Abstract
The indigenous microbiota impact mucosal, as well as systemic, immune responses, but whether the microbiota are involved in stressor-induced immunomodulation has not been thoroughly tested. A well characterized murine stressor, called social disruption (SDR), was used to study whether the microbiota are involved in stressor-induced enhancement of macrophage reactivity. Exposure to the SDR stressor enhanced the ability of splenic macrophages to produce microbicidal mediators (e.g., inducible nitric oxide synthase (iNOS), superoxide anion, and peroxynitrite) and to kill target Escherichia coli. Exposure to the SDR stressor also increased cytokine production by LPS-stimulated splenic macrophages. These effects, however, were impacted by the microbiota. Microbicidal activity and cytokine mRNA in splenic macrophages from Swiss Webster germfree mice that lack any commensal microbiota were not enhanced by exposure to the SDR stressor. However, when germfree mice were conventionalized by colonizing them with microbiota from CD-1 conventional donor mice, exposure to the SDR stressor again increased microbicidal activity and cytokine mRNA. In follow up experiments, immunocompetent conventional CD-1 mice were treated with a cocktail of antibiotics to disrupt the intestinal microbiota. While exposure to the SDR stressor enhanced splenic macrophage microbicidal activity and cytokine production in vehicle-treated mice, treatment with antibiotics attenuated the SDR stressor-induced increases in splenic macrophage reactivity. Treatment with antibiotics also prevented the stressor-induced increase in circulating levels of bacterial peptidoglycan, suggesting that translocation of microbiota-derived peptidoglycan into the body primes the innate immune system for enhanced activity. This study demonstrates that the microbiota play a crucial role in stressor-induced immunoenhancement.
Keywords: Microbiota, Stress, Social Disruption, Macrophage, Microbicidal Activity, Peroxynitrite, IL-1β, TNF-α, iNOS, Superoxide, Peptidoglycan
Introduction
The mammalian body is populated by immense and complex consortia of microbes generally referred to as the microbiota. Bacteria are by far the most predominant and beneficial members of the microbiota, but some archaea and eukarya can also be found residing on the body (O’Hara and Shanahan, 2006). In the healthy host, the vast majority of the microbiota are found within the lower sections of the gastrointestinal tract (i.e., distal ileum and colon) where they reside in stable climax communities as a result of ecological successions involving the selection of microbes best adapted for their given niche (Huffnagle, 2010). Although these climax communities are relatively resistant to change (Allison and Martiny, 2008), many factors, such as diet or antibiotic use, can cause transient alterations in community structure (Antonopoulos et al., 2009; Dethlefsen et al., 2008). We have shown that exposure to experimental stressors, such as prolonged restraint (Bailey et al., 2010) as well as social disruption (SDR) (Bailey et al., 2011), also impact the stability of the intestinal microbiota. For example, exposure to the SDR stressor significantly changed the relative abundance of 7 out of the top 30 genera of bacteria residing as the microbiota (Bailey et al., 2011). Whether these stressor-induced effects on the microbiota impact the health of the host is not yet known.
The microbiota are intimately associated with mucosal surfaces, and are well known to influence the development of the mucosal immune system. This is most evident in germfree mice, i.e., mice born and housed under completely sterile conditions, that have smaller Peyer’s patches, fewer intraepithelial lymphocytes, and lower levels of secretory IgA (Macpherson and Uhr, 2004). Of importance to this study, colonizing the germfree mice with microbiota from conventional mice leads to the normal development of these immune compartments, thus emphasizing the importance of intestinal bacteria in mucosal immune system development (Macpherson and Uhr, 2004). Fewer studies have focused on the importance of the microbiota for development of systemic immune compartments, but the ability of bone marrow-derived neutrophils to kill target microbes was found to be dependent upon an intact intestinal microbiota (Clarke et al., 2010). This previous study led us to test the hypothesis that the microbiota are also necessary for stressor-induced increases in microbicidal activity in splenic macrophages.
Macrophage activation is an essential component of innate resistance to microbial pathogens due to their production of cytokines and chemokines that initiate the inflammatory response, as well as their ability to phagocytose and subsequently kill invading pathogens. The ability of macrophages to kill bacterial pathogens is largely dependent upon the production of reactive oxygen and nitrogen intermediates (ROI and RNI, respectively). Reactive oxygen intermediates are formed from superoxide anions that are produced from the reduction of molecular oxygen using the electrons derived from NADPH oxidase (Geiszt and Leto, 2004; Lambeth, 2004). Macrophage activation also leads to the expression of inducible nitric oxide synthase (iNOS) which converts arginine into nitric oxide and citrulline (Nathan and Xie, 1994). Both superoxide and nitric oxide are essential components for the synthesis of microbicidal compounds, and when they are both elevated within a cell they rapidly react to form peroxynitrite (Beckman and Koppenol, 1996; McLean et al., 2010). Because the microbiota have been shown to enhance the ability of neutrophils to kill bacteria (Clarke et al., 2010), and because bacterial killing (by both macrophages and neutrophils) is largely dependent upon peroxynitrite production (McLean et al., 2010; Weatherby et al., 2003), an additional goal of this study was to determine whether the microbiota are necessary for the production of peroxynitrite by splenic macrophages.
The physiological stress response is well known to affect the functioning of the immune system. While the common view has been that stress suppresses immune system activity due to the suppressive effects of stress-induced glucocorticoid hormones (Padgett and Glaser, 2003), there are now multiple studies demonstrating that stressor exposure can also enhance the immune response (Fleshner et al., 1998; Johnson et al., 2002; Johnson et al., 2003; O’Connor et al., 2003). However, the mechanisms by which stressor exposure enhances immune system activity have not been as extensively studied. We have been using the murine social stressor SDR to study stressor-induced immune enhancement. Exposure to the SDR stressor increases both innate and adaptive components of the immune system (Avitsur et al., 2005; Bailey et al., 2007; Bailey et al., 2009a; Mays et al., 2010; Quan et al., 2001; Stark et al., 2001). Of particular relevance for the current study, exposure to the SDR stressor primes splenic macrophages for enhanced reactivity to antigenic stimulation. For example, the production of cytokines (e.g., TNF-α and IL-1β) is increased in LPS-stimulated splenic macrophages from mice exposed to the SDR stressor (Avitsur et al., 2005; Bailey et al., 2009b; Engler et al., 2008). In addition, the ability of splenic macrophages to kill target Escherichia coli is significantly increased by exposure to the stressor (Bailey et al., 2007). This increased microbicidal activity is associated with a significant increase in iNOS gene expression and gene expression of the regulatory subunits of the NADPH oxidase complex (Bailey et al., 2007). How the stressor primes these cells for enhanced activity, however, is not understood. Thus, germ-free mice lacking any microbiota, as well as conventional mice treated with broad spectrum antibiotics, were exposed to the SDR stressor to test the hypothesis that the microbiota are necessary for the stressor-induced enhancement of macrophage reactivity to bacterial challenge.
Methods
Animal Handling
Outbred male Swiss-derived CD-1 mice between the ages of 6 to 8 weeks were purchased from Charles River Laboratories (Wilmington, MA). Prior to experimentation, the mice were housed 3 – 5 per cage and allowed to adjust to the animal vivarium for 1 week. They were kept on a 12 hr light:dark schedule with lights on at 06:00 and food and water available ad libitum. The Ohio State University’s Animal Care and Use Committee approved all experimental procedures. The pathogen status of all mice maintained at The Ohio State University is routinely monitored. The mice used in these studies were maintained in colonies that were confirmed to be free of opportunistic pathogens (See Supplemental Data).
Social Disruption
The murine social disruption (SDR) stressor involves repeated social defeat from agonistic interactions between an aggressive intruder male mouse and the resident male mice. The SDR stressor began at approximately 17:00, and continued for a 2 hr period that was repeated on 6 consecutive days. The aggressive intruders used to defeat the home cage residents were previously identified as aggressive toward their cage-mates and isolated from the rest of the colony. SDR began by placing the aggressive mouse into the home cage of the resident mice. The mice were observed for the first 10 min to ensure that the aggressor fought with and defeated all of the residents. If after 5 min the aggressor had not fought and defeated all of the residents, a different aggressor was added until fighting occurred. After each 2 hr SDR session the aggressor was removed and the mice left undisturbed until the next day. The morning following the 6th consecutive day of SDR, the defeated home cage residents were euthanized. Although fighting may result in wounding, the mice used in this study had only superficial wounds to the back and tail; mice with wounds penetrating the cutaneous layer were not used.
