Abstract
Ascidians (tunicates) are rich sources of structurally elegant, pharmaceutically potent secondary metabolites and more recently, potential biofuels. It has been demonstrated that some of these compounds are made by symbiotic bacteria and not by the animals themselves, and for a few other compounds evidence exists supporting a symbiotic origin. In didemnid ascidians, compounds are highly variable even in apparently identical animals. Recently, we have explained this variation at the genomic and metagenomic levels and have applied the basic scientific findings to drug discovery and development. This review discusses what is currently known about the origin and variation of symbiotically derived metabolites in ascidians, focusing on Family Didemnidae, where most research has occurred. Applications of our basic studies are also described.
INTRODUCTION
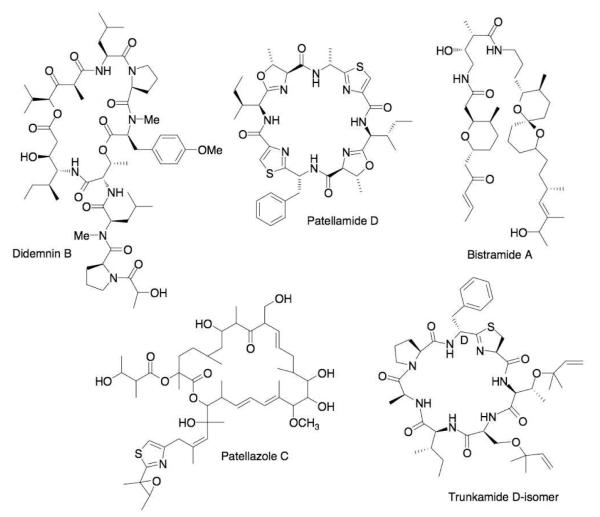
Over a course of nearly 40 years, marine natural products chemists have isolated approximately 1,080 compounds from ascidians, including clinically used compounds, preclinical leads, and other potently bioactive compounds.1,2 Probably the earliest identified didemnid chemicals were the cyclic peptides ulicyclamide and ulithiacyclamide, reported by Ireland and Scheuer in 1980.3 Since that time, Didemnidae has been the most chemically prolific ascidian family, being responsible for at least 375 (~35%) of known ascidian compounds. Didemnidae includes 578 species (21% of total species), while the 25 remaining families contain a total of 2237 described species.4 Didemnid chemicals have attracted widespread attention because of their biological activities and their intricate and elegant chemical structures. A short list includes anticancer leads such as the didemnins,5 the bistramides,6,7 the patellazoles,8-10 and trunkamide,11,12 and multidrug-resistance blockers such as some of the patellamides,13 among others (Figure 1). Ascidians are thus excellent sources of active compounds with novel structural motifs, and didemnids are prominent within this group.
Figure 1.
Bioactive compounds isolated from didemnid ascidians. The natural products shown here have unique mechanisms of action against diverse targets, and some are potent to ~100 pM against cancer-relevant cell lines. (Note: trunkamide has been reported to be both the L- and D-Phe isomer from the natural source. We have found that the pathway naturally produces the L-Phe isomer, which slowly isomerizes to the D-form.)
Even though Didemnidae is a rich source for pharmaceutical discovery, natural products chemists sometimes shy away from collecting representatives of this family. The animals take a compound colonial form, meaning that many — sometimes perhaps thousands — of individuals live in a common tunic.14 The resulting animals vary greatly in size, from tiny colonies ~1 mm in diameter to enormous sheets that can drape square meters of reef surfaces. These colonies are capable of fusing and dividing in the course of a few days,15 further complicating this situation. Even comparatively distant relatives can combine, sometimes preserving their individual genomic signatures. Similarly, often neighboring colonies appear visually identical, yet their chemical components vary greatly.16 This fact makes it difficult to collect sufficient material for natural products discovery reliably, and it can be all but impossible to supply these materials for later clinical development.
Despite these limitations, this colony and chemical variation has one great benefit: it increases the natural products potential of Didemnidae. For example, it was shown that adjacent colonies are often essentially genetically identical, except that natural product biosynthetic pathways vary in discrete ways.17 This variation leads to a natural combinatorial library, where individual, tiny colonies contain different natural products. Quite often, these products form large families of metabolites that vary in subtle structural details. The didemnids are also exceptionally rich hosts to numerous types of individual biosynthetic pathways.18 Although chemical variation is not unique to Didemnidae and is instead a very common theme in marine invertebrates, there are some genetic aspects that almost certainly reflect the unique life history of the animals and their symbiotic bacteria.
We set out to identify the biosynthetic origin of ascidian metabolites and the underlying genetic causes of this variation. Results from these studies were expected to provide unique scientific insights, but also to be immediately applicable to practical issues in marine drug discovery. Recently, we identified a novel mechanism of biosynthetic gene exchange that goes a long way toward explaining the chemical variation observed in many animals. Underlying this mechanism, specific symbiotic bacteria have been identified that live in ascidians and are responsible for producing complex “ascidian” molecules.17-21 Since these molecules originate in bacteria, a simple explanation for their variation would be to cite horizontal gene transfer, or perhaps variation in bacterial species. However, this is not what was found. Instead, we found that the bacteria Prochloron didemni in these animals represent a population, in which identical biosynthetic genes are found mixed and matched across animals in the ocean. In short, this is a departure from simple expectations as to how pathways should vary. These basic studies impact our understanding of biodiversity, but they also have direct practical outcomes in biotechnology, including improvements to the discovery, supply, and bioengineering of marine natural products.
