Abstract
Simultaneous activation of signaling pathways requires dynamic assembly of higher-order protein complexes at the cytoplasmic domains of membrane-associated receptors in a stimulus-specific manner. Here, using the paradigm of cellular activation through cytokine and innate immune receptors, we demonstrate the proof-of-principle application of small molecule probes for the dissection of receptor-proximal signaling processes, such as activation of the transcription factor NF- B and the protein kinase p38.
Keywords: NF-kB pathway, Toll-like-receptor, Selective inhibitor, Innate immunity, Tumor Necrosis Factor
An intricate network of protein-protein interactions (PPIs) is involved in many essential cellular processes, such as receptor-dependent signaling pathways.1–2 In order to understand the mechanisms by which the same downstream signaling pathway is activated through different receptors, selective inhibition of PPIs involved in these processes could be a useful tool for basic research as well as drug discovery.3 A recent development in the field of PPI studies is the use of small molecules that exhibit specificity and affinity to crucial dominant protein-protein interfaces, also known as hot-spots.4–6 We examined this tact for the dissection of receptor-mediated signaling mechanisms with focus on the NF- B pathway as model system of biomedical importance.7
Upon engagement of cytokine and innate immune receptors, activation of the classical NF- B pathway is initiated through transient assembly of a large multiprotein signalsome that contains the key components of NF- B signaling, such as the I B (inhibitor of NF- B) kinase (IKK) complex.8–10 Despite the involvement of distinct receptor proteins, NF- B stimuli induce signaling within the IKK-NF- B module as well as parallel activation of other signaling pathways, including the p38 mitogen- (or stress-) activated protein kinase (MAPK/SAPK) cascade.11
For example, bacterial lipopolysaccharide (LPS) activates the IKK and MAPK cascades through the Toll-like receptor 4 (TLR4), a member of the innate immune receptor family,12 while tumor necrosis factor alpha (TNF) signals through the TNF receptor (TNFR), member of the TNFR superfamily.13
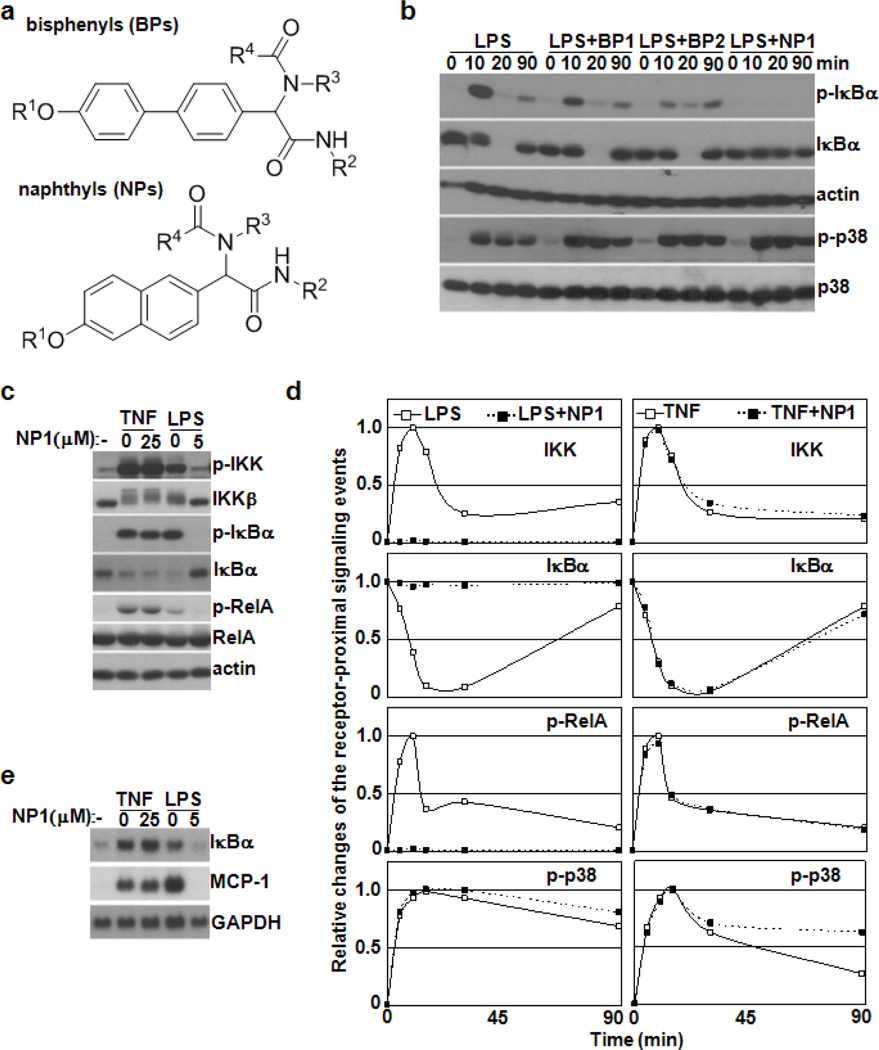
These observations prompted our investigations into whether a small molecule-based strategy might allow for dissection of the receptor-proximal signaling processes induced by different NF-B-activating stimuli, namely LPS and TNF. As a starting point an initial screening was undertaken using our non-peptidic small molecule library, two scaffolds contained within the library are depicted in Figure 1a.14 Thus, using a cell line carrying an NF- B reporter system and LPS as stimulus resulted in the identification of both napthyl- (NP1) and bisphenyl-based (BP1 and BP2) scaffolds as potential modulators