Abstract
Motivation: We investigate and quantify the generalizability of the white blood cell (WBC) transcriptome to the general, multiorgan transcriptome. We use data from the NCBI's Gene Expression Omnibus (GEO) public repository to define two datasets for comparison, WBC and OO (Other Organ) sets.
Results: Comprehensive pair-wise correlation and expression level profiles are calculated for both datasets (with sizes of 81 and 1463, respectively). We have used mapping and ranking across the Gene Ontology (GO) categories to quantify similarity between the two sets. GO mappings of the most correlated and highly expressed genes from the two datasets tightly match, with the notable exceptions of components of the ribosome, cell adhesion and immune response. That is, 10 877 or 48.8% of all measured genes do not change >10% of rank range between WBC and OO; only 878 (3.9%) change rank >50%. Two trans-tissue gene lists are defined, the most changing and the least changing genes in expression rank. We also provide a general, quantitative measure of the probability of expression rank and correlation profile in the OO system given the expression rank and correlation profile in the WBC dataset.
Contact: vvaltchinov@partners.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
One of the reasons that the classification of malignancy was one of the earliest human applications of microarray transcriptional profiling (Golub et al., 1999) was that the tissue to be characterized, the tumor, was extracted as a matter of a routine surgical oncological care. Other clinical domains have lagged because the tissue involved in a pathophysiology may not be reasonably obtained from a living individual (e.g. in behavioral or psychiatric disorders) or the organ specificity of a disease is unclear (e.g. type II diabetes mellitus or hypertension). Increasingly, investigators have explored the possibility of classifying, prognosticating and characterizing the mechanism of diseases using the gene expression patterns measured in white blood cells (WBCs) (Coppola et al., 2008; Padmos et al., 2008; Scherzer et al., 2007; Washizuka et al., 2009).
For those studies seeking to find the mechanisms/genes dysregulated in the tissue of interest, the large majority have been structured very similarly: a direct comparison is made between expression profiling of peripheral blood and the specific tissue where the disease is known to develop. An alternative method is to establish the degree of isomorphism between the peripheral blood transcriptome and the overlap in expression profiles from a fixed number of representative human tissues (e.g. brain, colon, heart, kidney, etc., altogether nine tissue types) (see Liew et al., 2006). To the surprise of many, these investigations have proven to show considerable shared transcriptome across these tissues. We adopt here a systematic survey approach using the ever-growing mountain of public microarray expression data of both WBC and dozens of other tissues, under a variety of conditions and pathophysiological states. Our goal is to provide a quantitative estimate of the generalizability of gene expression findings in WBC to those in other tissues. We seek to identify the most robust similarities and corresponding differences in a genome-wide expression profiles, and to quantify them appropriately. We hypothesize there will be robust correlations that survive the well-known variability of the multiply sourced gene expression database such as the Gene Expression Omnibus (GEO) (Barrett et al., 2005), that allow large fractions of the peripheral blood transcriptome to be reflected in other organs and tissues.
This hypothesis (heretofore referred to as the WBC relevance hypothesis) entails the following three questions: (i) to what extent do those genes with the highest levels of expression in the WBC transcriptome also have high levels of expression in other organ systems? Specifically, can we quantify how the rank of the gene in WBC informs us of the rank in non-WBC tissues? (ii) How do correlations between pairs of genes in WBC inform us of their correlation in non-WBC tissues? (iii) How does the overall correlation structure [e.g. Relevance Network (Butte et al., 1999)] of WBC compare to those of other tissues? To the extent that the WBC relevance hypothesis is supported we explore whether the informativeness of WBC gene expression is broad or varies by specific functional categories. We look at both the broad categories of gene annotation (Ashburner et al., 2000; Dennis et al., 2003), such as apoptosis, as well as examining individual genes. We used the NCBI's GEO to find experiments measuring WBC expression and a large number of experiments on non-WBC tissues that we lump into the Other Organ (OO) category. To minimize the noise from inter-platform comparisons (Nimgaonkar et al., 2003), we used only those data obtained on the GEO's GPL96 platform for both the WBC and OO categories. This resulted in a total of 1463 samples for the OO and 81 samples for the WBC set. The OO set consists of about 95% solid tissue samples and about 8% are cancer-related tissues (see Section 2.2).