Germfree and Conventionalized Mice
Germfree Swiss Webster/Tac/UNC mice were originally obtained from the National Gnotobiotic Rodent Resource Center (University of North Carolinas), and subsequently bred and raised in sterile germfree isolators (Park BioServices, Groveland, MD). These germfree mice were fed sterile food and water, and therefore, contained no endogenous bacterial populations. Germfree status was confirmed prior to each experiment by culturing stool samples from all mice to detect living bacteria. In addition, germfree sentinel mice were euthanized every 3 months for quality assurance necropsy (see Supplemental Data Table 2). There was no evidence of microbial contamination in any of the germfree mice used in these studies (Supplemental Data Fig. 1).
When germfree mice were 6–8 wks of age, they were removed from the germfree isolator, placed in sterile cages with closeable lids, and transported to the laboratory. The sterile cages were kept in laminar flow hoods to help maintain sterility. Three groups of mice were used in the initial experiments to assess the role of the intestinal microbiota in stressor-enhanced macrophage activity. The first group consisted of germfree mice that were kept germfree by continuous housing in sterile cages in the laminar flow hood. SDR in germfree mice was accomplished by placing germfree retired breeders, into the residents’ cage using sterile gloves. The SDR was performed in sterile cages in the laminar flow hood. Germfree status was confirmed in HCC Control and SDR Stressor germfree mice by culturing the cecal contents to detect living bacteria (Supplemental Data Fig. 1).
The second group, referred to as conventionalized mice, consisted of germfree mice that were orally gavaged with intestinal microbiota from conventionally housed CD-1 mice. The process of conventionalizing the mice involved administering 100 μl of diluted cecal stool (300 mg/5 ml of pre-reduced PBS, diluted 1:500) from conventionally housed mice via oral gavage. Conventionalized mice were given the stool suspension approximately 30 hrs prior to the first exposure to the SDR stressor (i.e., 7 days prior to euthanasia). Like the germfree mice, the conventionalized mice were housed in the laboratory, but were kept separate from the germfree mice to prevent contamination. The success of conventionalizing the mice was confirmed by culturing the cecal contents (Supplemental Data Fig. 1). Results from germfree and conventionalized mice were compared to results from mice in the third group (i.e., conventional mice) that were housed in conventional cages and contained an intact intestinal microbiota.
Antibiotic Administration
Conventional mice were treated with an antibiotic cocktail consisting of vancomycin (0.5 mg/ml; Sigma-Aldrich, St. Louis, MO), metronidazole (1 mg/ml; Sigma-Aldrich), and neomycin trisulfate (1 mg/ml; Sigma-Aldrich) in sterile water similarly to previously described studies (Bailey et al., 2011; Ochoa-Reparaz et al., 2009; Rakoff-Nahoum et al., 2004). The antibiotic cocktail was administered by oral gavage (200 μl) once per day at 1600 for 3 days prior to the start of SDR and throughout the 6-day course of SDR. The vehicle control mice were orally gavaged with 200 μl of sterile water on the same schedule as the antibiotic-treated mice. This antibiotic treatment reduced the total microbiota by approximately 10 fold (Supplemental Fig. 2). To confirm whether the effects of the antibiotics were due to effects on the microbiota, rather than direct effects on leukocytes themselves, a separate experiment was conducted in which the antibiotic cocktail was injected intraperitoneally at the identical time points (Supplemental Data Fig. 3).
Splenocyte Processing
Mice were euthanized via CO2 asphyxiation and cardiac blood immediately taken. The spleens from individual mice were weighed and processed as previously reported (Bailey et al., 2007). Briefly, spleens were removed and macerated with a Stomacher 80 Biomaster Lab System (Seward, Bohemia, NY) in cold HBSS. The resultant cell suspensions were washed via centrifugation. Following centrifugation, the supernatants were removed and 1 ml of RBC lysis buffer was added to the cell pellet for 2 min to lyse red blood cells. To stop the lysis reaction, HBSS containing 10% heat-inactivated FBS was added to the cells. After washing, the cells were filtered using 70 μm filters, washed again, and re-suspended in CTLL RPMI 1640 (containing 0.075% sodium bicarbonate, 10 mM HEPES buffer, 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, 1.5 mM L-glutamine, and 0.00035% 2-mercaptoethanol) + 10% FBS. A Z2 Coulter Particle Counter and Size Analyzer (Beckman Coulter, Brea, CA) was used to count the cells and each sample was adjusted to contain 5×106 cells per ml. 2.5×105 cells were then stained with 1 μl of FITC-labeled anti-Gr1 and APC-labeled anti-CD11b antibodies to identify cell subsets in the spleen using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Monocytes/macrophages (which will collectively be referred to as macrophages) and granulocytes were identified by forward- and side-scatter characteristics and differences in CD11b and Gr-1 expression (Lagasse and Weissman, 1996). Forward- and side-scatter and absence of CD11b and Gr-1 staining was used to identify lymphocytes. The cell suspension was then adjusted so that the final concentration of macrophages was 5×104 per ml. The cells were cultured in 1 ml per well in a 24-well tissue culture plate for 2 hr at 37°C with 5% CO2 to allow the macrophages to adhere. There was no difference in tissue adherence between cells from non-stressed HCC Control mice and SDR Stressor mice as determined via microscopy (61±17 vs. 52±14 cells per field of view respectively). We have previously reported that over 98% of these adherent cells are mononuclear (Bailey et al., 2007), indicating that few neutrophils adhere to the culture plates.
Bacteria
Escherichia coli strain K12 (ATCC 10798), originally obtained from the American Type Culture Collection, was grown from frozen stocks in trypticase soy broth overnight at 37°C.
Microbicidal Assay
5×104 splenic CD11b+ macrophages were adhered to 24-well tissue culture plates in duplicate so that phagocytosis and bacterial killing could be determined after 20 and 90 min, respectively. After removing nonadherent cells, 1 ml of a mixture containing 5×107 CFU of E. coli in RPMI was added to each well. The E. coli were opsonized by adding 5% fresh serum (pooled from non-stressed HCC mice and stressed SDR mice) to the bacterial suspension. After adding the bacteria, the plates were incubated for 20 min, and then extracellular bacteria were washed away by pipetting 1 ml of RPMI into the well for 3 washes. After the final wash, 1 ml of 1% Triton X was added to one of the wells to lyse the splenocytes. This lysate was collected into a sterile tube for pour plate enumeration of phagocytized bacteria. The duplicate well was washed 3 times with RPMI+10% FBS. After the final wash, 1 ml of RPMI+10% FBS was added to the duplicate wells and the plates incubated for an additional 70 min. After 70 min, any extracellular bacteria were washed from the cells and the cells lysed with 1% Triton X to quantify bacteria remaining alive within the macrophages via standard pour plate analysis.
RNA isolation and cDNA synthesis
Two duplicate sets of 5×104 adherent macrophages per ml were plated in 24-well tissue culture plates at 37°C and 5% CO2. After a 2 hr incubation, the nonadherent cells were removed by washing 3X with RPMI. Following the final wash, 1 ml of RPMI + 10% FBS was added to half of the wells and the other half of the cells were stimulated with 1 ml of RPMI + 10% FBS containing 1 μg/ml of E. coli-derived LPS. After a 90 min incubation, the cells were washed with RPMI and then collected into 1 ml of Trizol reagent (Gibco, Rockville, MD). RNA was isolated according to the Trizol protocol provided by the manufacturer (Gibco). Total RNA was reverse transcribed using a commercially available kit (Promega, Madison, WI) according to manufacturer’s instructions. Briefly, 2 μg of total RNA was combined with 5 mM MgCl2, 1 mM of each dNTP, 1X RT buffer, 1 U/μl RNasin, 15 U/μg AMV reverse transcriptase, and primed with 0.5 μg random hexamers and DEPC H2O to a volume of 40 μl. The reaction was first incubated for 10 min at room temperature, and then at 42°C for 1 h. This was then followed by a 5 min incubation in boiling water and a cooling period of 5 min on ice. The volume was adjusted to 50 μl by adding DEPC water.
