory cytokine and chemoattractant for immune effector and progenitor cells. Previously, we have shown that the IKKβ and IKKα-dependent NF-κB signaling pathways are simultaneously required for cell migration to HMGB1. The IKKβ-dependent canonical pathway is needed to maintain expression of RAGE, the ubiquitously expressed receptor for HMGB1, but the target of the IKKα non-canonical pathway was not known. Here we show that the IKKα-dependent p52/RelB non-canonical pathway is critical to sustain CXCL12/SDF-1 production in order for cells to migrate towards HMGB1. Utilizing both mouse bone marrow-derived macrophages and mouse embryo fibroblasts (MEFs) it was observed that neutralization of CXCL12 by a CXCL12 monoclonal antibody completely eliminated chemotaxis to HMGB1. In addition, the HMGB1 migration defect of IKKα KO and p52 KO cells could be rescued by adding recombinant CXCL12 to cells. Moreover, p52 KO MEFs stably transduced with a GFP retroviral vector that enforces physiological expression of CXCL12 also showed near normal migration toward HMGB1. Finally, both AMD3100, a specific antagonist of CXCL12's G-protein coupled receptor CXCR4, and an anti-CXCR4 antibody blocked HMGB1 chemotactic responses. These results indicate that HMGB1-CXCL12 interplay drives cell migration towards HMGB1 by engaging receptors of both chemoattractants. This novel requirement for a second receptor-ligand pair enhances our understanding of the molecular mechanisms regulating HMGB1-dependent cell recruitment to sites of tissue injury.
INTRODUCTION
High Mobility Group Box 1 (HMGB1) is a non-histone, chromatin architectural protein ubiquitously expressed by all mammalian cells; but functions outside cells as a potent cytokine and chemoattractant. In vivo, HMGB1 is passively released by necrotic cells and actively secreted by immune effector cells (1–4). Extracellular HMGB1 signals through the Receptor for Advanced Glycation End-products (RAGE), Toll-Like Receptor 2 (TLR 2) and TLR 4 (3–9). In this capacity HMGB1 acts as an alarmin or damage-associated molecular pattern (DAMP) that senses tissue damage and elicits a variety of pro-inflammatory responses {reviewed in (3, 4, 6, 10, 11)}. Moreover, HMGB1's chemotactic activity is an important initiating aspect of the wound healing response and how cells migrate to repair damaged tissues (12, 13).
Cell migration to HMGB1 requires the action of several interconnected signal transduction pathways. RAGE ligand-induced cell migration requires RAGE interaction with Diaphanous-1 (Dia-1), which is required for Rac-1 and Cdc42 regulated cell movement (14). We have previously shown that cellular chemotaxis towards HMGB1 in vitro requires canonical Nuclear Factor κB (NF-κB) activation in a variety of cell types (fibroblasts, mesoangioblasts, macrophages and neutrophils) in vitro and also for the respective migration of neutrophils and mesoangioblasts in in vivo mouse models of HMGB1-elicited peritonitis and muscle damage (15, 16). HMGB1 induction of canonical NF-κB signaling and fibroblast chemotaxis requires ERK (extracellular signal-regulated kinase) activation (15) and SFKs (Src family kinases), which re-organize the cellular cytoskeleton and induce Src, FAK and Paxillin phosphorylation (17). Time-lapse video microscopy experiments have revealed the IKKβ and IKKα signaling pathways are essential for cells to become polarized to an HMGB1 gradient, indicative of critical functional roles in the initial steps of directed cell movement (16). Finally, we have also reported that the activity of IKKβ-dependent canonical NF-κB signaling is mechanistically essential for cells to maintain RAGE expression for their HMGB1 migratory response, while the IKKα-driven non-canonical NF-κB p52-RelB signaling pathway is simultaneously critical for HMGB1 elicited chemotaxis for a different reason (16).
Here, we have defined the mechanism of action of the IKKα-driven NF-κB RelB/p52 signaling pathway for HMGB1 chemotaxis. Surprisingly, for cells to migrate in response to HMGB1, the NF-κB non-canonical pathway is solely required to maintain an autocrine loop of CXLC12, also known as stromal cell-derived factor-1 (SDF-1). A neutralizing CXCL12 monoclonal antibody completely blocks the HMGB1 migration responses of fibroblasts and macrophages. In addition, incubating IKKα or NF-κB p52 deficient cells with a limiting amount of recombinant CXCL12 rescued their directed migration response to HMGB1; and NF-κB p52 KO fibroblasts engineered to express near physiological levels of CXCL12 migrate in response to HMGB1 akin to WT cells. Moreover, AMD3100, a specific antagonist of CXCL12's G-protein coupled receptor CXCR4 (18–20) and a anti-CXCR4 monoclonal antibody both prevented HMGB1 migration responses, indicating that the CXCL12 receptor CXCR4 in addition to HMGB1's receptor RAGE is also an essential requirement for cell migration towards HMGB1. Taken together our results reveal that cell migration towards HMGB1 requires the IKKα/non-canonical NF-κB pathway to ensure that migrating cells continuously secrete CXCL12/SDF-1, which would either interact and/or co-signals with HMGB1 via their respective receptor CXCR4 and RAGE for cells to migrate in response to HMGB1.
MATERIALS AND METHODS
Conditional IKKα KO mice
Mice with IKKα alleles flanked by LoxP recombination sites (IKKαf/f mice) that express Cre recombinase under the control of the macrophage lysozyme (MLys) promoter only in mature macrophages (MΦ) and neutrophils (IKKαf/f:MLysCre mice) have been previously described (16). All animal work was approved by Stony Brook University's IACUC committee in accordance with USA NIH grant guidelines.