2 MATERIALS AND METHODS
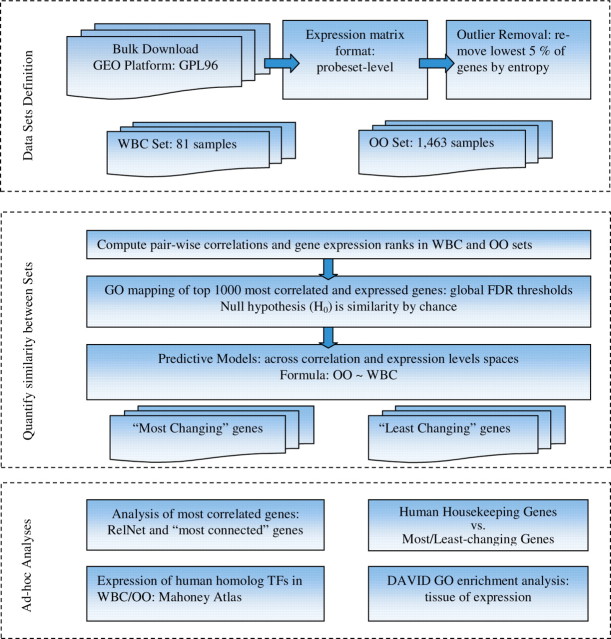
The overall flow of the analysis is summarized in Figure 1. The major steps are listed in the figure caption and are also explained in detail.
Fig. 1.
A diagram representing the workflow of the analysis. (i) ‘Data Sets Definition’ includes bulk download of the GEO archive files for the GPL96 platform, parsing out all GSM sample files with similar probeset-averaged expression matrix, detecting and removing outlier genes and constructing two sets for comparison, WBC and OO sets. (ii) ‘Quantify Similarity between sets’ include: use GO mapping of the most correlated (highly expressed) genes (as defined by their FDR q-value thresholds) to quantify change across sets; the null hypothesis H0 is overlap between corresponding sets from WBC and OO by chance); use a linear model and general least squares and PCA to quantify relationships between OO and WBC across expression and correlation profiles, respectively; define two list of ‘most-changing’ and ‘least-changing’ from WBC to OO genes, across expression profiles. (iii) In ‘Ad-hoc Analyses’, we first construct the RelNets of the 200 most correlated WBC genes and their corresponding OO pairs ranks changes. We next looked at the TFs human homologs from the Mahoney Atlas as expressed in WBC. Another step is looking at how the most-changing and least-changing genes from step (ii) are represented in the list of human housekeeping genes. Finally, we run GO enrichment analysis of the ‘least-changing’ genes with respect to ‘tissue-of-expression’, in DAVID.
2.1 GEO datasets
We have selected the NCBI's public archive GEO as a source of the microarray expression data (Barrett et al., 2005). To minimize the noise across platforms (Nimgaonkar et al., 2003), we have elected to use only datasets that were taken by the GPL96 platform, corresponding to the HG-U133A chip by Affymetrix, and only included probeset-averaged expression levels. A copy of the GEO database was downloaded on April 5, 2006. An outlier detection and removal analysis was performed (Butte, 1999). There are 1463 and 81 samples in the OO and WBC dataset, respectively.
2.2 Quantify tissue types in the OO set
To quantify the types of tissues across the OO set, we utilize the R package GEOquery to download and parse out the annotation for each of the 1463 samples assigned to the set. Four of the fields in the GSM annotation were used: ‘title’, ‘organism’, ‘source’ and ‘description’; the list is presented in the Supplementary Table S1.
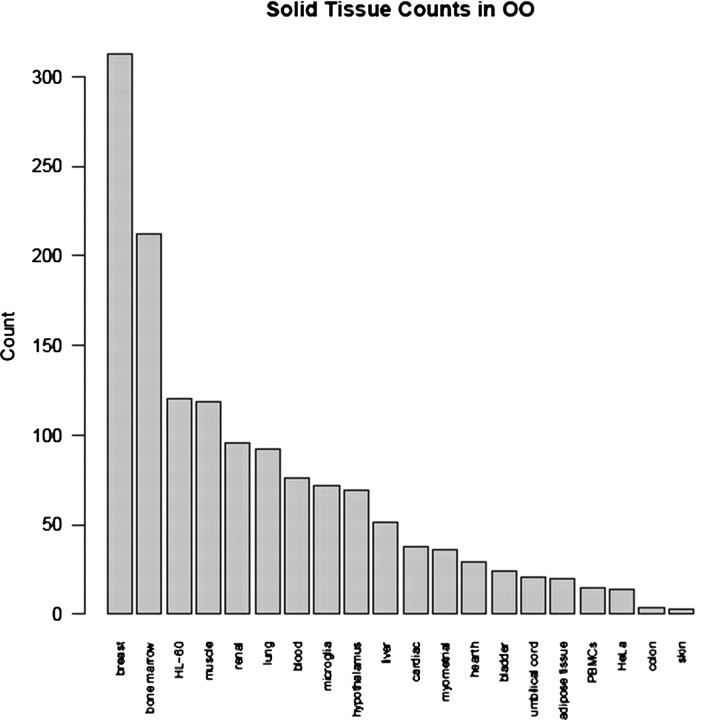
Overall, 27 GSM samples are no longer available from GEO and thus cannot be included in the statistics. There are 1387 samples that are from solid-tissue organs, or ∼ 95% of all samples in the OO set. One hundred and sixteen (or ∼8%) are samples that come from malignant tissues. The resulting tissue categories distribution is shown in Figure 2.