RT-PCR
Sequences for the primers and probes were previously published and were synthesized by Applied Biosystems. The 5′ – 3′ sequences are: iNOS (forward: CAG CTG GGC TGT ACA AAC CTT, reverse: TGA ATG TGA TGT TTG CTT CGG, probe: CGG GCA GCC TGT GAG ACC TTT GA), TNF-α (forward: CTGTCTACTGAACTTCGGGGTGAT, reverse: GGTCTGGGCCATAGAACTGATG, probe: ATGAGAAGTTCCCAAATGGCCTCCCTC), IL-1β (forward: GGCCTCAAAGGAAAGAATCTATACC, reverse: GTATTGCTTGGGATCCACACTCT, probe: ATGAAAGACGGCACACCCACCCTG), and 18S as an internal housekeeping gene (forward: CGCTACCACATCCAAGGAA, reverse: GCTGGAATTACCGCGGCT, probe: TGCTGGCACCAGACTTGCCCTC). The PCR reaction mixture consisted of 2.5 μl of cDNA, 2.5 μl primer mix (900 nM), 2.5 μl probe, 5 μl sterile dH2O, and 12.5 μl Taqman Universal Master Mix (PE Applied Biosystems, Foster City, CA) for a final volume of 25 μl. Following an initial 2-min cycle at 50 °C followed by 10 min at 95 °C, the reaction ran for an additional 40 total cycles consisting of a 15-s denaturing phase (90 °C) and a 1-min anneal/extension phase (60 °C). The change in fluorescence was measured using an Applied Biosystems 7000 Prism Sequence Detector (PE Applied Biosystems) and analyzed using Sequence Detector version 1.0. The relative amount of transcript was determined using the comparative Ct method as described by the manufacturer. In these experiments, gene expression in unstimulated spleen cells from non-stressed HCC mice was used as the calibrator. Gene expression, therefore, is expressed as the fold increase in expression over non-stressed control mice.
Fluorescence Assay (Peroxynitrite and Superoxide)
Splenocytes were collected and processed, and the CD11b+ cells were magnetically separated from the total cells using CD11b+ microbeads and MACS MS separation columns according to the manufacturer’s directions (Miltenyi Biotec, Auburn, CA). Consistent with previous reports (Bailey et al., 2007), the purity of the separated cells was high and the cultures were composed of >95% CD11b+ cells (data not shown). The separated CD11b+ macrophages were adjusted to 3×106/ml in CTLL RPMI + 10 % FBS and 200 μl/well were plated in 2 sets of triplicates on opaque 96-well tissue culture plates. Nonadherent cells were removed and 100 μl of HBSS without phenol red containing 25 μM of 1,2,3-dihydrorhodamine (Invitrogen, Eugene, OR), a peroxynitrite detecting dye, or 5 μM dihydroeithidium (Sigma, St. Louis, IL), a superoxide detecting dye, was added. Additionally, the cells in half the wells were stimulated with 5 ng/ml phorbol myristate (PMA) (Sigma), 1 μg/ml E. coli LPS (Sigma), and 5 ng/ml recombinant mouse IFNγ (Thermo Scientific, Rockford, IL). Peroxynitrite was measured using a fluorescence plate reader (Synergy HT Biotek, Winooski, VT) every 15 min for 90 min using excitation and emission wavelengths of 485 nm and 530 nm. Superoxide anion levels were measured every 15 min using excitation and emission wavelengths of 530 nm and 610 nm.
Cytokine Measurement in Culture Supernatants
To determine whether antibiotics would attenuate the stressor-induced increase in LPS-induced cytokine levels in culture supernatants, splenocytes were collected and processed and the CD11b+ cells were separated using magnetic beads (Miltenyi Biotec). The CD11b+ cells were adjusted to 5×106 cells per ml and 200 μl of cells were plated in 96 well flat bottom plates with 1 μg/ml of E. coli-derived LPS. Cultures were incubated for 18 hr at 37°C prior to collecting supernatants. Supernatants were kept frozen at −80°C until TNF-α and IL-1β levels were determined using OptEIA Set Mouse ELISA’s per manufacturer’s instructions (BD Pharmingen, San Diego, CA).
Corticosterone Measurement
Whole blood was collected from non-stressed and stressed mice via retro-orbital sampling immediately following SDR (~19:00). All blood samples were obtained from non-anesthetized mice within 5 min of disturbing the cage. Serum was separated from the whole blood and frozen at −80°C until use. Serum corticosterone was measured using a commercially available kit (Corticosterone EIA, Enzo Life Sciences) per manufacturer’s directions.
Lipopolysaccharide and Peptidoglycan Measurement
Blood samples were collected via cardiac puncture from HCC Control and SDR Stressor mice that were either untreated or treated with vehicle or antibiotics. Serum was separated from the whole blood and frozen at −80°C until use. Levels of lipopolysaccharide were assessed using the limulus amebocyte lysate (LAL) gel clot method according to manufacturer’s directions (Associates of Cape Cod, East Falmouth, MA). This is the preferred method of detecting endotoxin in the blood, and has a sensitivity of 0.03 EU/ml. Levels of peptidoglycan were assessed using the SLP (silk worm larvae plasma) Reagent Kit (Wako Chemicals, Richmond, VA). This is a colorimetric assay with a limit of detection of 1.5 ng/ml. Peptidoglycan levels were determined through a standard curve generated with peptidoglycan derived from Staphylococcus aureus.
Statistics
Two factor analysis of variance (ANOVA) was used to detect differences in cytokine mRNA with group (i.e., either conventional vs. germfree vs. conventionalized or vehicle vs. antibiotic) as one between subjects factor and stimulation (i.e., unstimulated vs. LPS stimulated) as the second between subjects factor. A three factor ANOVA was used to assess cytokine protein levels with condition (i.e., HCC Control vs. SDR Stressor), group (i.e., vehicle vs. antibiotic), and stimulation (i.e., unstimulated vs. LPS stimulated) as the between subjects factors. A mixed factor ANOVA was used to determine statistical significance in the number of bacteria remaining alive in co-culture with splenic cells, with time (i.e., 20 min vs. 90 min) as the repeated variable and group (i.e., HCC Control vs. SDR Stressor) as the between subjects factor. A mixed factor ANOVA was also used to determine statistical significance for the superoxide and peroxynitrite measurements with time (i.e., 0, 15, 30, 45, 60, 75, and 90 min) as the within subjects factor and group (i.e., HCC Control vs. SDR Stressor) and condition (i.e., conventional vs. germfree vs. conventionalized or vehicle vs. antibiotic) as the between subjects factor. Differences in serum corticosterone levels, spleen mass, splenic cellular composition, and peptidoglycan levels were determined using either ANOVA with the between-subjects factors being the treatment groups (i.e. Vehicle vs. Antibiotics) and condition (i.e., HCC Control vs. SDR Stressor) or independent samples t-test with HCC Control vs. SDR Stressor as the independent samples. Protected t-tests were used as post-hoc tests. In all cases, the level of significance was set at p<.05. All statistics were calculated using SPSS for Windows version 17 (SPSS, Chicago, IL).