Reagents
Full length LPS-free recombinant HMGB1 protein was obtained from HMGBiotech (Turin, Italy) and was rigorously tested to be LPS free by the manufacturer. Recombinant murine SDF-1) was from PeproTech (Rocky Hill, NJ). Other reagents were obtained from the following suppliers: 8 μm pore size cellulose nitrate filters (for macrophages) and PVP-free polycarbonate filters (for fibroblasts) for 48 well microchemotaxis (Boyden-type) chambers (Neuroprobe Inc., Cabin John, MD); fibronectin (Roche), human recombinant PDGF (R&D Systems, Minneapolis, MN); Human recombinant complement C5a, 4-hydroxytamoxifen (4-OHT), AMD3100 (Sigma, St. Louis, MO), alexa flour 488 conjugated streptavidin (Molecular probes, Eugene, OR), alexa flour 647 conjugated anti-CXCR4 antibody (BioLegend) and biotin conjugated anti-RAGE antibody, anti-CXCR4 neutralizing monoclonal antibody, mouse IgG2a and rat IgG2b isotype control antibodies (all from R&D systems).
Cells and tissue culture
Immortalized WT, IKKα KO and p52 KO MEFs were maintained as previously described in Dulbecco's Modified Eagle's Medium (DMEM) with 10% Fetal Bovine Serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin. Bone marrow progenitors from the femurs of IKKα WT (IKKαf/f) and IKKαf/f:MLysCre and adult mice were differentiated to MΦ in M-CSF conditioned DMEM/10%FBS for 7 days as previously described (16).
Retroviral transduction
NF-κB p52 KO MEFs were stably transduced by a diluted stock of a Moloney murine retroviral vector containing a human CXCL12 cDNA expressed as part of a bi-cistronic IRES-GFP (21). A murine CXCR4 cDNA was subcloned upstream of an IRES-puromycin cassette in the BIP murine Moloney retroviral vector (22, 23). The generation of amphotyped viruses, infection of cells and selection of stable GFP or puromycin resistant cell populations have been previously described (16, 22, 23).
In vitro chemotaxis assays
Microchemotaxis chamber assays with MEFs and MΦ were performed as previously described (16). MEF and MΦ migration assays were performed with 5 × 104 and 1 × 105 cells respectively per well of a 48 well microchemotaxis (Boyden-type) chamber. Briefly, chemotaxis of MEFs utilized 8 μm pore size PVP-free polycarbonate filters pre-coated with fibronectin (50 μg/ml) and chambers were incubated for 3 hours at 37°C. MEFs were quantified as the number of migrated cells per high power field (400×), and the background control (serum-free media) was subtracted so data are expressed as net migrated cells. Macrophage chemotaxis employed cellulose nitrate filters (8 μm pore size) and chambers were incubated for 3 hours at 37°C. Distance migrated into the filter was measured by the leading front method as previously described (24). Macrophage migration data is presented as net cell movement per 3 hours.
CXCL12/SDF-1 ELISA Assays
WT, p52 KO and p52 KO + CXCL12-GFP mouse embryonic fibroblasts were plated at 200,000 cells per well in 6 well plates in 2 ml of complete growth media. Because CXCL12 is constitutively secreted by cells at very low levels, to accurately quantify the relative levels of the cytokine produced by each cell type, we prepared conditioned cell supernatants in overnight incubations for CXCL12 ELISA analysis. Thus, cells were incubated at 37 °C and 5% CO2 for 18–19 hours in complete growth media, after which their supernatants were collected and assayed for CXCL12 with a SDF-1α ELISA (R&D systems) according to the manufacturer's protocol. To account for variations in cell division rates during the overnight incubation, cells were counted at the end of the 18–19 hr incubation; and CXCL12 values were normalized to 200,000 cells per well.
FACS purification and analysis
NF-κB p52 KO MEFs, which had been stably tranduced with a diluted stock of a CXCL12-GFP expressing retrovirus, were expanded for four days and GFP positive cells were purified by flow cytometry (shown in Supplementary Figure 1). FACS data were also analyzed with FlowJo software (Tree Star Inc.).
Statistical analysis
P values were determined with either Prism V4.0 Software or InStat. (both GraphPad Inc., San Diego, CA) to 4 significant figures by one way ANOVA with Tukey's multiple comparisons post test.
RESULTS
Neutralizing the CXCL12/SDF-1 actively produced by cells completely blocks their migration response to HMGB1
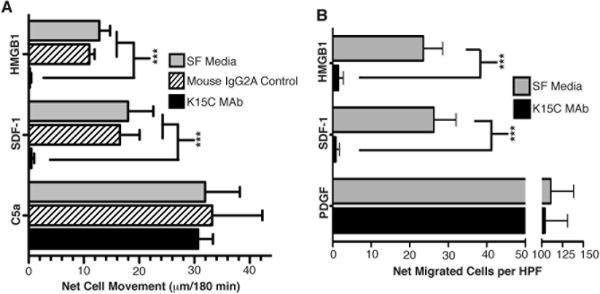
Recently, we reported conclusive in vitro and in vivo evidence that the IKKβ-dependent canonical and IKKα-dependent non-canonical signaling pathways were simultaneously essential for cells to migrate towards HMGB1. Multiple lines of evidence point to RAGE, HMGB1's ubiquitously expressed receptor, as the required target of the IKKβ-dependent RelA/p5 pathway; but the critical target or targets of IKKα-dependent RelB/p52 pathway remained undefined. Basal and induced transcription of CXCL12/SDF-1 directly depends on NF-κB p52/RelB heterodimers (25, 26) and we found that CXCL12 mRNA was modestly induced by HMGB1 with a dependency on IKKα (16). Thus, we investigated the possibility that cells may need to actively produce CXCL12/SDF-1 for their migratory response to HMGB1. CXCL12 actively secreted by HMGB1 responding cells was neutralized using the K15C monoclonal antibody that specifically targets an amino terminal epitope in CXCL12 (27). As shown in Figure 1, K15C completely blocked both fibroblast and primary macrophage chemotactic responses to HMGB1. As expected K15C also completely blocked cell migration to CXCL12 itself as a positive control, but it had no effect on MEF and macrophage chemotaxis to their respective positive controls PDGF or C5a (Figure 1). Moreover, cell migration assays performed with an irrelevant mouse IgG2a antibody as an isotype-matched negative control showed no effect on cell migration in response to HMGB1 or CXCL12/SDF-1 (Figure 1A).