In addition to variation within the P. didemni population, ascidians also harbor other symbiotic bacteria that make natural products. Beyond Didemnidae, this phenomenon has also been explored with the non-didemnid ascidian, Ecteinascidia turbinata, the producer of the clinically used ecteinascidin-743.22,23 Within didemnids, sometimes individual ascidians contain multiple types of symbiotic bacteria that contribute chemicals to the organism. In comparison to the wealth of data available for P. didemni, the stories are just beginning to emerge for these other bacteria, and essentially nothing is yet known about their variation or symbiosis.
In this review, we will focus on our work with the metagenome of Family Didemnidae, providing specific examples that address major problems in the science and biotechnology of marine natural products. Recent advances in ascidian symbiosis, as it relates to natural products chemistry, will also be discussed.
DIDEMNID HOUSES FOR BACTERIA
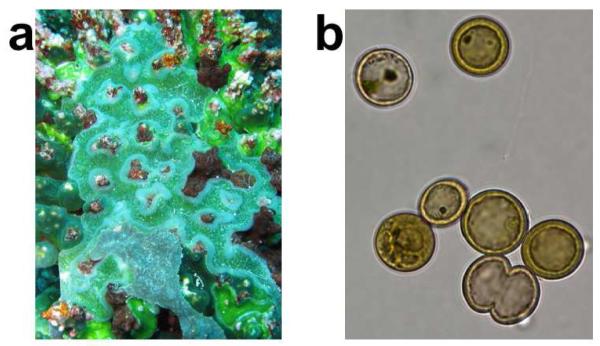
Didemnid ascidians form colonies comprising up to thousands of individual animals embedded in a common tunic.24,25 In addition to the individual animals and various tunic cells, didemnids are often crowded with cyanobacteria from the single genus, Prochloron (Figure 2).26,27 These fascinating bacteria exhibit unique properties, such as an unusual photosynthetic apparatus.28 Importantly, Prochloron can provide nearly all of the necessary carbon and nitrogen for animal survival via photosynthesis and nitrogen recycling.29-31 The cyanobacteria thus enable didemnids to thrive in otherwise nutrient-limited tropical environments.
Figure 2.
Didemnid ascidians and their symbiotic cyanobacteria, P. didemni. (A) L. patella. (B) P. didemni.
So far, there are no confirmed reports of stable cultivation of Prochloron, meaning that studies have involved examining the bacteria by removing them alive from their hosts or via metagenomic techniques. Recently, we completed an analysis of the Prochloron didemni genome, from the ascidian Lissoclinum patella.18 As expected, the genome reflected key features of the interaction between cyanobacteria and host. However, it is not obvious why the bacteria are not easily cultivated, since the metabolic pathways appear to be identical to the situation found in free-living cyanobacteria.
Other bacteria also occupy didemnid ascidians, but they have been relatively less studied than Prochloron. These mostly include other cyanobacteria,32-36 but also reports about other types of bacteria, especially proteobacteria.37-40 In the course of sequencing the P. didemni genome, we obtained an immense amount of data concerning other bacteria as well.18 These bacteria also contribute to the metabolic function of the symbiosis. Thus, even Prochloron-bearing ascidians are complex, multi-partner microbiomes.
In addition, there are reports of specific symbiotic bacteria aside from the cyanobacteria in non-didemnid ascidians. As one example that is pertinent to natural products research, the Family Perophoridae includes the ascidian E. turbinata, which contains potent peptide alkaloids, the ecteinascidins.22,23 E. turbinata also harbors many different bacteria. The ascidian’s microbiome is dominated by proteobacteria and includes the intracellular γ-proteobacterial symbiont, Candidatus Endoecteinascidia frumentensis.
BACTERIA MAKE SOME ASCIDIAN COMPOUNDS
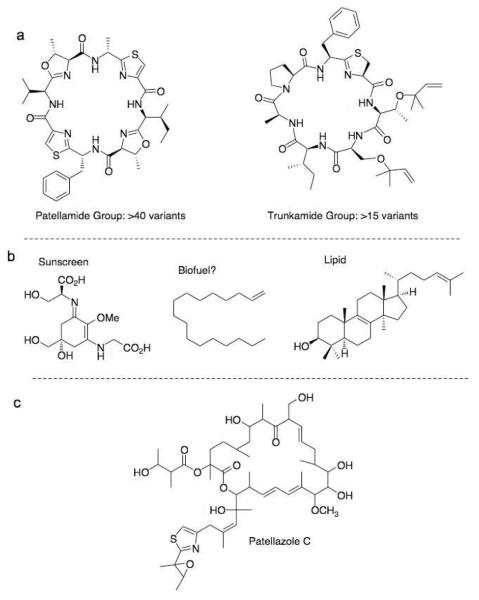
It has long been proposed that marine animal compounds originate in symbiotic bacteria. Recent reviews summarize the existing evidence for such synthesis in ascidians and in animals in general.41-44 In addition, several classes of compounds are now known to be symbiotic in origin, but these have not been described in previous reviews (Figure 3).18 Here, we describe all cases for which definitive evidence exists.