Results
Exposure to the SDR Stressor Increases the Ability of Splenic Macrophages to Kill E. coli
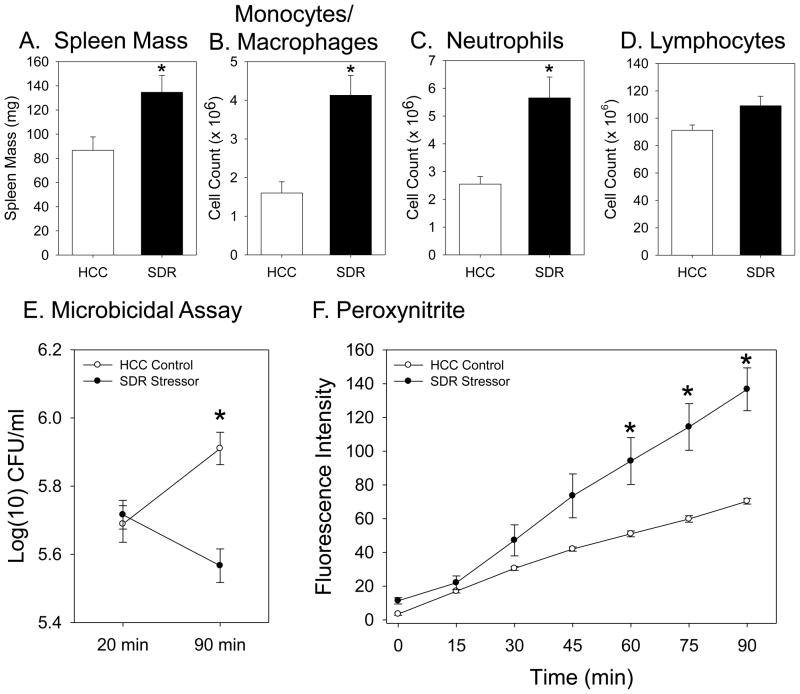
Exposing mice to the SDR stressor significantly increased spleen mass (t(4) = 2.70, p = 0.05; Fig. 1A), which was associated with a significant increase in the number of splenic macrophages (t(4) = 4.28, p < 0.05; Fig. 1B) and neutrophils (t(4) = 3.88, p < 0.05; Fig. 1C), but not lymphocytes (t(4) = 2.27, p = 0.09 (not significant); Fig. 1D), which is in agreement with previous studies (Bailey et al., 2007; Engler et al., 2004; Stark et al., 2001). Exposure to the SDR stressor also increased the ability of splenic macrophages to kill E. coli (F(1,4) = 39.70, p < 0.01). While exposure to the stressor did not significantly affect the ability of the macrophages to phagocytose the E. coli, as indicated by similar numbers of E. coli within the cells at the 20 min time point (Fig. 1E), by 90 min there was a significantly lower number of living E. coli within the macrophages from mice exposed to the SDR stressor (p < 0.05; Fig. 1E). This enhanced microbicidal activity is consistent with a previous study (Bailey et al., 2007), and was associated with a significant increase in peroxynitrite production (F(6, 24) = 21.13, p < 0.01; Fig. 1F). Splenic macrophages from mice exposed to the SDR stressor produced significantly higher levels of peroxynitrite than did cells from non-stressed HCC Control mice after 60, 75, or 90 min of stimulation (p < 0.05; Fig. 1F).
Figure 1.
Splenic macrophages from mice exposed to the SDR stressor kill more E. coli and produce more peroxynitrite than macrophages from non-stressed control mice. Spleen mass (Fig 1A), the number of splenic monocytes/macrophages (Fig. 1B), and the number of splenic neutrophils (Fig. 1C) were increased in conventional mice exposed to the SDR stressor (*p<0.05 vs. HCC Control). Exposure to the SDR stressor did not affect the number of splenic lymphocytes (Fig. 1D). The number of E. coli remaining alive within splenic macrophages from SDR stressor-exposed mice was significantly lower than the number remaining alive within macrophages from non-stressed HCC Control mice (*p<0.05 vs. HCC Control at 90 min) (Fig. 1E). Splenic macrophages from mice exposed to the SDR stressor produce significantly higher levels of peroxynitrite after stimulation in comparison to levels produced by cells from nonstressed HCC Control mice (*p<.05 vs. HCC Control at 60, 75, and 90 min) (Fig. 1F). Values are means ± SE [n=3 HCC and 3 SDR].
The Enhancive Effects of the SDR Stressor on the Functioning of Splenic Macrophages are Attenuated in Germfree Mice
To determine whether indigenous microbiota were necessary for the SDR stressor-induced increases in macrophage reactivity to manifest, we exposed germfree mice to the SDR stressor. To be sure any significant effects were due to a lack of microbiota, and not due to an inherent underdevelopment of innate immunity in germfree mice, the germfree mice were conventionalized by colonizing them with stool from conventional mice that had an intact microbiota. The effects of the SDR stressor on spleen mass (F(1, 30) = 4.16, p < 0.05; Fig. 2A), splenic macrophages (F(1, 30) = 11.98, p < 0.05; Fig. 2B), and neutrophils (F(1, 30) = 6.78, p < 0.05; Fig. 2C) were manifest in both germfree mice and conventionalized mice (as indicated by main effects for stress). But, conventionalizing the germfree mice with microbiota led to larger spleen masses (F(1, 30) = 5.45, p < 0.05) and increased numbers of splenic macrophages (F(1, 30) = 11.48, p < 0.05) as indicated by main effects for condition. The stressor did not affect the number of lymphocytes in the spleen (F(1, 30) = 0.78, p = 0.38 (not significant); Fig. 2D).
Figure 2.
Intestinal microbiota are necessary for SDR stressor-induced enhancement in bacterial killing and peroxynitrite production. Fig 2A–C: Spleen mass, the number of splenic monocytes/macrophages, and number of splenic neutrophils were increased in both germfree and conventionalized mice exposed to the SDR stressor (*p<0.05 vs. HCC Control). Fig. 2D: Stressor exposure did not affect the number of splenic lymphocytes. Fig. 2E: Splenic macrophages from SDR stressor-exposed germfree mice that lack any endogenous microbiota failed to kill more E. coli than cells from nonstressed HCC Control germfree mice. Conventionalizing the Germfree mice with stool from conventionally-housed mice recapitulated the SDR stressor-enhanced bacterial killing. Splenic macrophages from conventionalized mice exposed to the SDR Stressor killed significantly more E. coli than did cells from nonstressed HCC Control conventionalized mice (*p<0.05). Fig 2F: Splenic macrophages from conventionalized mice exposed to SDR produced significantly more peroxynitrite at 45, 60, 75, 90 min compared to macrophages from mice in all other groups (*p<0.05). Values are means ± SE [n=9(HCC Control Germfree), n=8(SDR Stressor Germfree), n=9(HCC Control Conventionalized), n=8(SDR Stressor Conventionalized), *p<0.05)].
As predicted, exposure to the SDR stressor did not enhance the microbicidal activity of splenic macrophages from germfree mice (F(1, 15) = 0.15, p = 0.70 (not significant); Fig. 2E). Splenic macrophages from conventionalized mice that were exposed to the SDR stressor killed significantly more E. coli than did splenic macrophages from conventionalized mice that were left undisturbed as HCC Controls (F(1, 15) = 18.66, p < 0.001; Fig. 2E).
Peroxynitrite levels differed depending upon the presence of an intact microbiota (F(6, 180) = 16.92, p < 0.001; Fig. 2F). Splenic macrophages from conventionalized mice that were exposed to the SDR stressor produced significantly higher levels of peroxynitrite than did splenic macrophages from mice in all other groups after 45, 60, 75, and 90 min of stimulation (p < 0.05). While peroxynitrite levels were higher in germfree mice exposed to the stressor in comparison to non-stressed HCC Control germfree mice (Fig. 2F), the levels of peroxynitrite in germfree mice were still significantly lower than levels found in the conventionalized mice exposed to the stressor (p < 0.05; Fig. 2F).