of NF- B activity (see Supplementary Material, Figure 1). To investigate whether these compounds indeed affect NF- B signaling, the effects of BP1, BP2 and NP1 on the kinetics of LPS- or TNF-induced I B phosphorylation, degradation and resynthesis in macrophages were studied. As specificity control, the phosphorylation of p38 kinase (p-p38), a marker of a MAPK cascade,15 was also assayed. Similar levels of p-p38 were observed in response to LPS alone or in combination with the inhibitors. The temporal patterns of NF- B signaling were also unaffected in the presence of BP1 or BP2; in contrast, addition of NP1 resulted in substantial inhibition of LPS-induced I B phosphorylation and degradation shown in Figure 1b (and Supplementary Material, Figure 2). Notably, neither BP1/2 nor NP1 had any effect on TNF-induced p-p38 and activation of NF- B signaling (Supplementary Material, Figure 3).
Figure 1.
NP1 acts as a specific modulator of NF- B signaling. (a) Generic structures of bisphenyl (BP)- and naphthyl (NP)-based members of the “credit card” library. (b) Western blot analysis of I B p and their phosphorylated forms (p-I B and p-p38, respectively) in extracts prepared from bone-marrow derived macrophages (BMDMs) after treatment with LPS alone or in combination with BP1, BP2 or NP1 (50 M of each). (c) Western blot analysis of IKK , I B , RelA and their phosphorylated forms in BMDM extracts after treatment with TNF and LPS for 5 min in the absence or presence of NP1 as indicated. (d) The profiles of IKK and p38 activities as well as I B protein concentration and RelA phosphorylation in BMDMs stimulated with LPS, TNF or in combination with NP1 as indicated. Quantitated and normalized results from three independent experiments are shown. (e) Northern blot analysis of total RNA prepared from BMDMs monitors the effect of NP1 on the mRNA expression levels of I B and MCP-1 genes induced by LPS or TNF.
The phosphorylation-dependent activation of IKK (also known as IKK-2), a subunit of the IKK complex, is essential for induction of NF- B activity by TNF or LPS; once activated IKK phosphorylates I B proteins and RelA subunit of NF- B. The functional consequence of these processes is transcriptional induction of B and other NF- B target genes.10 Therefore, to better define the specificity of NP1, its effects on activation of the -I B -RelA signaling module in response to TNF or LPS was investigated. LPS-induced phosphorylation of IKK , I B and RelA was substantially disrupted in the presence of NP1. However, functional integrity of the IKK -I B -RelA signaling module remained intact in cells stimulated with TNF in the presence of NP1 shown in Figure 1c-d. Thus, the core components of NF- B signaling are not affected by NP1. Northern blot analysis independently confirmed that NP1 selectively blocked the induction of NF- B target genes in LPS- but not TNF-treated macrophages (Figure 1e). These findings are consistent with the fact that TNFR and TLR4 use distinct adapter protein components to assemble signaling complexes essential for the activation of receptor-proximal IKK cascades.12–13