Figure 1. HMGB1 Chemotaxis of MEFs and primary mature macrophages requires their production of CXCL12.
Migration assays primary WT MΦ (A) and immortalized WT MEFs (B) were performed in 48 well microchemotaxis chambers as previously described (16) in response HMGB1 (50 ng/ml), PDGF (10 ng/ml) (MEF migration positive control) or C5a (2 nM) (MΦ migration positive control) for 3 hrs. Neutralizing monoclonal antibody against CXCL12 (K15C) was added to the cells where indicated at 30 μg/ml, which effectively blocks CXCL12 activity (27). Migration results with 30/μg/ml of a mouse IgG2A isotype control antibody are also shown in Panel A. Bars are distances migrated through filter pores (WT MΦ in A) or net cell movement per High Power Field (HPF) after subtracting basal migration in serum free media (WT MEFs in B). Experiments were repeated 3–5 times each in duplicate. All error bars are standard error of the mean. Mean values of WT MΦ basal migration distances towards serum free media controls were 35 ± 2.6 and 35 ± 3.3 μm with and without K15C respectively and 31 ± 1.6 μm for mouse IgG2a controls. Mean values of WT MEF basal migration towards serum free media controls were 26 ± 1.1 and 22 ± 1.4 cells per HPF with and without K15C respectively. P values were determined by two (Panel A) or one (Panel B) way ANOVA with Tukey's post test (***p = <0.001).
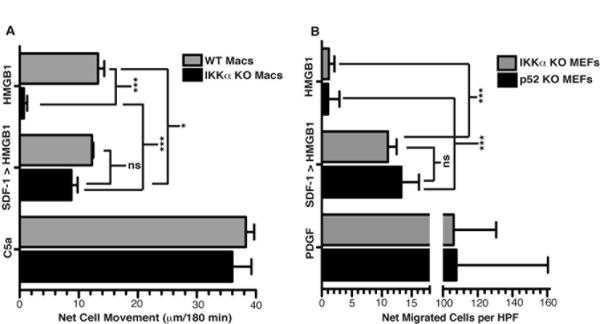
Recombinant CXCL12/SDF-1 added to either IKKα or p52 deficient cells rescues their defective HMGB1 migration responses up to WT levels
Next, in light of the above results, we asked if CXCL12/SDF-1 was the sole IKKα/p52/RelB target whose production by cells was essential for their HMGB1 migration response. To test this idea, we mixed recombinant CXCL12/SDF-1α with either IKKα or p52 KO cells and assessed their migration response to an HMGB1 gradient. Indeed, supplementing IKKα and p52 KO MEFs (Figure 2A) or IKKα conditional KO primary macrophages, (differentiated from the bone marrow progenitors of IKKαf/f;MLysCre mice), (Figure 2B) with only 5 ng/ml of CXCL12/SDF-1, (1/10th the necessary concentration for CXCL12/SDF-1 migration assays) rescued their HMGB1 chemotactic responses. In addition, a dose response experiment with IKKα conditional KO macrophages showed that as little as 0.5 ng/ml of CXCL12/SDF-1 was sufficient to rescue their HMGB1 migratory response (Suppl. Fig. 2). Although this type of ectopic addition experiment provides reasonable suggestive evidence that CXCL12 may be the target of the IKKα/non-canonical NF-κB pathway that is required for cell migration to HMGB1, this strategy alone is not sufficient to make this conclusion for two main reasons: (1) the very low level of CXCL12 produced by WT cells in the context of short term cell migration assays performed in serum free media precludes its quantification by ELISA to determine if this is comparable to the effective ectopic doses of CXCL12; and (2) the localized concentration of CXCL12 produced by WT cells is likely to be considerably higher at the cell surface before it diffuses into the surrounding media, and this can not be replicated by ectopic CXCL12 addition to cells.
Figure 2. Supplementing IKKα or p52 KO cells with recombinant CXCL12/SDF-1 rescues their migration responses to HMGB1.
IKKα conditional KO primary MΦ (A) or immortalized IKKα or p52 KO MEFs (B) with or without supplementation by 5 ng/ml of recombinant CXCL12/SDF-1 were subjected to HMGB1 migration assays as indicated; and PDGF served as a positive migration control for the KO cells. Experiments were performed 3–5 times (3× for WT and 5× for all KO samples) each in duplicate. Mean values of WT and IKKα KO MΦ basal migration distances towards serum free media controls were 30 ± 1.0 and 31 ± 2.5 μm respectively. Mean values of IKKα KO and p52 KO MEF basal migration towards serum free media controls were 11 ± 4.0 and 6 ± 2.0 cells per HPF respectively. Basal migration in serum free media with independent preparations of WT MΦ and MEFs routinely varied between 20 to 25 microns and 15 to 20 cells per HPF respectively over the 3 hour chemotaxis assay. Error bars are standard error of the mean. P values were determined by one way ANOVA with Tukey's post test as follows: ***p = <0.001; *p = <0.05; ns = not significant.