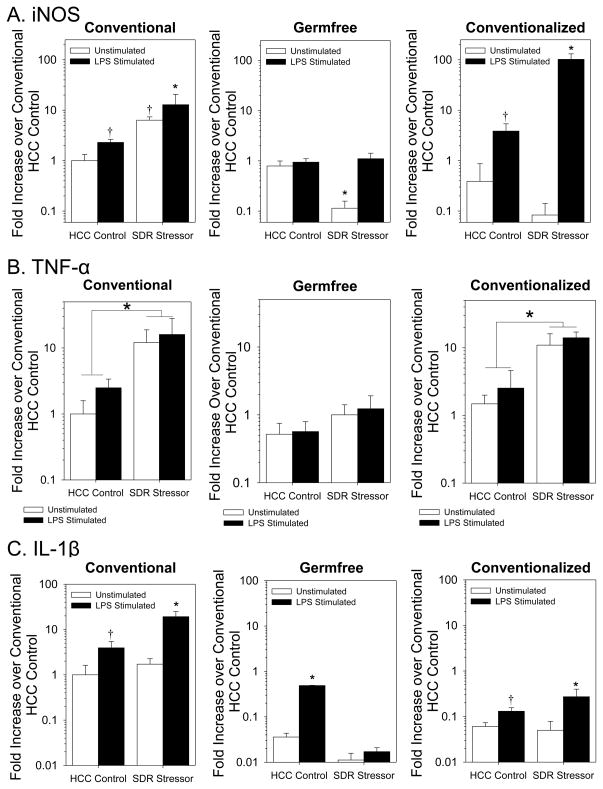
Nitric oxide is necessary for peroxynitrite production and is produced when the enzyme iNOS reacts with arginine. Therefore, iNOS gene expression was assessed in unstimulated and LPS stimulated splenic macrophages, and was found to be significantly higher in splenic macrophages from the SDR Stressor mice compared to HCC Control mice (F(1, 16) = 9.58, p < 0.05; Fig. 3A). This effect was evident in both unstimulated and stimulated cells (p < 0.05; Fig. 3A). Exposing germfree mice to the SDR stressor failed to increase iNOS gene expression in either unstimulated or stimulated splenic macrophages (Fig. 3A). In fact, iNOS gene expression was significantly lower in unstimulated splenic macrophages from germfree mice exposed to the SDR stressor (F(1, 29) = 26.38, p < 0.001; Fig. 3A). While iNOS gene expression remained low in unstimulated macrophages from conventionalized mice, exposure to the SDR stressor restored the stressor-induced increase in iNOS in splenic macrophages stimulated with LPS (F(1, 25) = 29.03, p < 0.001; Fig. 3A).
Figure 3.
Exposure to SDR increases iNOS and proinflammatory cytokine gene expression in LPS-stimulated splenic macrophages from mice with intact intestinal microbiota. Fig 3A: iNOS mRNA expression was significantly increased in LPS-stimulated splenic macrophages from stressor-exposed conventionally-housed and conventionalized mice in comparison to cells from nonstressed controls (*p<0.05 vs. all other groups; †p<0.05 vs. unstimulated HCC Control). Fig 3B: Stressor exposure increases TNF-α mRNA expression in splenic macrophages from conventional and conventionalized mice regardless of stimulation (*p<0.05 main effect for stress). Stressor exposure failed to enhance TNF-α mRNA in splenic macrophages from germfree mice. Fig 3C: IL-1β mRNA expression is higher in LPS-stimulated macrophages (†p<0.05 vs. unstimulated HCC Control). Exposing mice with microbiota to the SDR stressor further increased IL-1β mRNA expression (*p<0.05 vs. all other groups). Stressor exposure failed to increase IL-1β mRNA expression in macrophages from germfree mice even with LPS-stimulation. Values are means ± SE [n=5(HCC Control Conventional), n=5(SDR Stressor Conventional), n=9(HCC Control Germfree), n=8(SDR Stressor Germfree), n=9(HCC Control Conventionalized), n=6(SDR Stressor Conventionalized)].
Exposure to the SDR stressor increased both TNF-α (F(1, 16) = 74.67, p < 0.001; Fig. 3B) and IL-1β (F(1, 16) = 10.34, p < 0.005; Fig. 3C) gene expression in conventional mice. This effect of the SDR stressor was absent in germfree mice. TNF-α gene expression was unaffected by stressor exposure in germfree mice (F(1, 30) = 0.08, p = 0.78 (not significant); Fig. 3B), whereas IL-1β gene expression was lower in germfree mice exposed to the SDR stressor compared to nonstressed HCC Control germfree mice (F(1, 30) = 22.80, p < 0.05; Fig. 3C). Conventionalizing the germfree mice restored the effects of the stressor on TNF-α gene expression (F(1, 30) = 55.47, p < 0.001; Fig. 3B). Conventionalizing also led to a significant increase in IL-β gene expression in LPS stimulated splenic macrophages from mice exposed to the SDR stressor (F(1, 30) = 10.30, p < 0.005; Fig. 3C), but overall IL-1β levels were lower in conventionalized mice in comparison to conventional mice (Fig. 3C).
Antibiotic Administration Attenuates the Ability of the SDR Stressor to Enhance the Functioning of Splenic Macrophages
In a follow up experiment, the intestinal microbiota were disrupted by orally administering an antibiotic cocktail to conventional mice. Antibiotic administration did not abrogate the effects of the SDR stressor on spleen mass (Fig. 4A), the number of splenic monocytes/macrophages (Fig. 4B) or the number of splenic neutrophils (Fig. 4C) as indicated by significant main effects for stress (F(1, 26) = 31.11, p < 0.05; F(1, 26) = 14.76, p < 0.05; F(1, 26) = 19.29, p < 0.05, respectively), but no significant main effects for condition (i.e., vehicle vs. antibiotics) or significant interactions in the two-way ANOVA. Unlike previous studies, however, stressor exposure enhanced the number of splenic lymphocytes in the spleens of vehicle and antibiotic-treated mice (F(1, 26) = 5.13, p < 0.05; Fig. 4D). Repeated oral gavage of water as a vehicle control significantly affected the ability of splenic macrophages from SDR Stressor mice to phagocytose E. coli (F(1, 14) = 47.15, p < 0.001; Fig. 4E). However, the increased capacity of these cells to kill E. coli was still evident by a significant reduction in the number of E. coli remaining alive within the macrophages at the 90 min time point (Fig. 4E, p < 0.05). This stressor-induced increase in phagocytosis and of bacterial killing was not evident in mice treated with the antibiotics (F(1, 14) = 2.89, p = 0.11 (not significant); Fig. 4E). In addition, antibiotics disrupted the production of peroxynitrite (F(6, 168) = 4.17, p < 0.001; Fig. 4F). Peroxynitrite production was significantly higher in splenic macrophages from vehicle-treated SDR Stressor mice compared to all other groups after 60, 75, and 90 min of stimulation (p < 0.05, Fig. 4F).
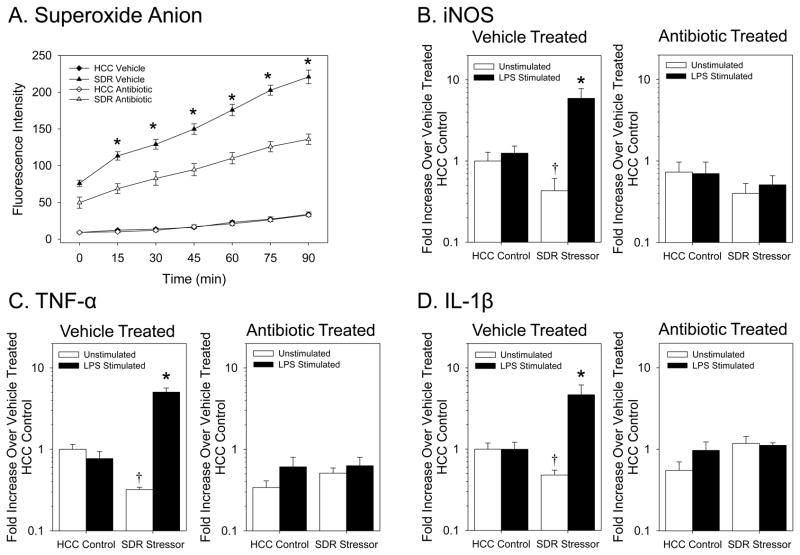
Figure 4.
Antibiotic reduction of the intestinal microbiota abrogated the SDR stressor-induced increase in bacterial killing and peroxynitrite production. Fig. 4A–D: Exposure to the SDR stressor increased spleen mass, the number of splenic monocytes/macrophages, the number of splenic neutrophils, and the number of splenic lymphocytes in both vehicle- and antibiotic-treated mice (*p<0.05 vs. HCC Control). Fig 4E: Exposing vehicle-treated mice to the SDR stressor significantly increased phagocytosis (*p<0.05 vs. HCC Control at 20 min) and bacterial killing (*p<0.05 vs. HCC Control and 90 min). However, SDR stressor exposure failed to enhance bacterial killing by macrophages from antibiotic-treated mice. Fig 4F: Splenic macrophages from vehicle-treated mice exposed to the SDR stressor produced significantly higher levels of peroxynitrite (*p<0.05 vs. all other groups at 60, 75, and 90 min of stimulation). Values are means ± SE [n=8(HCC Control Vehicle), n=8(SDR Stressor Vehicle), n=8(HCC Control Antibiotics), n=8(SDR Stressor Antibiotics)].