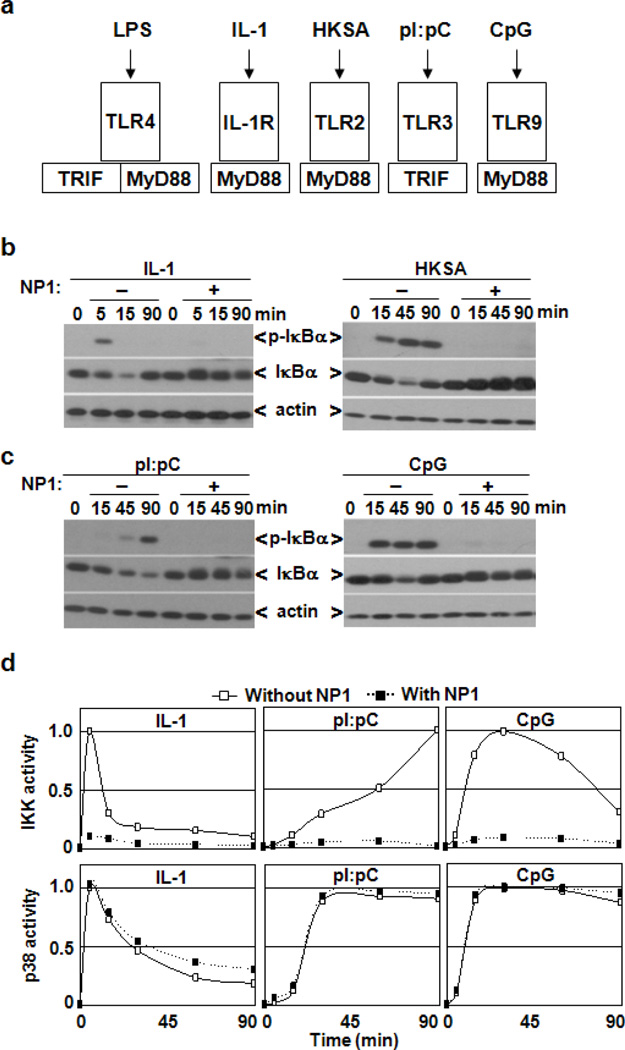
TLR4 engagement initiates assembly of multicomponent receptor-associated signaling complexes that activate NF- B and p38 pathways12. Several of these components are also recruited by the interleukin-1 receptor (IL-1R) and other TLRs; namely, TLR3 signals through TRIF adaptor protein, while the remaining known TLRs as well as IL-1R use MyD88 as adaptor protein; notably both adaptors are utilized by TLR4 (Fig 2a). To better understand the signaling mechanisms targeted by NP1, we examined the effect of NP1 on activation of the NF- B and p38 pathways in response to IL-1R-, TLR2-, TLR3- or TLR9-specific ligands. As seen in Figure 2a-b, NP1 substantially blocked I B phosphorylation and degradation induced by IL-1R and TLR2 ligands, suggesting a role of NP1 in targeting MyD88-dependent processes. However, a complementation assay demonstrated that NP1 had no effect on the direct interaction between the cytoplasmic domains of TLR4 and MyD88 (Supplementary Material, Figure 4). Moreover, although NP1 blocked MyD88-dependent TLR9-mediated activation of the NF-B pathway, it also disrupted the MyD88-independent signaling mediated through TLR3-TRIF interactions shown in Figure 2c-d. Interestingly, in contrast to cell surface-expressed IL-1R, TLR2 and TLR4, both TLR3 and TLR9 are intracellular proteins.12, 16
Figure 2.
NP1 selectively impairs IL-1R/TLR-mediated activation of the NF- B pathway. (a) The scheme shows recruitment of MyD88 and TRIF adaptor proteins to IL-1R and TLRs in response to receptor-specific ligands, such as LPS, IL-1, HKSA (heat-killed Staphylococcus aureus), pI:pC (polyI:polyC) and CpG (DNA oligonucleotide containing unmethylated CpG motifs). (b and c) Western blot analysis of total protein extracts prepared from BMDMs monitors the effect of NP1 on I B degradation and resynthesis as well as I B phosphorylation induced by IL-1 and HKSA (b) or pI:pC and CpG (c) as indicated. (d) The profiles of IKK and p38 activities in BMDMs stimulated with the indicated stimuli alone or in combination with NP1. Quantitated and normalized results from three experiments are shown.
Thus, these data indicate that NP1 acts in the cytoplasm to affect IL-1R/TLR-mediated activation of NF- B signaling. Although further investigations are required to identify the exact mechanisms involved, it is unlikely that NP1 interferes with primary assembly of IL-1R/TLR-associated signaling complexes, because receptor-proximal activation of the p38 pathway still occurred in the presence of NP1 shown in Figure 2a-d (and Supplementary Material, Figure 5).