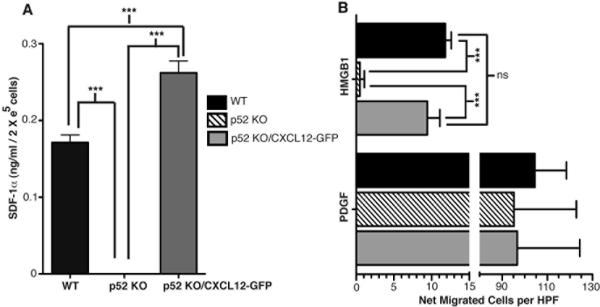
Next, as a more physiologically rigorous experiment that does not have the inherent caveats of adding CXCL12 to cells, we enforced constitutive CXCL12 expression in p52 KO MEFs by stably transducing them with a murine Moloney retrovirus harboring a CXCL12-IRES-GFP bicistronic expression cassette (21). We used a diluted stock of CXCL12-GFP retrovirus, so that p52 KO target cells would be infected by one retroviral particle per cell to avoid over-expressing the CXCL12 transgene. Under these limiting retroviral infection conditions only ~4% of p52 KO cells were clearly GFP positive, and the latter cells were FACS purified for our migration assays (see Suppl. Fig. 3). Next, we employed a quantitative CXCL12 ELISA assay to compare the relative levels of CXCL12 produced by WT, p52 KO and p52 KO CXCL12-GFP cells. To accurately quantify the levels of CXCL12 produced by each cell type, we prepared 18–19 hour conditioned cell supernatants from 2 × 105 cells seeded in multiple wells of 6 well plates. These quantitative ELISA assays showed that FACS purified p52 KO CXCL12-GFP positive cells produced CXCL12 at levels only somewhat higher than WT control MEFs, while p52 KO MEFs were completely deficient for CXCL12 secretion (Figure 3A). We employed the above strategy to validate that our engineered p52 KO CXCL12-GFP cells produced comparable levels of CXCL12 as WT MEFs, because the very low level of the cytokine constitutively produced by WT cells precluded its direct quantification by ELISA in the context of our cell migration assays, which were initiated with freshly prepared cells only 30–60 minutes after their resuspension in serum-free media. Importantly, p52 KO/CXCL12-GFP cells migrated towards HMGB1 with an efficiency that was statistically comparable to WT MEFs, while in comparison their p52 KO counterparts were completely negative for HMGB1-induced migration (Figure 3B). Taken together, the above experiments definitively demonstrate that CXCL2/SDF-1 is the sole target of the IKKα/non-canonical NF-κB pathway that is essential for cell migration in response to HMGB1.
Figure 3. NF-κB p52 KO cells engineered to express near physiological levels of CXCL12 migrate in response to HMGB1 akin to WT cells.
(A) CXCL12 ELISA assays of supernatants of WT MEFs, NF-κB p52 KO MEFs and FACS purified CXCL12-GFP expressing p52 KO MEFs; (B) Boyden chamber cell migration assays of WT MEFs compared to p52 KO and p52 KO/CXCL12-GFP cells. ELISA assays (A) were performed 6 times. Migration assays (B) were performed 3–4 times (3× for WT and 4× for p52 KO and p52 KO/CXCL12-GFP cells) each in duplicate. Mean values of WT, p52 KO and p52 KO + CXCL12 MEF basal migration towards serum free media controls were 7 ± 1.0, 11 ± 2.1 and 9 ± 1.8 cells per HPF respectively. P values were derived by one way ANOVA with Tukey's post test as follows: ***p = <0.001; ns = not significant.
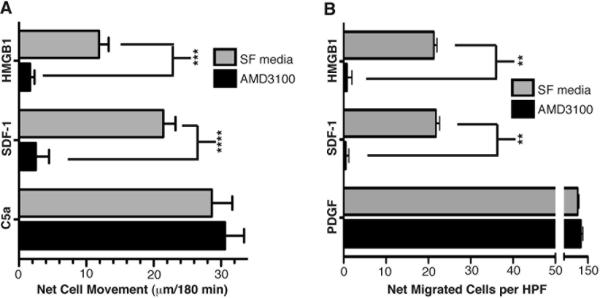
Cellular chemotactic responses to HMGB1 are blocked by AMD3100, a specific CXCR4 antagonist and also by a monoclonal anti-CXCR4 antibody
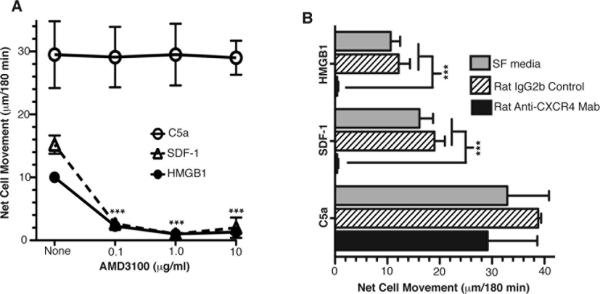
CXCR4, a ubiquitously expressed G-protein coupled receptor, uniquely promotes CXCL12-mediated chemotaxis (28–31); and it was previously reported that in addition to HMGB1's ubiquitously expressed receptor RAGE, cellular chemotaxis towards HMGB1 also requires the activity of at least one G-protein coupled receptor because pertussis toxin treatment abolishes the HMGB1 chemotactic response (12). Since our data clearly shows that non-canonical NF-κB regulated CXCL12 production is critical for cell migrations towards HMGB1, we next began to investigate the potential role of CXCR4 in HMGB1-mediated chemotactic responses. To this end we performed HMGB1 migration assays in the presence of AMD3100, a highly specific bicyclam non-peptide antagonist of CXCR4 that has no inhibitory effects on other G protein coupled receptors at concentrations up to 50 μg/ml (18–20). AMD3100 (10 μg/ml = 12.5 μM) completely ablated the HMGB1 migration responses of primary macrophages (Figure 4A) and MEFs (Figure 4B). As expected this same dose of AMD3100 extinguished cell migration to CXCL12/SDF-1 (as a positive control); but it had no effect on cell migration to the positive controls PDGF or C5a (Figure 4A&4B). To further demonstrate that the inhibitory effect of AMD3100 was specific for CXCR4, we performed additional experiments with several other independently prepared batches of in vitro differentiated primary mature macrophages. First, we exposed WT macrophages to different concentrations of AMD3100 and found that a drug dose as low as 0.1 μg/ml (0.125 μM) was sufficient to inhibit cell migration to either HMGB1 or CXCL12/SDF-1 to the same degree (Figure 5A). Importantly, 0.125 μM is just above the 50% inhibitory concentration (IC50) of AMD3100 previously shown to inhibit CXCL12/SDF-1 induced calcium flux in a variety of different cell types (20).