Because of the importance of superoxide for the formation of peroxynitrite, we also assessed superoxide anion levels in this follow up experiment, and found that antibiotics also affected superoxide anion levels (F(6, 96) = 34.83, p < 0.001; Fig. 5A). While levels of superoxide anion were significantly higher in the macrophages from vehicle-treated SDR Stressor mice compared to all other groups (p < 0.05), antibiotic administration significantly reduced superoxide anion in macrophages from stressed mice (p < 0.05 after 15, 30, 45, 60, 75 and 90 min of stimulation). Overall superoxide anion levels, however, were still elevated in the stressor-exposed mice treated with anitibiotics compared to the non-stressed controls (Fig. 5A). In addition, iNOS gene expression, which was significantly increased in LPS stimulated macrophages from vehicle-treated mice exposed to the SDR stressor (F(1, 16) = 7.03, p < 0.001; Fig. 5B) was significantly affected by antibiotic treatment. Macrophages from antibiotic-treated SDR Stressor mice expressed similar levels of iNOS mRNA as did nonstressed controls (F(1, 16) = 0.07, p = 0.79 (not significant); Fig. 5B).
Figure 5.
Antibiotic treatment attenuates the SDR stressor-induced increase in splenic macrophage reactivity to stimulation. Fig 5A. Splenic macrophages from vehicle-treated mice exposed to the SDR stressor produce significantly higher levels of superoxide anion (*p<0.05 vs. all other groups at 15, 30, 45, 60, 75, and 90 min). Fig. 5B. iNOS gene expression is significantly higher in LPS-stimulated macrophages from vehicle-treated mice exposed to the SDR stressor (*p<0.05 vs. all other groups). SDR exposure results in significantly lower iNOS gene expression in unstimulated macrophages from vehicle-treated mice (†p<0.05 vs. unstimulated HCC Control). Antibiotic administration prevented these effects. Fig 5C: TNF-α mRNA expression is increased in LPS-stimulated macrophages from vehicle-treated stressor-exposed mice in comparison to macrophages from nonstressed controls. Antibiotic treatment prevented this increase. Fig 5D: IL-1β mRNA expression was significantly increased in LPS-stimulated macrophages from vehicle-treated stressor-exposed mice compared to macrophages from nonstressed HCC Controls (*p<0.05 vs. all other groups). Vehicle treatment significantly reduced IL-1β gene expression in unstimulated splenic macrophages from mice exposed to the SDR stressor (†p<0.05 vs. unstimulated HCC Controls). Values are means ± SE [n=5 for all groups].
Treatment with antibiotics also significantly affected cytokine gene expression. TNF-α gene expression (F(1, 16) = 48.43, p < 0.001; Fig. 5C) and IL-1β gene expression (F(1, 16) = 11.57, p < 0.005; Fig. 5D) were significantly elevated in the LPS-stimulated macrophages from vehicle-treated SDR Stressor mice. However, administration of antibiotics attenuated these stressor-induced increases (Fig. 5C and 5D) (F(1, 16) = 0.03, p = 0.86 (not significant) and F(1, 16) = 0.94, p = 0.35 (not significant), respectively).
Stimulating splenic CD11b+ cells with LPS for longer periods (i.e., 18 hrs) increased TNF-α (Fig. 6A) and IL-1β levels (Fig. 6B), but the effects were dependent upon whether the mice were exposed to the SDR Stressor or were treated with antibiotics as evidenced by the 3-way interaction in the ANOVA (TNF-α: (F(1, 32) = 8.68, p < 0.01; IL-1β: (F(1, 32) = 10.40, p < 0.01)). Post-hoc testing determined that cells from vehicle-treated mice exposed to the SDR stressor produced higher levels of TNF-α and IL-1β than did cells from mice in any other condition (p < 0.05). Treatment with antibiotics reduced the cytokine levels (p < 0.05), but cytokine production by cells from antibiotic-treated mice exposed to the SDR stressor was still higher than from antibiotic-treated, HCC Control mice (p < 0.05; Fig. 6A & B).
Figure 6.
Stressor-induced increases in TNF-α and IL-1β production by splenic CD11b+ cells were attenuated in mice treated with antibiotics. Splenic CD11b+ macrophages were stimulated with 1 μg of E. coli-derived LPS. After 18 hr in culture, TNF-α (Fig. 6A) and IL-1β (Fig. 6B) levels were measured via ELISA. **p< 0.05 vs. all other groups. *p<0.05 vs. HCC Antibiotic+LPS and vs. SDR Vehicle+LPS. Values are means± SE [n=5 for all groups].
Antibiotic Administration Does Not Block SDR Stressor-Induced Corticosterone Levels
To confirm that treatment with antibiotics did not affect the physiological stress response, we assessed circulating corticosterone, which is well known to be affected by exposure to the SDR Stressor. Corticosterone levels were significantly elevated in both the vehicle and antiobiotic-gavaged mice (F(1,15) = 7.68, p < 0.05; Table 1), confirming that the SDR stressor activated the HPA axis in these animals.
Table 1.
Circulating Corticosterone
Exposure to the SDR Stressor increases circulating corticosterone. Data are the mean S.E. in ng/ml.
main effect of stress, p < 0.05
Stressor-Induced Increases in Serum Peptidoglycan is Attenuated by Antibiotic Administration
To begin to understand the mechanisms by which the microbiota could impact the innate immune system, we determined whether bacterial products likely derived from the microbiota could be detected in the serum. Gram-negative bacterial LPS was below the detectable limit of 0.03 EU/ml in the serum of non-stressed HCC Control mice and of mice exposed to the SDR stressor (data not shown). In contrast, mice had detectable levels of peptidoglycan (Table 2), and the levels of peptidoglycan were significantly higher in the stressor exposed mice in comparison to the non-stressed HCC Control (F(1, 20) = 9.20, p < 0.05; Table 2). Treating mice with the antibiotic cocktail significantly reduced the stressor-induced increase in circulating peptidoglycan (p < 0.05; Table 2).
Table 2.
Circulating Peptidoglycan
Exposure to the SDR Stressor increases circulating peptidoglycan. Antibiotic administration reduces the stressor-induced increase Data are the mean S.E. in ng/ml.
p < 0.05 vs. all other groups.
p<.05 vs. HCC Control+Antibiotics
Discussion
The results of this study demonstrate that the microbiota contribute to stressor-induced increases in macrophage cytokine production and microbicidal activity. This was first evident in germfree mice that were born and raised in sterile environments, and thus were not colonized by any microbiota. Exposing germfree mice to the SDR stressor failed to increase the microbicidal activity of splenic macrophages. Even though germfree mice are known to have an underdeveloped immune system (Macpherson and Uhr, 2004), it is not likely that the inability of the SDR stressor to enhance microbicidal activity was due to aberrant development of the innate immune system. This is based on the finding that conventionalizing the mice by colonizing them with microbiota from conventional mice was sufficient to restore the effects of the stressor on macrophage activity. However, to more fully understand the importance of intestinal bacteria on stressor-induced immunomodulation, follow up experiments were conducted in conventional immunocompetent mice treated with antibiotics. Consistent with findings in germfree mice, administering broad spectrum antibiotics reduced the stressor-induced increase in splenic macrophage microbicidal activity. Although vancomycin and neomycin are poorly absorbed from the gut (Phongsamran et al., 2010; Wilhelm and Estes, 1999), metronidazole can be absorbed and can impact the phagocyte oxidative burst (Miyachi, 2000). However, it is not likely that antibiotic administration abrogated the stressor-induced increase in splenic macrophage activity through direct effects on splenic macrophages, because administering the antibiotic cocktail through intraperitoneal injection (rather than oral administration) did not prevent the stressor-induced increase in macrophage microbicidal activity or peroxynitrite production (Supplemental Data, Fig. 3). This suggests that antibiotic effects on the microbiota, rather than on leukocytes themselves, attenuated the stressor-induced increases in splenic macrophage reactivity.