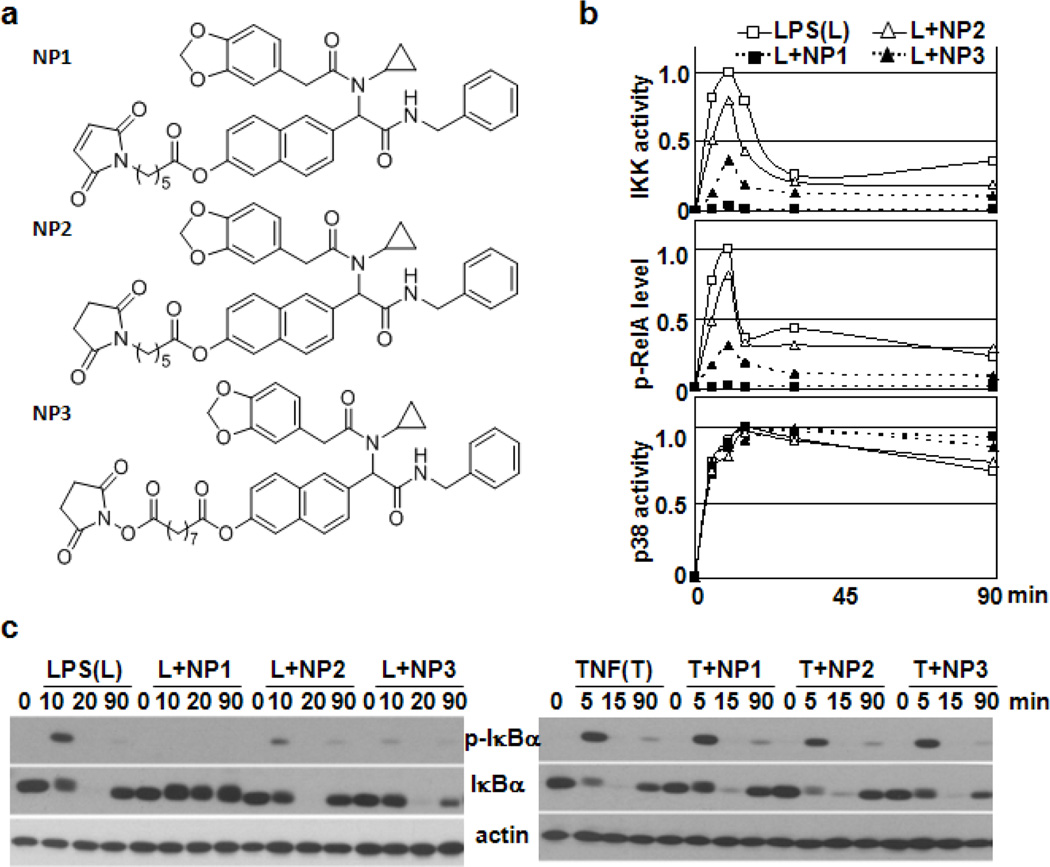
Several thiol-reactive molecules have been shown to inhibit phosphorylation and degradation of I B in cells stimulated with TNF and LPS by blocking IKK kinase activity via a Michael addition reaction with a specific cysteine residue within the activation loop of IKK 17–19 Similar to N-ethyl maleimide,20 these inhibitors also target a free cysteine residue in the NF- B subunit RelA, and in turn such modification results in reduction of NF-B/RelA-mediated transcription.21–22 NP1 contains a maleimide moiety; however, our data clearly demonstrate that NP1 did not alter the signal processing within the -I B -RelA signaling module. Nevertheless, the inhibitory effect of NP1on LPS-induced NF- B signaling required the maleimide group (Supplementary Material, Fig. 6), suggesting that covalent target modification contributes to the observed signaling inhibition. To ascertain whether chemical reactivity and inhibitory functions of NP1 are linked, we synthesized compounds NP2 and NP3 depicted in Figure 3a, representing a chemically-inert as well as a reversibly reactive derivative of NP1, respectively (see Supplementary information). Comparison of NP1, NP2 or NP3 effects on LPS-induced NF- B signaling shown in Figure 3b revealed that NP2 was a very week inhibitor, whereas NP3-mediated inhibition was comparable with the effect seen for NP1. Importantly, both NP1 derivatives retain selectivity for TLR-dependent induction of the NF- B pathway, as NP2 or NP3 had no effect on TNFR-mediated activation of the -I B -RelA signaling module. However, the induction of the p38 cascade through cytokine and immune receptors still occurred in the presence of the compounds as evident in Figure 3c (and Supplementary Material, Figure 7). Thus, potential covalent protein reactivity of NP1 and NP3 substantially improves the efficacy of their planar-aromatic motif as inhibitor of IL-1R/TLR-mediated activation of the IKK signaling cascade.