Figure 4. AMD3100, a CXCR4 antagonist, blocks WT MEF and MΦ migration in response to HMGB1.
WT MΦ (A) and WT MEF (B) migrations were performed with or without the CXCR4 inhibitor AMD3100 (10 μg/ml) as indicated. Experiments were performed 4× in Panel A and 3× in B, each in duplicate. Mean values of WT MΦ basal migration distances towards serum free media controls were 28 ± 3.0 and 29 ± 3.0 μm with and without AMD3100 respectively. Mean values of WT MEF basal migration towards serum free media controls were 21 ± 3.0 and 16 ± 2.0 cells per HPF with and without AMD3100 respectively. All error bars are standard error of the mean. P values were determined by one way ANOVA with Tukey's post test as follows: ****p = <0.0001; ***p = <0.001, **p = 0.0035.
Figure 5. Exposure of WT MΦ to either low doses of AMD3100 or to an anti-CXCR4 neutralizing antibody show that CXCR4 activity is essential for cell migration in response to HMGB1.
(A) Effects of different doses of AMD3100 on WT MΦ migrations as indicated. (B) WT MΦ migrations were done in the presence and absence of a neutralizing anti-CXCR4 monoclonal antibody (15 μg/ml) as indicated. Migration results with 15/μg/ml of a rat IgG2B isotype control antibody are also shown in Panel B. Experiments were performed 3–5×, each in duplicate. Mean values of WT MΦ basal migration distances towards serum free media controls were 24 ± 4.0 μm and 30 ± 1.2 μm for rat IgG2b controls. All error bars are standard error of the mean. P values were determined by one way ANOVA with Tukey's post test as follows: ***p = <0.001.
Finally, WT macrophage chemotaxis assays were also done in the presence of a rat anti-mouse CXCR4 neutralizing monoclonal antibody, which completely blocked migration responses to either HMGB1 or CXCL12/SDF-1 but had no effect on their migration to C5a (Figure 5B). In addition, cell migration assays performed with an irrelevant rat IgG2b antibody as an isotype-matched negative control showed no effect on cell migration in response to HMGB1 or CXCL12/SDF-1 (Figure 5B).
DISCUSSION
Cell migration is a sophisticated, multi-step phenomenon critical for animal development, innate and adaptive immunity and the recognition and repair of damaged tissue. Inappropriate and/or dysregulated cell migration contributes to the progression of cancers, poor wound healing, inflammatory-induced tissue injury and several other maladies {reviewed in (32, 33)}. In cell nuclei, HMGB1 functions as an essential chromosome architectural protein, but remarkably, outside cells it acts as a cytokines and chemoattractant. Moreover, extracellular HMGB1 allows the body to initially recognize and subsequently begin to repair damaged tissue by first recruiting pro-inflammatory cells which are followed by cells with the ability to orchestrate tissue regeneration and scar formation (3, 4, 13, 34).
HMGB1-mediated receptor engagement activates several signaling pathways including NF-κB, Erk and Src, that are each required to induce and maintain cell migration towards HMGB1 (15–17); but the downstream targets of these pathways, whose activity are required for HMGB1 chemotactic responses, remain unclear. Although many targets of the NF-κB signaling pathways encode cytokines, chemokines and receptors, which are systemically important for cell migration responses involved in immunity {reviewed in (35–37)}, NF-κB activity is not intrinsically necessary inside cells for their directed movement towards a variety of chemotactic stimuli (15, 16). Thus, because both major arms of NF-κB signaling (IKKβ-driven canonical and IKKα-driven non-canonical) are simultaneously critical for eliciting HMGB1-mediated cell migration responses, the cell signaling machinery and regulation of HMGB1 chemotactic responses are expected to be considerably more complex than observed for most if not all other chemoattractants. Here we have shown that sustained production of CXCL12/SDF-1 by the IKKα driven non-canonical NF-κB signaling pathway is essential for cell movement towards HMGB1. Thus, HMGB1 chemotactic responses are unique by requiring the IKKβ-driven canonical NF-κB pathway to maintain expression of HMGB1's receptor RAGE in responder cells (16); and as shown here the IKKα/non-canonical NF-κB regulated, functional interplay between cell produced CXCL12 and extracellular HMGB1.