The stressor-induced increase in microbicidal activity was associated with a significant increase in iNOS gene expression and superoxide anion levels. These stressor-induced increases were partly dependent upon the microbiota, and likely led to the enhanced microbicidal activity. Nitric oxide and superoxide rapidly react to form peroxynitrite (Beckman and Koppenol, 1996), which has been shown to mediate the ability of macrophages to kill microbial pathogens, including E. coli (McLean et al., 2010; Weatherby et al., 2003). In our study, peroxynitrite levels were highest in splenic macrophages from mice exposed to the SDR stressor that also had an intact microbiota. These data suggest that the microbiota are involved with priming splenic macrophages for enhanced microbicidal activity by enhancing peroxynitrite production.
In addition to priming for enhanced microbicidal activity, exposure to the SDR stressor also primes splenic macrophages for enhanced cytokine production (Avitsur et al., 2005; Bailey et al., 2009b; Engler et al., 2008; Quan et al., 2001; Stark et al., 2002). In the current study, the stressor-induced increases in TNF-α and IL-1β mRNA only occurred when the macrophages were taken from mice with an intact microbiota; splenic macrophages from germfree or antibiotic-treated mice that were exposed to SDR did not have elevated cytokine gene expression. While antibiotics blocked the SDR stressor-induced increase in cytokine mRNA in short (i.e., 90 min) cultures, treating mice with antibiotics only attenuated the stressor-induced increase in cytokine levels in cultures stimulated for longer periods (i.e., 18 hrs). This suggests that additional factors, such as danger associated molecular patterns or even activation of the sympathetic nervous system contribute to the stressor-induced increase in cytokine production (Campisi and Fleshner, 2003; Fleshner et al., 2007; Johnson et al., 2005). However, even small differences in TNF-α and IL-1β due to alterations in the microbiota could be biologically meaningful, because both cytokines have been shown to increase phagocyte production of superoxide anion and iNOS gene expression (Brown et al., 2004; El-Benna et al., 2008; Guha and Mackman, 2001).
It is not yet clear how the intestinal microbiota can impact systemic immunity, but the effects likely involve the translocation of bacteria, or their products, from the lumen of the intestines to the interior of the body. Exposure to a variety of different types of stressors increases intestinal permeability (Gareau et al., 2007; Saunders et al., 2002; Soderholm et al., 2002), and alterations in the community structure of the intestinal microbiota can facilitate the translocation of intestinal bacteria from the lumen of the intestines to the interior of the body (Berg, 1999). We have previously shown that exposure to the SDR stressor significantly affects the microbial composition in the gut and also increases bacterial translocation (Bailey et al., 2006; Bailey et al., 2010; Bailey et al., 2011). In previous studies, living bacteria were identified in the spleens of approximately 40% of mice exposed to the SDR stressor (Bailey et al., 2006), but SDR stressor-induced enhancement of splenic macrophage microbicidal activity is evident in nearly all mice. Thus, we tested whether the translocation of bacterial products could also be detected in the circulation of mice exposed to the SDR stressor.
Mice exposed to the SDR stressor had higher levels of bacterial cell wall peptidoglycan in their serum than did non-stressed control mice. This was detected with the SLP reagent that has high reactivity with peptidoglycan. But, it must be noted that the SLP reagent is not selective for bacterial peptidoglycan and can also react with 1→3 β-glucan, which is often found in fungi (Tsuchiya et al., 1996) In our model, it is likely that the SLP reagent is reacting with bacterial peptidoglycan and not 1→3 β-glucan, because the LAL reagent (used to detect bacterial endotoxin) can also reacts with 1→3 β-glucan (Roslansky and Novitsky, 1991). None of the serum samples from HCC Control mice or from SDR Stressor-exposed mice reacted with the LAL reagent, suggesting that the serum does not contain detectable levels of endotoxin or 1→3 β-glucan.
The finding of bacterial cell wall peptidoglycan in the serum of stressor-exposed mice is important, because once within the host, microbial cell wall components can prime the innate immune system for enhanced reactivity to microbial stimuli. For example, others have shown that microbiota prime the innate immune system by activating the nucleotide-binding, oligomerization domain-containing protein-1 (Nod1) receptor, which recognizes meso-diaminopimelic acid-containing peptidoglycan found in the cell wall of Gram-negative, but not Gram-positive, bacteria (Clarke et al., 2010). Future studies will confirm whether bacterial peptidoglycan is a critical link between the microbiota and stressor-induced enhancement of innate immunity.
Our findings of stressor-induced enhancement of microbicidal activity are consistent with others (Campisi et al., 2002; Campisi and Fleshner, 2003), and illustrate the importance of considering the microbiota in studies within the field of PsychoNeuroImmunology. Bidirectional communication exists between the brain and gut microbiota such that alterations in one can impact the functioning of the other (Rhee et al., 2009). While we have focused on “top-down” communication and implications on immune activity (Bailey et al., 2010; Bailey et al., 2011), an increasing number of studies demonstrate “bottom-up” communication and have shown that the microbiota impact animal behavior, such as learning and memory (Gareau et al., 2011; Li et al., 2009), anxiety (Neufeld et al., 2011), and exploratory behavior (Bercik et al., 2011). This may have implications in human diseases, since bacterial translocation, as assessed by an increased occurrence of circulating antibodies to microbiota, has been linked to mood disorders, such as depression (Maes, 2008; Maes et al., 2008). Our studies have revealed an important physiological function of the microbiota, particularly during stressful periods, and indicate that the microbiota are interactively involved in stressor-induced immunoenhancement.
Supplementary Material
Research Highlight.
The indigenous microbiota are important contributors to social stressor-induced increases in splenic macrophage microbicidal activity and cytokine production.
Acknowledgments
Rebecca Allen was supported by a T32 training grant DE014320. This work was supported by NIH RO3AI069097-01A1 and Ohio State University start up funds to M.T.B as well as NIH RO1AI073971 to B.M.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105(Suppl 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitsur R, Kavelaars A, Heijnen C, Sheridan JF. Social stress and the regulation of tumor necrosis factor-alpha secretion. Brain Behav Immun. 2005;19:311–317. doi: 10.1016/j.bbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Parry NM, Galley JD, Schauer DB, Lyte M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun. 2010;78:1509–1519. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Engler H, Powell ND, Padgett DA, Sheridan JF. Repeated social defeat increases the bactericidal activity of splenic macrophages through a Toll-like receptor-dependent pathway. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1180–R1190. doi: 10.1152/ajpregu.00307.2007. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Engler H, Sheridan JF. Stress induces the translocation of cutaneous and gastrointestinal microflora to secondary lymphoid organs of C57BL/6 mice. J Neuroimmunol. 2006;171:29–37. doi: 10.1016/j.jneuroim.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Kierstein S, Sharma S, Spaits M, Kinsey SG, Tliba O, Amrani Y, Sheridan JF, Panettieri RA, Haczku A. Social stress enhances allergen-induced airway inflammation in mice and inhibits corticosteroid responsiveness of cytokine production. J Immunol. 2009a;182:7888–7896. doi: 10.4049/jimmunol.0800891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Kinsey SG, Padgett DA, Sheridan JF, Leblebicioglu B. Social stress enhances IL-1beta and TNF-alpha production by Porphyromonas gingivalis lipopolysaccharide-stimulated CD11b+ cells. Physiol Behav. 2009b;98:351–358. doi: 10.1016/j.physbeh.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- Berg RD. Bacterial translocation from the gastrointestinal tract. Adv Exp Med Biol. 1999;473:11–30. doi: 10.1007/978-1-4615-4143-1_2. [DOI] [PubMed] [Google Scholar]