Fig. 2.
Distribution of solid tissues in the samples of the OO set. A total of 1436 GSM samples annotations were parsed out and reviewed with 27 samples from the original 2006 download that was no longer available in GEO.
2.3 Pair-wise correlation computation
For each of the datasets we have about ∼250 million pair-wise correlations between the 22 283 REF_IDs on the GPL96. Computations were carried out on an HPC Cluster at Partners HealthCare System, Inc.
2.4 ‘Top N’ most highly expressed and correlated gene lists as false discovery rate cutoff parameters
The selection of the top N used for the GO categories mapping statistics was based on a false discovery rate (FDR) (Benjamini, 1995; Schweder, 1982; with the specific q-value thresholds described below.
For the most highly expressed gene list, we have transformed the expression levels averaged over all samples to a ‘standard score’ (z-score), followed by a call to the ‘fdrtool’ R package to determine the corresponding q-values (Strimmer, 2008). Both OO and WBC cases were computed and the minimum of the two cutoff parameters was taken to be the global FDR cutoff parameter. The specific choice of N=1000 used in the analysis below corresponds to an FDR cutoff q-value = 0.02.
Similarly for the case of top N most correlated gene lists, we have transformed to z-scores for the Mutual Information Content (MIC) (Strimmer, 2008) and also used | r| for the case of the Pearson's correlation coefficient. Q-values were computed for both similarity measure cases and the minimum of the two was taken. The most correlated pairs list was transformed to gene lists by adding the two members of all pairs less the overlap set of genes between the members. Here, an FDR q-value of 0.005 corresponds approximately to N = 1000 (most correlated genes) parameter, for which comparison results are listed below.
2.5 Definition of change scores
Given two ranked lists of genes, we have defined a change score of each gene as the signed integer equal to the number of positions its rank changes between the WBC and OO datasets.
2.6 Test for independence of intersection of GO categories
We have utilized the χ2 test of independence to analyze the results listed in Table 2 for the WBC and OO overlap. Each of the lines in Table 2 was fed into chisq.test() in R (ver. 2.10.1) with the parameter ‘simulate.p.value = TRUE’ set so that a Monte Carlo sampling simulation of the P-value is performed. The average P=0.0004998 is based on 2000 MC simulations.
Table 2.
Representation of GO categories in 1000 most expressed (‘E’, in last column) or correlated (‘C’) genes
| Intersection | WBC | OO | E/C | |
|---|---|---|---|---|
| Cell | 116 | 53 | 65 | C |
| MF | 258 | 179 | 238 | C |
| BP | 280 | 220 | 233 | C |
| Cell | 144 | 18 | 87 | E |
| MF | 303 | 246 | 233 | E |
| BP | 322 | 267 | 257 | E |
2.7 General least squares and principal component analysis fitting procedures
For the expression profile modeling procedure, we first applied the low entropy filer and then have removed the ribosome-related genes by removing all genes that are annotated to belong to these two GO categories: (i) 5840, annotation=“ribosome” and (ii) 5842, annotation=“cytosolic large ribosomal subunit (sensu Eukaryota)”. We have retained the more restrictive low-entropy condition (for the WBC case) to keep the list of genes equal in the OO and WBC expression sets. Both low-entropy and the removal of ribosome genes procedures amounted to take 300 probes off the genes list. For the list of remaining genes, we have averaged the expression levels for each of them over the number of the samples in their corresponding dataset. The resulting arrays were fed into the R lm() function and a linear model was used for the functional relationship between OO and WBC sets.
For the correlation profiles case, a similar procedure was used for removing outliers (low-entropy filter) and the ribosomal-related set of genes. For each correlation metric, we have transformed to standard z-scores and computed the FDR q-values for those pairs, and only retained q-value below a threshold of α<0.05. The unification of the OO and WBC sets was performed and this enlarged set of correlation coefficients between the gene pairs was used as the basis for performing both general least squares and principal component analysis (PCA) in the samples versus OO and WBC gene-pair correlation space. The corresponding sizes of the data matrices analyzed were 30 619 × 2 and 61 190 × 2 for the MIC and Pearson's correlation coefficients, respectively. We used a standard singular value decomposition method from the ‘pcaMethods’ R package and lm() function for the linear least squares. Results are summarized in Table 3.
Table 3.