As RAGE is the receptor for HMGB1, our results herein suggest that CXCL12 interplay with HMGB1 provides cells with an essential accessory or co-receptor signal via its receptor CXCR4 to enable directed cell movement towards HMGB1. Interestingly, secretion of HMGB1 by macrophages and dendritic cells was recently shown to enhance their chemotaxis towards recombinant CXCL12/SDF-1 (38), suggesting that the combined action of HMGB1 and CXCL12/SDF-1 signaling may collaborate to enhance specific cell migration responses. Moreover multiple reports have shown that in addition to evidence of HMGB1 interacting with CXCL12/SDF-1 (38), HMGB1 also forms specific, functional complexes with a selected number of endogenous and exogenous effectors (including CpG-ODNS, LPS, IL-1β and nucleosomes) to enhance pro-inflammatory cytokine production by HMGB1 responding cells (39–46). Herein our results with AMD3100, a highly specific CXCR4 antagonist, and a neutralizing anti-CXCR4 monoclonal antibody both reveal that the activity of CXCL12's receptor CXCR4 is critical for HMGB1-mediated cell migration responses (Figures 4 and 5). Recently we demonstrated that IKKβ KO macrophages, MEFs and neutrophils were incompetent for HMGB1 migration and that this defect could be fully corrected by enforced over-expression of RAGE, a direct target of IKKβ-dependent NF-κB p65/p50 signaling (16), indicating that IKKβ-driven canonical NF-κB signaling was necessary to maintain RAGE expression for cells to migrate towards HMGB1. Interestingly, we find that IKKβ KO-RAGE MEFs, akin to their WT counterparts, require CXCR4 activity to migrate in response to HMGB1, as AMD3100 blocks their HMGB1-induced migration (Suppl. Fig. 3A). Other experiments with IKKβ KO-RAGE MEFs show that the K15C anti-CXCL12 monoclonal antibody also blocks their HMGB1 migration (data not shown). Thus, even RAGE over-expression is not sufficient to supplant the need for the critical, additional requirement of CXCL12-CXCR4 engagement for cell migration to HMGB1. However, unlike RAGE, enforcing CXCR4 in IKKβ KO cells failed to rescue their HMGB1 migration response (Suppl. Fig. 3B); and CXCR4 over-expression did not alter the barely detectable levels of cell surface RAGE on IKKβ KO cells (Suppl. Fig. 3C–D). Thus, on the basis of our collective data, we propose that CXCL12-mediated engagement of CXCR4 works as an essential co-receptor signal for RAGE receptor-dependent HMGB1 migration responses. Interestingly, CXCL12/SDF-1 binding to CXCR4 can also functionally couple to the output of other receptors. For instance, CXCL12-CXCR4 engagement is also known to induce CXCR4-TCR heterodimerization, which is required for a variety of signaling outcomes in T cells including enhanced gene transcription and cytokine production, increased calcium ion concentrations in conjunction with CXCR4 endocytosis and CXCL12-mediated cell migration (47–49). Although we can not formally rule out the possibility that CXCL12 engagement of CXCR4 might have other indirect effects such as enhancing HMGB1 binding to RAGE, we consider this latter possibility less likely because HMGB1 has been previously shown to directly bind to RAGE with higher affinity than other RAGE ligands (5, 50). Rather, we envision that the essential second cell signal delivered by CXCL12-CXCR4 engagement may either be necessary to collaborate with HMGB1-mediated RAGE signaling for cells to sense and move towards an HMGB1 gradient, or cell secreted CXCL12 may be interacting with HMGB1 in a “piggy-backing” fashion to facilitate secreted CXCL12's delivery to cell surface CXCR4 thereby permitting cell migration towards HMGB1. Future work will be directed towards the further elaboration of the remaining details of this novel mechanism.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Drs. Fernando Arenzana-Seisdedos and Patricia Rueda and for providing us with sufficient quantities of K15C anti-CXCL12 monoclonal antibody for our experiments. We also thank Dr. Andrew Zannettino for his kind gift of CXCL12-GFP expressing murine retroviral vector, and Drs. Yuelei Shen and Dan Littman for providing us with their pMIGR-MuCXCR4 expression construct. We also are indebted to Ms. Mahalakshmi (Maha) Ramadass for her help with PCR-based mycoplasma testing of all cell stocks. The expert technical help of the Stony Brook Flow facility is also gratefully acknowledged for their assistance in the FACS purification of GFP positive cells.
Grant support: The initial phase of this work was supported by the MAIN FP6 European Union Network of Excellence (MP and KBM) and subsequently by USA NIH grants GM066882 awarded to KBM and GM063769 to RRK and 5T32 GM008468 (for support of DMH) awarded to Stony Brook University's Molecular and Cellular Biology graduate training program.
Non-standard abbreviations
- C5a
complement component 5a
- HMGB1
Dia-1 (diaphanous 1); High Mobility Group Box 1
- HPF
high power field
- IKK
Inhibitor of NF-κB kinase
- MEFs
Mouse Embryo Fibroblasts
- MΦ
mature macrophages
- PDGF
platelet derived growth factor
- RAGE
receptor for advanced glycation end products
- SDF-1/CXCL12
Stromal Cell Derived Factor 1/C-X-C motif ligand 12
- SF
serum free media
- SFK
(Src family kinases)
- WT
wild type
- GFP
green fluorescent protein
Footnotes
RRK and KBM contributed equally to this work
Author current addresses: MP's current address: Department of Experimental Pathology, Alma Mater Studiorum Universita' di Bologna, 40126 Bologna, Italy
The authors declare no direct financial interest.