Figure 3.
Didemnid “ascidian” compounds made by symbiotic bacteria. (A) Cyclic peptides were demonstrated to originate in P. didemni. (B) Other compounds from P. didemni. (C) Compounds from other types of bacteria.
Within ascidians, symbiotic bacteria were shown to make a single group of cyclic peptides, but the vast majority of ascidian compounds have yet to be studied thoroughly. This group, termed “cyanobactins”, embraces macrocyclic compounds such as patellamides, trunkamide, patellins and many other small molecules that comprise ~6% of known metabolites from ascidians.45,46 Two strategies were used to determine the origin of cyanobactins in the ascidian, Lissoclinum patella. A strategy by Long et al. involved construction of a large-insert DNA library from L. patella samples.47 This library led to synthesis of patellamides in E. coli. Our strategy used metagenome sequencing to obtain the complete sequence of the patellamide pathway, which was then expressed in E. coli.19 The advantage of the former method is its elegant simplicity. The advantage of the latter is that the massive amount of available sequencing data quickly defined P. didemni as the only likely patellamide source in the host organism. In addition, we believed that metagenome sequencing was the most efficient way to find biosynthetic pathways and that it would become the standard method once the costs of sequencing decreased. In 2005, when our first studies were reported, the method was still expensive and scientifically very challenging, but it has become widely accessible.
In all, four metagenomes of L. patella have been sequenced thus far. These whole genomes, as well as genome fragments from many organisms, have helped to show that P. didemni makes many other compounds isolated from whole ascidians,18 including numerous cyanobactin derivatives.17,18,20,21 P. didemni almost certainly makes the mycosporine amino acids that were experimentally shown to shield the animals from ultraviolet damage.18,48 It also likely makes lipid components, including the potential biofuel component heptadec-1-ene, which is one of the major lipids found in the whole organism.18 Finally, P. didemni has the genetic capacity to make many other natural products that are as yet undetected.17
Beyond P. didemni, other bacteria almost certainly make natural products in didemnid ascidians.18 In the same ascidian in which P. didemni makes trunkamide, we found the potent antitumor polyketides, patellazoles. However, the P. didemni genome did not encode patellazole production. Instead, other bacteria harbored genes that might be responsible for synthesizing patellazoles. These genes were from the trans-acyltransferase (trans-AT) polyketide synthase group,49 a subclass of polyketide synthases that has been associated with the production of several polyketides found in marine animals.50,51 These trans-AT genes were reminiscent of those discovered by Baker’s group in examining another didemnid ascidian, the producer of polyketides known as palmerolides.40 The palmerolide producer occurs in Antarctica, indicating that tropical P. didemni is not the producer of these compounds either. Based upon 16S gene sequence analysis, it was proposed that the producer could be proteobacterial. As a result of structural similarity between the palmerolide and patellazoles structures, we propose that there may be specific natural products symbioses involving proteobacteria in ascidians. Additionally, trans-AT clusters are important in the synthesis of natural products by sponge and bryozoan symbiotic bacteria, as shown in groundbreaking studies.50,51
In addition to these trans-AT genes, we found an unprecedented absolute number and diversity of biosynthetic genes in single ascidian animals.18 These results indicate that ascidians have an unusually broad chemical capacity and suggest avenues of research into how molecular abundance is controlled in animals.
The early suspicion that symbiotic bacteria produce “ascidian” natural products was based on structural similarity. Indeed, a recent review documents 18 different families of ascidian compounds that very closely resemble bacterial natural products, and in some cases the compounds are even identical to their bacterial homologues.41 One recent advance adds strongly to this picture. The orphan anticancer drug aplidine is part of a group of compounds known as didemnins, isolated from didemnid ascidians.5 These depsipeptides look very much like they could be made by bacteria via nonribosomal synthesis, but they did not share many chemical features with characterized bacterial metabolites. Very recently, the Tsukimoto group isolated the α-proteobacterium Tistrella mobilis from marine sediments and found that it produced didemnin B in culture.52 While this does not demonstrate that the ascidian compounds are derived from these (or other) bacteria, it provides compelling indirect evidence that the compounds may have a bacterial origin.
Another recent advance involves synthesis of ecteinascidin-743 in E. turbinata. The ecteinascidins are isoquinoline alkaloids that very strongly resemble bacterial molecules such as saframycins and safracins, which are produced by bacteria via the nonribosomal peptide synthetase (NRPS) pathway.53 Using a diversity of approaches including metagenomics, metaproteomics, and metabolomics, the Sherman group identified a saframycin-like NRPS pathway in E. turbinata.23,54 Bioinformatics analysis indicated that this pathway was bacterial, and that it likely originated from E. frumentensis, a previously identified intracellular endosymbiont. Part of the cluster was expressed, and using a synthetic substrate analogue, it was shown to possess the predicted functionality. These and other data provided strong evidence implicating E. frumentensis in the production of the “ascidian” metabolites, ecteinascidin-743 and relatives.