Figure 3.
Inhibitory effects of NP1-based scaffolds depend on their chemical reactivity. (a) Chemical structures of NP1 and its analogs. (b) The profiles of IKK and p38 activities and RelA phosphorylation in BMDMs stimulated with LPS alone or in combination with NP1 or its analogs as indicated. Quantitated and normalized results from three experiments are shown. (c) Western blot analysis of total protein extracts prepared from BMDMs monitors the effect of NP1 and its analogs on I B degradation and resynthesis as well as I B phosphorylation induced by LPS (left panel) and TNF (right panel) as indicated.
Our current understanding of signal transduction through growth factor, cytokine and innate immune receptors suggests that both assembly and dissociation of multiprotein signaling complexes at/from intracellular receptor domains are required for the regulation of receptor-proximal MAPK and IKK cascades.11–13, 23–24 For example, it has been reported that CD40, a TNFR superfamily member, activates IKK through a receptor-associated complex while the MAPK cascade is triggered after translocation of the multicomponent signaling complex from the intracellular CD40 domain into the cytosol.24 Our findings strongly support this model of spatial and temporal separation of MAPK and IKK signaling cascades, and show how the principles of chemical biology can be applied to begin to dissect receptor-proximal signaling events.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (AI88229 to Y.J.K., AI080715 to G.F.K., and AI077644 to K.D.J.), the Worm Institute for Research and Medicine (K.D.J.) and the Skaggs Institute for Chemical Biology (K.D.J.). This is manuscript # 21546 from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keskin O, Gursoy A, Ma B, Nussinov R. Chem Rev. 2008;108:1225. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 2.McNeill H, Woodgett JR. Nat Rev Mol Cell Biol. 2010;11:404. doi: 10.1038/nrm2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson AJ. Chem Soc Rev. 2009;38:3289. doi: 10.1039/b807197g. [DOI] [PubMed] [Google Scholar]
- 4.Bogan AA, Thorn KS. J Mol Biol. 1998;280:1. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 5.Clackson T, Wells JA. Science (New York, N.Y. 1995;267:383. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 6.Workman P, Collins I. Chemistry & biology. 2010;17:561. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltimore D. Nature immunology. 2011;12:683. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 8.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. Nature. 1997;388:548. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 9.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. Science (New York, N.Y. 1997;278:860. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 10.Oeckinghaus A, Hayden MS, Ghosh S. Nature immunology. 2011;12:695. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 11.Baud V, Karin M. Trends Cell Biol. 2001;11:372. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 12.Beutler B. Nature. 2004;430:257. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Goeddel DV. Science (New York, N.Y. 2002;296:1634. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. Bioorg Med Chem. 2006;14:2660. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 15.Zarubin T, Han J. Cell Res. 2005;15:11. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 16.Nishiya T, Kajita E, Miwa S, Defranco AL. The Journal of biological chemistry. 2005;280:37107. doi: 10.1074/jbc.M504951200. [DOI] [PubMed] [Google Scholar]
- 17.Hehner SP, Heinrich M, Bork PM, Vogt M, Ratter F, Lehmann V, Schulze-Osthoff K, Droge W, Schmitz ML. The Journal of biological chemistry. 1998;273:1288. doi: 10.1074/jbc.273.3.1288. [DOI] [PubMed] [Google Scholar]
- 18.Hehner SP, Hofmann TG, Droge W, Schmitz ML. J Immunol. 1999;163:5617. [PubMed] [Google Scholar]
- 19.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Nature. 2000;403:103. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 20.Toledano MB, Leonard WJ. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:4328. doi: 10.1073/pnas.88.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Pineres AJ, Castro V, Mora G, Schmidt TJ, Strunck E, Pahl HL, Merfort I. The Journal of biological chemistry. 2001;276:39713. doi: 10.1074/jbc.M101985200. [DOI] [PubMed] [Google Scholar]
- 22.Ha KH, Byun MS, Choi J, Jeong J, Lee KJ, Jue DM. Biochemistry. 2009;48:7271. doi: 10.1021/bi900660f. [DOI] [PubMed] [Google Scholar]
- 23.Schlessinger J. Cell. 2000;103:211. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 24.Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, Vignali DA, Gallagher E, Karin M. Science (New York, N.Y. 2008;321:663. doi: 10.1126/science.1157340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.