REFERENCES
- 1.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 2.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. Embo J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86:573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 5.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D, Morser J, Stern D, Schmidt AM. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 6.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83:876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, Gamboni-Robertson F, He Q, Svetkauskaite D, Kim JY, Strassheim D, Sohn JW, Yamada S, Maruyama I, Banerjee A, Ishizaka A, Abraham E. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am J Physiol Cell Physiol. 2006;290:917–924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- 8.Yu M, Wang H, Ding A, Golenbock DT, Latz E, Czura CJ, Fenton MJ, Tracey KJ, Yang H. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock. 2006;26:174–179. doi: 10.1097/01.shk.0000225404.51320.82. [DOI] [PubMed] [Google Scholar]
- 9.Kokkola R, Andersson A, Mullins G, Ostberg T, Treutiger CJ, Arnold B, Nawroth P, Andersson U, Harris RA, Harris HE. RAGE is the major receptor for the proinflammatory activity of HMGB1 in rodent macrophages. Scand J Immunol. 2005;61:1–9. doi: 10.1111/j.0300-9475.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 10.Andersson U, Erlandsson-Harris H, Yang H, Tracey KJ. HMGB1 as a DNA-binding cytokine. J Leukoc Biol. 2002;72:1084–1091. [PubMed] [Google Scholar]
- 11.Herold K, Moser B, Chen Y, Zeng S, Yan SF, Ramasamy R, Emond J, Clynes R, Schmidt AM. Receptor for advanced glycation end products (RAGE) in a dash to the rescue: inflammatory signals gone awry in the primal response to stress. J Leukoc Biol. 2007;82:1–9. doi: 10.1189/jlb.1206751. [DOI] [PubMed] [Google Scholar]
- 12.Palumbo R, Sampaolesi M, De Marchis F, Tonlorenzi R, Colombetti S, Mondino A, Cossu G, Bianchi ME. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J Cell Biol. 2004;164:441–449. doi: 10.1083/jcb.200304135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palumbo R, Bianchi ME. High mobility group box 1 protein, a cue for stem cell recruitment. Biochemical Pharmacology. 2004;68:1165–1170. doi: 10.1016/j.bcp.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 14.Hudson BI, Kalea AZ, del Mar Arriero M, Harja E, Boulanger E, D'Agati V, Schmidt AM. Interaction of the RAGE Cytoplasmic Domain with Diaphanous-1 Is Required for Ligand-stimulated Cellular Migration through Activation of Rac1 and Cdc42. J. Biol. Chem. 2008;283:34457–34468. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palumbo R, Galvez BG, Pusterla T, De Marchis F, Cossu G, Marcu KB, Bianchi ME. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-{kappa}B activation. J. Cell Biol. 2007;179:33–40. doi: 10.1083/jcb.200704015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penzo M, Molteni R, Suda T, Samaniego S, Raucci A, Habiel DM, Miller F, Jiang H.-p., Li J, Pardi R, Palumbo R, Olivotto E, Kew RR, Bianchi ME, Marcu KB. Inhibitor of NF-{kappa}B Kinases {alpha} and {beta} Are Both Essential for High Mobility Group Box 1-Mediated Chemotaxis. J Immunol. 2010;184:4497–4509. doi: 10.4049/jimmunol.0903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palumbo R, De Marchis F, Pusterla T, Conti A, Alessio M, Bianchi ME. Src family kinases are necessary for cell migration induced by extracellular HMGB1. J Leukoc Biol. 2009;86:617–623. doi: 10.1189/jlb.0908581. [DOI] [PubMed] [Google Scholar]
- 18.Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, Moore JP. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 19.Matthys P, Hatse S, Vermeire K, Wuyts A, Bridger G, Henson GW, De Clercq E, Billiau A, Schols D. AMD3100, a potent and specific antagonist of the stromal cell-derived factor-1 chemokine receptor CXCR4, inhibits autoimmune joint inflammation in IFN-gamma receptor-deficient mice. J Immunol. 2001;167:4686–4692. doi: 10.4049/jimmunol.167.8.4686. [DOI] [PubMed] [Google Scholar]
- 20.Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS letters. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- 21.Diamond P, Labrinidis A, Martin SK, Farrugia AN, Gronthos S, To LB, Fujii N, O'Loughlin PD, Evdokiou A, Zannettino ACW. Targeted Disruption of the CXCL12/CXCR4 Axis Inhibits Osteolysis in a Murine Model of Myeloma-Associated Bone Loss. Journal of Bone and Mineral Research. 2009;24:1150–1161. doi: 10.1359/jbmr.090210. [DOI] [PubMed] [Google Scholar]
- 22.Massa PE, Li X, Hanidu A, Siamas J, Pariali M, Pareja J, Savitt AG, Catron KM, Li J, Marcu KB. Gene expression profiling in conjunction with physiological rescues of IKKalpha null cells with wild type or mutant IKKalpha reveals distinct classes of IKKalpha/NF-kappaB-dependent genes. J. Biol. Chem. 2005;280:14057–14069. doi: 10.1074/jbc.M414401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penzo M, Massa PE, Olivotto E, Bianchi F, Borzi RM, Hanidu A, Li X, Li J, Marcu KB. Sustained NF-kappaB activation produces a short-term cell proliferation block in conjunction with repressing effectors of cell cycle progression controlled by E2F or FoxM1. Journal of Cellular Physiology. 2009;218:215–227. doi: 10.1002/jcp.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zigmond SH, Hirsch JG. Leukocyte locomotion and chemotaxis. New methods for evaluation, and demonstration of a cell-derived chemotactic factor. J Exp Med. 1973;137:387–410. doi: 10.1084/jem.