In summary, although only a few ascidian groups have been studied in any detail, it is clear that symbiotic bacteria make many of the natural products isolated from the whole animals. This evidence is conclusive in the case of Prochloron production of cyanobactin cyclic peptides, including trunkamide, patellins, patellamides, and many others. Strong genetic evidence favors Prochloron production of sunscreens and biofuels. Indirect, but compelling evidence hints that proteobacteria might synthesize a class of drug lead polyketides in didemnid ascidians. Strong evidence also exists for symbiotic production of ecteinascidins in non-didemnid ascidians, and much indirect evidence exists for other compounds such as the didemnins.
VARIATION OF NATURAL PRODUCTS
Having established that bacteria make some of the compounds that are the dominant components of ascidian extracts, we set out to examine questions about chemical variation. About 60 cyanobactin derivatives had been isolated from didemnid ascidians, with differences including changes to the amino acid sequences, changes in size of the macrocyclic ring, and wholesale differences in types of chemical modifications, such as prenylation or heterocyclization.45 These compounds varied between animals, even those collected in the same place from year to year. The first question we asked was, what underlies the cyanobactin sequence and structural variation? The second question we sought to answer involved the seemingly sporadic distribution of these and other compounds. Like several other natural products chemists, we noticed that many compounds, such as the antitumor lead patellazoles, vary from sample to sample, but the reason for this variation was not known.55 We applied numerous approaches to answer these questions, using hundreds of ascidian samples obtained from a transect of the tropical Pacific Ocean. These samples were analyzed by chemical, genetic, and many other methods.
As alluded to above, some of the chemical differences between ascidians can be ascribed to their different microbial content.18 For example, evidence suggests that polyketides such as palmerolides and patellazoles might be synthesized by proteobacteria that employ the trans-AT pathway, while peptides such as cyanobactins are certainly synthesized by the cyanobacterium, P. didemni. Much is now known about how chemicals vary in P. didemni, but little is known about the putative polyketide-producing symbionts. An especially pressing question under active investigation is what governs the odd and seemingly sporadic distribution of these symbiotic polyketide producers.
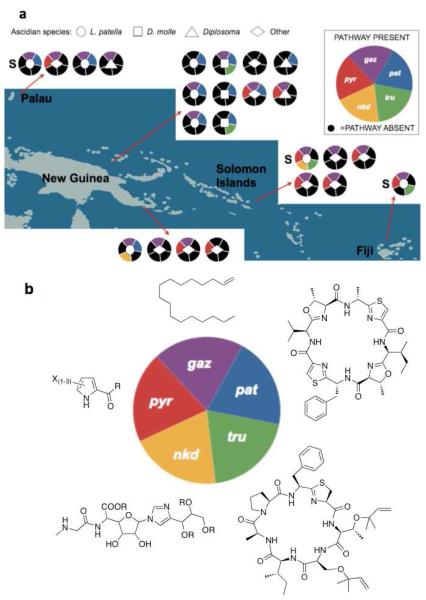
Within P. didemni, extensive research using metagenome sequencing, among other techniques, revealed three fundamental, genome-level changes that fully explain the observed chemical variation in ascidians (Figure 4).17 So far, we have sequenced the genomes and fully examined the chemical content of four uncultivated P. didemni strains from four L. patella samples collected in Palau, Papua New Guinea, the Solomon Islands, and Fiji. We also have examined more than 100 other ascidians from these and other locations using multiple approaches such as PCR and chemical analysis. Remarkably, despite the vast geographical distance between these four ascidians and great differences in chemical content, the P. didemni genomes are virtually identical and syntenic (with the genes in the same order on the chromosome), to a remarkable degree. To give an indication of the similarity, nearly all functionally assignable genes are >97% DNA sequence identical. Out of 126 pairwise differences in functionally assignable genes between three of the genomes, 116 differences were due to secondary metabolism. In answering the question of where these differences come from, we discovered three unanticipated new mechanisms underlying variation. These observations were further validated by PCR-based comparison with other ascidians.
Figure 4.
Genome level changes in P. didemni lead to a natural combinatorial library of bioactive natural products. (A) A map showing some of the samples examined, with each individual animal represented by a circle. Colors indicate some of the biosynthetic pathways identified in P. didemni. Pathways are basically identical when they are found, but the pathways are not universal. They are present sporadically, leading to mixtures of natural products. (B) Compounds made by the pathways shown in panel A. Chemical products have been identified for gaz, pat, and tru, but they have not yet been found and remain hypothetical for pyr and nkd.
Variation mechanism 1: sporadic presence or absence of biosynthetic gene clusters
At the first and broadest level of variation, within these conserved genomes whole biosynthetic pathways are either present or absent.17 When they are present, they are nearly identical (>99%) to each other, and they are located in the same chromosomal context. Thus, whole types of molecules may be either present or absent. These pathways sporadically co-occur. Seemingly at random, they are found in different combinations amongst 23 ascidians studied. New pathways continue to be found as more ascidians are examined. The lack of a pattern is of itself a pattern. It implies that pathways are required for different reasons and appear when needed to face different types of selection pressure, and that all pathways are available in principle from a large population of P. didemni. We proposed that these mixtures of pathways would result from a high rate of recombination between closely related P. didemni that form a single population in the tropical Pacific.