- Brown GE, Stewart MQ, Bissonnette SA, Elia AE, Wilker E, Yaffe MB. Distinct ligand-dependent roles for p38 MAPK in priming and activation of the neutrophil NADPH oxidase. J Biol Chem. 2004;279:27059–27068. doi: 10.1074/jbc.M314258200. [DOI] [PubMed] [Google Scholar]
- Campisi J, Fleshner M. Role of extracellular HSP72 in acute stress-induced potentiation of innate immunity in active rats. J Appl Physiol. 2003;94:43–52. doi: 10.1152/japplphysiol.00681.2002. [DOI] [PubMed] [Google Scholar]
- Campisi J, Leem TH, Fleshner M. Acute stress decreases inflammation at the site of infection. A role for nitric oxide. Physiol Behav. 2002;77:291–299. doi: 10.1016/s0031-9384(02)00861-2. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Benna J, Dang PM, Gougerot-Pocidalo MA. Priming of the neutrophil NADPH oxidase activation: role of p47phox phosphorylation and NOX2 mobilization to the plasma membrane. Semin Immunopathol. 2008;30:279–289. doi: 10.1007/s00281-008-0118-3. [DOI] [PubMed] [Google Scholar]
- Engler H, Bailey MT, Engler A, Sheridan JF. Effects of repeated social stress on leukocyte distribution in bone marrow, peripheral blood and spleen. J Neuroimmunol. 2004;148:106–115. doi: 10.1016/j.jneuroim.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Engler H, Bailey MT, Engler A, Stiner-Jones LM, Quan N, Sheridan JF. Interleukin-1 receptor type 1-deficient mice fail to develop social stress-associated glucocorticoid resistance in the spleen. Psychoneuroendocrinology. 2008;33:108–117. doi: 10.1016/j.psyneuen.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleshner M, Johnson JD, Friedman J. Extracellular Hsp72: A double-edged sword for host defense. In: Asea AA, De Maio A, editors. Heat Shock Proteins: Potent Mediators of Inflammation and Immunity. 1. New York: Springer; 2007. pp. 235–263. [Google Scholar]
- Fleshner M, Nguyen KT, Cotter CS, Watkins LR, Maier SF. Acute stressor exposure both suppresses acquired immunity and potentiates innate immunity. Am J Physiol. 1998;275:R870–R878. doi: 10.1152/ajpregu.1998.275.3.R870. [DOI] [PubMed] [Google Scholar]
- Gareau MG, Jury J, Perdue MH. Neonatal maternal separation of rat pups results in abnormal cholinergic regulation of epithelial permeability. Am J Physiol Gastrointest Liver Physiol. 2007;293:G198–G203. doi: 10.1152/ajpgi.00392.2006. [DOI] [PubMed] [Google Scholar]
- Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, MacQueen G, Sherman PM. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- Geiszt M, Leto TL. The Nox family of NAD(P)H oxidases: host defense and beyond. J Biol Chem. 2004;279:51715–51718. doi: 10.1074/jbc.R400024200. [DOI] [PubMed] [Google Scholar]
- Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- Huffnagle GB. The microbiota and allergies/asthma. PLoS Pathog. 2010;6:e1000549. doi: 10.1371/journal.ppat.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JD, Campisi J, Sharkey CM, Kennedy SL, Nickerson M, Greenwood BN, Fleshner M. Catecholamines mediate stress-induced increases in peripheral and central inflammatory cytokines. Neuroscience. 2005;135:1295–1307. doi: 10.1016/j.neuroscience.2005.06.090. [DOI] [PubMed] [Google Scholar]
- Johnson JD, O’Connor KA, Deak T, Stark M, Watkins LR, Maier SF. Prior stressor exposure sensitizes LPS-induced cytokine production. Brain Behav Immun. 2002;16:461–476. doi: 10.1006/brbi.2001.0638. [DOI] [PubMed] [Google Scholar]
- Johnson JD, O’Connor KA, Hansen MK, Watkins LR, Maier SF. Effects of prior stress on LPS-induced cytokine and sickness responses. Am J Physiol Regul Integr Comp Physiol. 2003;284:R422–R432. doi: 10.1152/ajpregu.00230.2002. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Weissman IL. Flow cytometric identification of murine neutrophils and monocytes. J Immunol Methods. 1996;197:139–150. doi: 10.1016/0022-1759(96)00138-x. [DOI] [PubMed] [Google Scholar]
- Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Li W, Dowd SE, Scurlock B, costa-Martinez V, Lyte M. Memory and learning behavior in mice is temporally associated with diet-induced alterations in gut bacteria. Physiol Behav. 2009;96:557–567. doi: 10.1016/j.physbeh.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann N Y Acad Sci. 2004;1029:36–43. doi: 10.1196/annals.1309.005. [DOI] [PubMed] [Google Scholar]
- Maes M. The cytokine hypothesis of depression: inflammation, oxidative & nitrosative stress (IO&NS) and leaky gut as new targets for adjunctive treatments in depression. Neuro Endocrinol Lett. 2008;29:287–291. [PubMed] [Google Scholar]
- Maes M, Kubera M, Leunis JC. The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuro Endocrinol Lett. 2008;29:117–124. [PubMed] [Google Scholar]
- Mays JW, Bailey MT, Hunzeker JT, Powell ND, Papenfuss T, Karlsson EA, Padgett DA, Sheridan JF. Influenza virus-specific immunological memory is enhanced by repeated social defeat. J Immunol. 2010;184:2014–2025. doi: 10.4049/jimmunol.0900183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean S, Bowman LA, Sanguinetti G, Read RC, Poole RK. Peroxynitrite toxicity in Escherichia coli K12 elicits expression of oxidative stress responses and protein nitration and nitrosylation. J Biol Chem. 2010;285:20724–20731. doi: 10.1074/jbc.M109.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyachi Y. Pharmacologic modulation of neutrophil functions. Clin Dermatol. 2000;18:369–373. doi: 10.1016/s0738-081x(99)00128-5. [DOI] [PubMed] [Google Scholar]
- Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- O’Connor KA, Johnson JD, Hansen MK, Wieseler Frank JL, Maksimova E, Watkins LR, Maier SF. Peripheral and central proinflammatory cytokine response to a severe acute stressor. Brain Res. 2003;991:123–132. doi: 10.1016/j.brainres.2003.08.006. [DOI] [PubMed] [Google Scholar]
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- Padgett DA, Glaser R. How stress influences the immune response. Trends Immunol. 2003;24:444–448. doi: 10.1016/s1471-4906(03)00173-x. [DOI] [PubMed] [Google Scholar]
- Phongsamran PV, Kim JW, Cupo AJ, Rosenblatt A. Pharmacotherapy for hepatic encephalopathy. Drugs. 2010;70:1131–1148. doi: 10.2165/10898630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Quan N, Avitsur R, Stark JL, He L, Shah M, Caligiuri M, Padgett DA, Marucha PT, Sheridan JF. Social stress increases the susceptibility to endotoxic shock. J Neuroimmunol. 2001;115:36–45. doi: 10.1016/s0165-5728(01)00273-9. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6:306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslansky PF, Novitsky TJ. Sensitivity of Limulus amebocyte lysate (LAL) to LAL-reactive glucans. J Clin Microbiol. 1991;29:2477–2483. doi: 10.1128/jcm.29.11.2477-2483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders PR, Santos J, Hanssen NP, Yates D, Groot JA, Perdue MH. Physical and psychological stress in rats enhances colonic epithelial permeability via peripheral CRH. Dig Dis Sci. 2002;47:208–215. doi: 10.1023/a:1013204612762. [DOI] [PubMed] [Google Scholar]
- Soderholm JD, Yates DA, Gareau MG, Yang PC, MacQueen G, Perdue MH. Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1257–G1263. doi: 10.1152/ajpgi.00314.2002. [DOI] [PubMed] [Google Scholar]
- Stark JL, Avitsur R, Hunzeker J, Padgett DA, Sheridan JF. Interleukin-6 and the development of social disruption-induced glucocorticoid resistance. J Neuroimmunol. 2002;124:9–15. doi: 10.1016/s0165-5728(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Stark JL, Avitsur R, Padgett DA, Campbell KA, Beck FM, Sheridan JF. Social stress induces glucocorticoid resistance in macrophages. Am J Physiol Regul Integr Comp Physiol. 2001;280:R1799–R1805. doi: 10.1152/ajpregu.2001.280.6.R1799. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M, Asahi N, Suzuoki F, Ashida M, Matsuura S. Detection of peptidoglycan and beta-glucan with silkworm larvae plasma test. FEMS Immunol Med Microbiol. 1996;15:129–134. doi: 10.1111/j.1574-695X.1996.tb00063.x. [DOI] [PubMed] [Google Scholar]
- Weatherby KE, Zwilling BS, Lafuse WP. Resistance of macrophages to Mycobacterium avium is induced by alpha2-adrenergic stimulation. Infect Immun. 2003;71:22–29. doi: 10.1128/IAI.71.1.22-29.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm MP, Estes L. Symposium on antimicrobial agents--Part XII. Vancomycin Mayo Clin Proc. 1999;74:928–935. doi: 10.4065/74.9.928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.