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejardin E, Droin NM, Delhase M, Haas E, Ca Y, Makris C, Karin M, Ware CF, Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 26.Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, Jegga AG, Aronow BJ, Ghosh G, Rickert RC, Karin M. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. Embo J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amara A, Lorthioir O, Valenzuela A, Magerus A, Thelen M, Montes M, Virelizier J-L, Delepierre M, Baleux F, Lortat-Jacob H, Arenzana-Seisdedos F. Stromal Cell-derived Factor-1-Alpha Associates with Heparan Sulfates through the First beta-Strand of the Chemokine. Journal of Biological Chemistry. 1999;274:23916–23925. doi: 10.1074/jbc.274.34.23916. [DOI] [PubMed] [Google Scholar]
- 28.Zou Y-R, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 29.Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- 30.Suratt BT, Petty JM, Young SK, Malcolm KC, Lieber JG, Nick JA, Gonzalo J-A, Henson PM, Worthen GS. Role of the CXCR4/SDF-1 chemokine axis in circulating neutrophil homeostasis. Blood. 2004;104:565–571. doi: 10.1182/blood-2003-10-3638. [DOI] [PubMed] [Google Scholar]
- 31.Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends in Immunology. 2007;28:299–307. doi: 10.1016/j.it.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13:R756–759. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 34.Dumitriu IE, Bianchi ME, Bacci M, Manfredi AA, Rovere-Querini P. The secretion of HMGB1 is required for the migration of maturing dendritic cells. J Leukoc Biol. 2007;81:1–8. doi: 10.1189/jlb.0306171. [DOI] [PubMed] [Google Scholar]
- 35.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 37.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 38.Campana L, Bosurgi L, Bianchi ME, Manfredi AA, Rovere-Querini P. Requirement of HMGB1 for stromal cell-derived factor-1/CXCL12-dependent migration of macrophages and dendritic cells. J Leukoc Biol. 2009;86:609–615. doi: 10.1189/jlb.0908576. [DOI] [PubMed] [Google Scholar]
- 39.Rouhiainen A, Tumova S, Valmu L, Kalkkinen N, Rauvala H. Analysis of proinflammatory activity of highly purified eukaryotic recombinant HMGB1 (amphoterin) J Leukoc Biol. 2007;81:49–58. doi: 10.1189/jlb.0306200. [DOI] [PubMed] [Google Scholar]
- 40.Tian J, Avalos AM, Mao S-Y, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov S, Dragoi A-M, Wang X, Dallacosta C, Louten J, Musco G, Sitia G, Yap GS, Wan Y, Biron CA, Bianchi ME, Wang H, Chu W-M. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110:1970–1981. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn JH, Oh YJ, Kim ES, Choi JE, Shin J-S. High Mobility Group Box 1 Protein Binding to Lipopolysaccharide Facilitates Transfer of Lipopolysaccharide to CD14 and Enhances Lipopolysaccharide-Mediated TNF-{alpha} Production in Human Monocytes. J Immunol. 2008;180:5067–5074. doi: 10.4049/jimmunol.180.7.5067. [DOI] [PubMed] [Google Scholar]
- 43.Sha Y, Zmijewski J, Xu Z, Abraham E. HMGB1 Develops Enhanced Proinflammatory Activity by Binding to Cytokines. J Immunol. 2008;180:2531–2537. doi: 10.4049/jimmunol.180.4.2531. [DOI] [PubMed] [Google Scholar]
- 44.Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, Bianchi ME, Kirschning C, Wagner H, Manfredi AA, Kalden JR, Schett G, Rovere-Querini P, Herrmann M, Voll RE. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hreggvidsdottir HS, Ostberg T, Wahamaa H, Schierbeck H, Aveberger A-C, Klevenvall L, Palmblad K, Ottosson L, Andersson U, Harris HE. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86:655–662. doi: 10.1189/jlb.0908548. [DOI] [PubMed] [Google Scholar]
- 46.Qin Y-H, Dai S-M, Tang G-S, Zhang J, Ren D, Wang Z-W, Shen Q. HMGB1 Enhances the Proinflammatory Activity of Lipopolysaccharide by Promoting the Phosphorylation of MAPK p38 through Receptor for Advanced Glycation End Products. J Immunol. 2009;183:6244–6250. doi: 10.4049/jimmunol.0900390. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Humphreys TD, Kremer KN, Bramati PS, Bradfield L, Edgar CE, Hedin KE. CXCR4 Physically Associates with the T Cell Receptor to Signal in T Cells. Immunity. 2006;25:213–224. doi: 10.1016/j.immuni.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 48.Patrussi L, Ulivieri C, Lucherini OM, Paccani SR, Gamberucci A, Lanfrancone L, Pelicci PG, Baldari CT. p52Shc is required for CXCR4-dependent signaling and chemotaxis in T cells. Blood. 2007;110:1730–1738. doi: 10.1182/blood-2007-01-068411. [DOI] [PubMed] [Google Scholar]
- 49.Kremer KN, Clift IC, Miamen AG, Bamidele AO, Qian N-X, Humphreys TD, Hedin KE. Stromal Cell-Derived Factor-1 Signaling via the CXCR4-TCR Heterodimer Requires Phospholipase C-β3 and Phospholipase C-γ1 for Distinct Cellular Responses. The Journal of Immunology. 2011;187:1440–1447. doi: 10.4049/jimmunol.1100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu R, Moric S, Wake H, Zhang J, Liub K, Izushib Y, Takahashib HK, Penga B, Nishiborio M. Establishment of in Vitro Binding Assay of High Mobility Group Box-1 and S100A12 to Receptor for Advanced Glycation Endproducts:Heparin's Effect on Binding. Acta Med. Okayama. 2009;63:203–211. doi: 10.18926/AMO/31812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.