Variation mechanism 2: homologous recombination within pathways
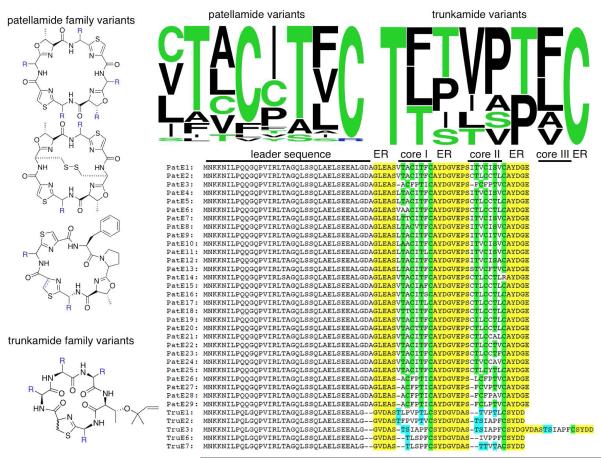
The remaining two levels of variation focus on the cyanobactin metabolites (Figure 5). In order to explain this variation, the patellamide biosynthetic pathway will be described briefly. Cyanobactins are ribosomal peptide natural products.56,57 A precursor peptide, exemplified by PatE, is synthesized on the ribosome and encodes both the mature natural products and enzyme recognition elements that are later cleaved and discarded. In the case of patellamides, PatE is modified first by heterocyclase PatD, which makes thiazoline or oxazoline out of cysteine, serine, or threonine.58,59 Thiazoline is oxidized to thiazole by the PatGOXI domain. Finally, two serine proteases, PatAPRO and PatGPRO, are responsible for removing the leader sequence and macrocyclizing the products.60,61
Figure 5.
Modifications in ribosomal peptide pathways. Natural genetic variations reveal that (A) identical cyanobactin enzymes accept extremely sequence-diverse substrates and (B) post-translational modifications can be moved between pathway types and are portable.
We sought to determine why patellamides contain oxazoline residues, while serine and threonine are instead modified by isoprenylation in trunkamide and relatives. It was ascertained that the trunkamide pathway is identical to the patellamide pathway in its flanking regions, but in the middle of the gene clusters, the DNA sequence similarity drops precipitously.20 While there are no new enzymes in the trunkamide cluster, there are differences in the heterocyclase. More importantly, there are vast differences in PatF homologs, which have no known relatives outside of cyanobactin pathways. Our group showed recently that these PatF variants represent a novel class of prenyltransferase enzymes that act specifically on diverse macrocycles.62 These prenyltransferases emphasize the “portability” of modification enzymes in ribosomal peptide natural products and their modular biosynthetic pathways.63
These sequence changes observed in ribosomal peptides are very fluid.17 For example, PatE and relatives encode two different products. Different mixtures of these products are found encoded on single PatE variants, indicating a constant exchange and mixing of these natural products. As yet another example, we have identified two different regions where the patellamide and trunkamide pathways have fused, indicating a dynamic exchange of these portable modification enzymes.
Variation mechanism 3: hypervariability of the precursor peptide
It was found that diverse patellamide derivatives are synthesized by different P. didemni strains.21 The genes encoding the biosynthetic enzymes and the intergenic sequences are virtually identical (>99%) between pathways. The only region that varies lies in the precise 21-24 nucleotide sequence region that directly encodes products. In an early study, 29 variants were discovered that could be synthesized by identical enzymes. This revealed enzymes of extremely broad substrate specificity.
Taken together, these results explain the mixtures and extreme variation that is observed in didemnid ascidians, even when moving between single colonies. Indeed, recently we have shown that adjacent colonies can encode wholly different biosynthetic pathways, even though animals look otherwise identical.16,17 Using genetic methods, our group has accessed new compounds from tiny colonies less than 1 cm in length. It can be predicted that the vast majority of pharmaceutically interesting ascidian products await discovery, and that individual colonies may harbor a heretofore “hidden majority” of natural products.
APPLICATIONS IN DRUG DISCOVERY AND DEVELOPMENT
One could anticipate many practical applications of these discoveries. For example, by understanding the source of compounds, potentially the supply problem of marine natural products would be eliminated, allowing immediate development of promising lead compounds. By knowing why individual colonies have different chemistry, new tools are suggested that allow access to new natural products for drug discovery. New enzymes might be discovered for various industrial applications. As yet another example, laboratory recapitulation of these precise natural evolutionary pathways might lead to new methods of combinatorial biosynthesis. Together, these new methods promise a new paradigm of marine natural products discovery and development.
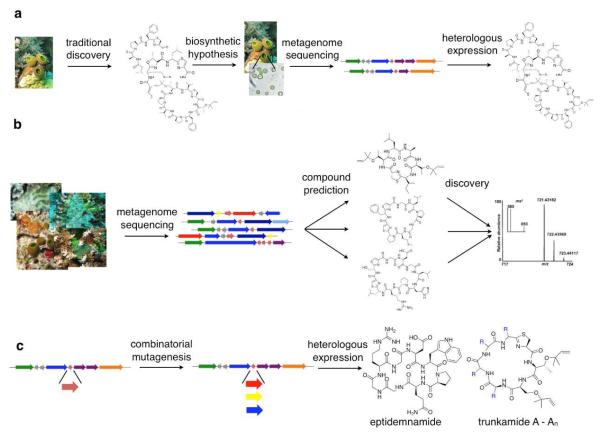
We are just at the start of these promising new approaches, and much remains to be done. However, in recent years we have provided proof-of-concept for each of these application areas, indicating that they are no longer speculative future events but that current technology is sufficient (Figure 6).
Figure 6.
Proof-of-concept for biotechnological applications. Using cyanobactin biosynthetic pathways, we have demonstrated that direct metagenomic methods can be used (A) to supply bioactive natural products, (B) to discover new compounds in genomes and in tiny coral reef animals, and (C) to synthesize single analogues and combinatorial analogue libraries.
In the area of drug supply, the initial reports of patellamide production from Long et al. and from our laboratory have demonstrated that, in principle, marine “animal” natural products could be supplied by recombinant technology.19,47 However, much work was required to move this to a practical level, and in addition the patellamides themselves are not necessarily considered as drug leads. Using more than 1000 individual engineering experiments, we have succeeded in obtaining sustained and robust yields of cyanobactins.16 A cyanobactin considered to be a potential drug, trunkamide, has been supplied in this manner.
We consider the cryptic colony variation of ascidians and sponges to represent a source potentially for the “hidden majority” of marine natural products. However, compounds from tiny colonies are often present in vanishingly small quantities. Direct genetic methods have been applied to: (1) identify these compounds; (2) show that they are major components of whole animals; and (3) express them directly in E. coli.16 This represents proof-of-concept for a directly genetic method to discover natural products important to whole animals, even from tiny colonies.
The genome prediction methods were also applied to cultivated cyanobacteria. Following the discovery of the patellamide biosynthetic gene cluster in ascidians, it became apparent that many cyanobacteria contain cyanobactin ribosomal peptide pathways. In fact, it has been reported that maybe close to one-third of cyanobacterial species encode cyanobactins.64,65 Two examples will be described briefly. An early example of the use of genome mining involved discovery of a new cyanobactin, trichamide, from Trichodesmium erythraeum. This cyanobacterial species is found globally in the open ocean, where it forms massive blooms that can be seen from space and that are critical to nitrogen fixation. Using the genome for T. erythraeum, we predicted the presence of the new compound, trichamide, and showed that it was indeed present in the bacteria using a robust mass spectrometry method.66 This method was later applied to the common supplement product, Spirulina, which is comprised of the cyanobacteria Arthrospira platensis. Using genome mining, cyanobactins were predicted to be present in these supplements, and indeed four new arthrospiramide derivatives were found in commercially available nutritional supplement products.63
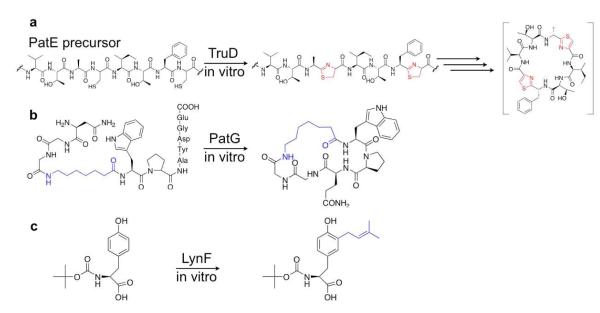
In the area of enzymes, we have used proteins discovered in these studies to decorate synthetic peptides with a variety of specific modifications (Figure 7).58,61,62 Finally, in the area of combinatorial biosynthesis, we have developed tools to synthesize diverse derivatives based upon recapitulating the natural evolutionary pathways.15,20 These tools have been used both to make analogues of known natural product drug leads, such as trunkamide, as well as to make wholly novel chemical entities, such as eptidemnamide. Recently, larger numbers of derivatives have been synthesized in vivo, including incorporation of non-proteinogenic amino acids into these complex, multistep pathways.67
Figure 7.
Enzymatic synthesis. Enzymes from cyanobactin pathways have been overexpressed and used in vitro for synthesis of (A) heterocycles, (B) macrocycles, and (C) prenylated amino acids.
FUTURE PERSPECTIVES
Above, the focus is on our work with specific classes of ascidian compounds. This represents a tiny fraction of the known chemical diversity in the ocean, much less the potential “hidden majority” of natural products. In addition, technological proof-of-concept experiments have been performed only with the cyanobactins family of natural products. Work on other groups of organisms, such as bryozoans harboring the bryostatins44 and sponges harboring onnamide and relatives,43 suggests that this is a very promising area for future research. Previously, many technical limitations and a lack of understanding about the basics of origin and variation made these studies very daunting and laborious. For example, molecular genetic work on bryostatin biosynthesis began in 1995, and our work on ascidians initiated in 2002, indicating the depth of the early challenges. Others working in this field have similar stories. Now, however, inexpensive DNA sequencing and robust synthetic biology tools are finally available, making this research more widely approachable.
Sometimes, simple explanations are provided that broadly describe natural product roles and the factors underlying natural product origin and variation. It is worth avoiding broad generalizations at this point, since in the context of the amazing diversity of the oceans, almost nothing has been done experimentally. Instead, interesting compounds in diverse organisms should continue to be examined without preconceived notions. It is certain that if the rich results described above can be obtained from looking at a simple, possibly obscure organism, marine symbiosis represents an incredible opportunity for discovery.
Acknowledgment
EWS wishes to thank his colleagues and students, the University of Utah, and excellent collaborators, especially Dr. Margo G. Haygood (OHSU). Dr. Chris M. Ireland (University of Utah) was instrumental in helping us to obtain samples used in this project, among many other important impacts. Our work described herein was funded by NIH (GM071425), NSF (EF-0412226), and the University of Utah.
Dedication This review is dedicated to two giants of natural products: the late D. John Faulkner, whose visionary insights into symbiosis are being fulfilled, and the late Matt Suffness, whose impact on developing natural products as anticancer agents continues to be realized.
Footnotes
Adapted from a Matthew Suffness award lecture, 51st Annual Meeting of the American Society of Pharmacognosy, St. Petersburg, Florida, July 10-14, 2010.
References
- (1).Blunt JW, Copp BR, Munro MH, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2010;27:165–237. doi: 10.1039/b906091j. [DOI] [PubMed] [Google Scholar]
- (2).Blunt JW, Copp BR, Munro MH, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2011;28:196–268. doi: 10.1039/c005001f. [DOI] [PubMed] [Google Scholar]
- (3).Ireland C, Scheuer PJ. J. Am. Chem. Soc. 1980;102:5688–5691. [Google Scholar]
- (4).Shenkar N, Swalla BJ. PLoS One. 2011;6:e20657. doi: 10.1371/journal.pone.0020657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rinehart KL, Jr., Gloer JB, Hughes RG, Jr., Renis HE, McGovren JP, Swynenberg EB, Stringfellow DA, Kuentzel SL, Li LH. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]
- (6).Gouiffes D, Juge M, Grimaud N, Welin L, Sauviat MP, Barbin Y, Laurent D, Roussakis C, Henichart JP, Verbist JF. Toxicon. 1988;26:1129–1136. doi: 10.1016/0041-0101(88)90297-8. [DOI] [PubMed] [Google Scholar]
- (7).Foster MP, Mayne CL, Dunkel R, Pugmire RJ, Grant DM, Kornprobst JM, Verbist JF, Biard JF, Ireland CM. J. Am. Chem. Soc. 1992;114:1110–1111. [Google Scholar]
- (8).Corley DG, Moore RE, Paul VJ. J. Am. Chem. Soc. 1988;110:7920–7922. [Google Scholar]
- (9).Zabriskie TM, Mayne CL, Ireland CM. J. Am. Chem. Soc. 1988;110:7919–7920. [Google Scholar]
- (10).Richardson AD, Aalbersberg W, Ireland CM. Anti-Cancer Drugs. 2005;16:533–541. doi: 10.1097/00001813-200506000-00009. [DOI] [PubMed] [Google Scholar]
- (11).Carroll AR, Coll JC, Bourne DJ, MacLeod JK, Zabriskie TM, Ireland CM, Bowden BF. Aust. J. Chem. 1996;49:659–667. [Google Scholar]
- (12).Wipf P, Uto Y. J. Org. Chem. 2000;65:1037–1049. doi: 10.1021/jo9914566. [DOI] [PubMed] [Google Scholar]
- (13).Williams AB, Jacobs RS. Cancer Lett. 1993;71:97–102. doi: 10.1016/0304-3835(93)90103-g. [DOI] [PubMed] [Google Scholar]
- (14).Monniot C, Monniot F, Laboute P. Coral Reef Ascidians of New Caledonia. Editions de l’ORSTOM; Paris: 1991. p. 247. [Google Scholar]
- (15).Cowan ME. Mar. Ecol. Prog. Ser. 1981;6:335–337. [Google Scholar]
- (16).Donia MS, Ruffner DE, Cao S, Schmidt EW. ChemBioChem. 2011;12:1230–1236. doi: 10.1002/cbic.201000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Donia MS, Fricke WF, Ravel J, Schmidt EW. PLoS One. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. Proc. Natl. Acad. Sci. U S A. 2011 doi: 10.1073/pnas.1111712108. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc. Natl. Acad. Sci. U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Donia MS, Ravel J, Schmidt EW. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- (22).Moss C, Green DH, Perez B, Velasco A, Henriquez R, McKenzie JD. Mar. Biol. 2003;143:99–110. [Google Scholar]
- (23).Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu F, Wolff JJ, Kweon HK, Christiansen MA, Hakansson K, Williams RM, Ehrlich GD, Sherman DH. ACS Chem. Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kott P. Mem. Queensl. Mus. 1980;20:1–47. [Google Scholar]
- (25).Kott P. Mem. Queensl. Mus. 2001;47:1–407. [Google Scholar]
- (26).Withers NW, Alberte RS, Lewin RA, Thornber JP, Britton G, Goodwin TW. Proc Natl Acad Sci U S A. 1978;75:2301–2305. doi: 10.1073/pnas.75.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lewin RA, Cheng L. Prochloron, a Microbial Enigma. Chapman and Hall; London: 1989. p. 129. [Google Scholar]
- (28).Bibby TS, Nield J, Chen M, Larkum AW, Barber J. Proc. Natl. Acad. Sci. U S A. 2003;100:9050–9054. doi: 10.1073/pnas.1532271100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Parry DL. Mar. Biol. 1985;87:219–222. [Google Scholar]
- (30).Odintsov VS. Endocyt. Cell Res. 1991;7:253–258. [Google Scholar]
- (31).Koike I, Yamamuro M, Pollard PC. Aust. J. Mar. Fresh. Res. 1993;44:173–182. [Google Scholar]
- (32).Lewin RA, Cheng L. Phycologia. 1975;14:149–152. [Google Scholar]
- (33).Bak RPM, Sybesma J, van Duyl FC. Mar. Ecol. Prog. Ser. 1981;6:43–52. [Google Scholar]
- (34).Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW. Proc. Natl. Acad. Sci. U S A. 2008;105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Larkum AWD, Cox GC, Hiller RG, Parry D, Dibbayawan TP. Mar. Biol. 1987;95:1–13. [Google Scholar]
- (36).Hirose E, Uchida H, Murakami A. Eur. J. Phycol. 2009;44:365–375. [Google Scholar]
- (37).Parry DL, Kott P. Bull. Mar. Sci. 1988;42:149–153. [Google Scholar]
- (38).Groepler W, Schuett C. Helgoland Mar. Res. 2003;57:139–143. [Google Scholar]
- (39).Schuett C, Doepke H, Groepler W, Wichels A. Helgoland Mar. Res. 2005;59:136–140. [Google Scholar]
- (40).Riesenfeld CS, Murray AE, Baker BJ. J. Nat. Prod. 2008;71:1812–1818. doi: 10.1021/np800287n. [DOI] [PubMed] [Google Scholar]
- (41).Schmidt EW, Donia MS. Curr. Opin. Biotechnol. 2010;21:827–833. doi: 10.1016/j.copbio.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Piel J. Nat. Prod. Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- (43).Piel J. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- (44).Schmidt EW. Nat. Chem. Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Donia MS, Schmidt EW. In: Comprehensive Natural Products II. Moore BS, Crews P, editors. Vol. 2. Elsevier; Oxford, UK: 2010. pp. 539–558. [Google Scholar]
- (46).Sivonen K, Leikoski N, Fewer DP, Jokela J. Appl. Microbiol. Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Long PF, Dunlap WC, Battershill CN, Jaspars M. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- (48).Maruyama T, Hirose E, Ishikura M. Biol. Bull. 2003;204:109–113. doi: 10.2307/1543546. [DOI] [PubMed] [Google Scholar]
- (49).Piel J. Nat. Prod. Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- (50).Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Proc. Natl. Acad. Sci. U S A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J. Nat. Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- (52).Tsukimoto M, Nagaoka M, Shishido Y, Fujimoto J, Nishisaka F, Matsumoto S, Harunari E, Imada C, Matsuzaki TJ. Nat. Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- (53).Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Nat. Chem. Biol. 2010;6:408–10. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- (54).Rath CM, Yu F, Janto B, Inai M, Williams R, Ehrlich G, Hakansson K, Sherman DH. American Society for Mass Spectrometry Annual Meeting; Salt Lake City. May 22-24, 2010. [Google Scholar]
- (55).Schmidt EW, Sudek S, Haygood MG. J. Nat. Prod. 2004;67:1341–1345. doi: 10.1021/np049948n. [DOI] [PubMed] [Google Scholar]
- (56).McIntosh JA, Donia MS, Schmidt EW. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Oman TJ, van der Donk WA. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).McIntosh JA, Donia MS, Schmidt EW. J. Am. Chem. Soc. 2010;132:4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).McIntosh JA, Schmidt EW. ChemBioChem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Lee J, McIntosh J, Hathaway BJ, Schmidt EW. J. Am. Chem. Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. J. Am. Chem. Soc. 2010;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).McIntosh JA, Donia MS, Nair SK, Schmidt EW. J. Am. Chem. Soc. 2011;133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Donia MS, Schmidt EW. Chem. Biol. 2011;18:508–519. doi: 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Leikoski N, Fewer DP, Sivonen K. Appl. Environ. Microbiol. 2009;75:853–857. doi: 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Leikoski N, Fewer DP, Jokela J, Wahlsten M, Rouhiainen L, Sivonen K. Appl. Environ. Microbiol. 2010;76:701–709. doi: 10.1128/AEM.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Sudek S, Haygood MG, Youssef DT, Schmidt EW. Appl. Environ. Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. J. Am. Chem. Soc. 2011 doi: 10.1021/ja208278k. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]