Linear least squares fit coefficients as computed for (i) expression profile, and linear squares and PCA with two PCs for (ii) cross-correlation using MIC, over the FDR q<0.05 truncated complete phase space of ∼250 million ‘points’ in the phase space of the pairs (WBC, OO) correlation numbers, and (iii) same as in (ii) for Pearson's correlation coefficient
| Linear least squares fitting: OO ∼ A + B × WBC Estimate SE; t-value; Pr(> |t|) | |
|---|---|
| Expression profiles | (A) 8.846590; 0.297784; 29.71 < 2e-16*** (B) 0.206404 0.001263; 163.48 < 2e−16*** |
| Residual SE | 40.47 on 21981 DF |
| Multiple R2 | 0.5487 |
| F-statistics | 2.672e+04 on 1 and 21981 DF, |
| P | <2.2e-16 |
| Correlation, MIC | (A) 5.73524; 0.01937; 296.2 < 2e-16*** (B) −1.42333; 0.00641; −222.1 < 2e−16*** |
| Residual SE | 0.3749 on 30617 DF |
| Multiple R2 | 0.6169 |
| Adjusted R2 | 0.6169 |
| F-statistics | 4.931e+04 on 1 and 30617 DF |
| P | <2.2e-16 |
| PCA: R2 | PC1: 0.926; PC2: 0.07403 |
| Correlation, Pearson | (A) 4.11674 0.01349 248.5 <2e-16*** (B) −0.42123; 0.00984 −309.3 <2e−16*** |
| Residual SE | 5.2748 on 60188 DF |
| Multiple R2 | 0.4751 |
| Adjusted R2 | 0.4751 |
| F-statistics | 1.721e+05 on 1 and 60188 DF |
| P | <2.2e-16 |
| PCA: R2 | PC1: 0.8314; PC2: 0.1686 |
See Section 2 for exact algorithmic steps. Significance level is encoded as ‘***’<0.0001.
2.8 Tissue of expression enrichment analysis in DAVID
A set of 48 REF_IDs for the GO category of sugar-binding genes (GO:5529) was submitted to DAVID. These probesets were mapped to 36 internal DAVID IDs. Only the ‘Tissue of Expression’ submenu options were selected. A functional annotation clustering was performed with the default options. Results are listed in Supplementary Table S6.
3 RESULTS
3.1 Fold and rank analysis
To first determine whether there was sufficient similarity between WBC and other tissues, we focused our analyses on those genes that had behaviors that were mostly likely to be consistent: genes with high levels of expression, with low entropies (i.e. low variances), and with high correlations with other genes across the entire GEO datasets on the GPL96 platform (see Section 2).
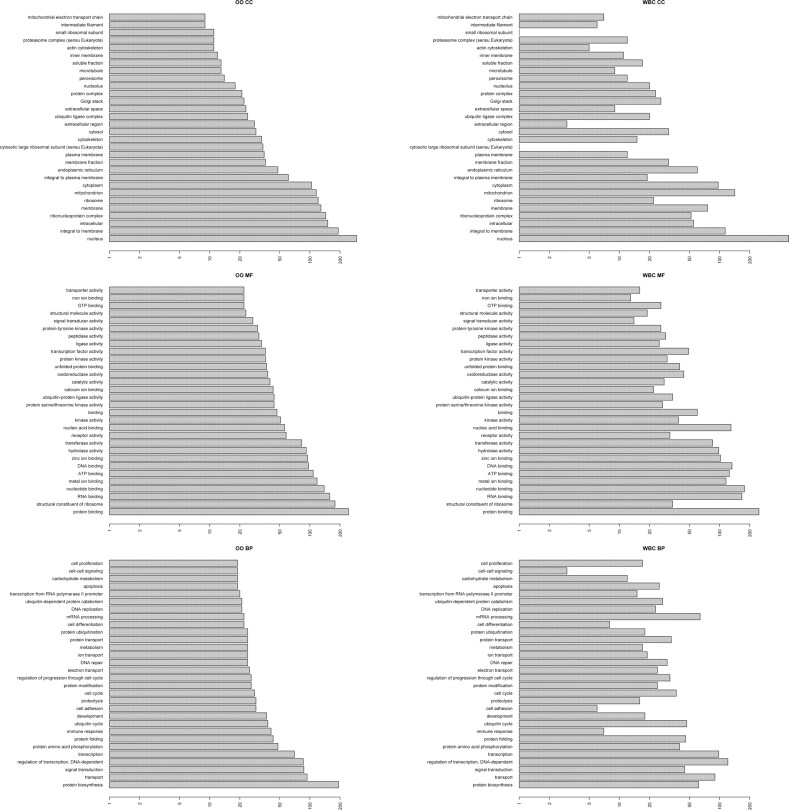
Solely reviewing the top N = 1000 most correlated genes (i.e. those with high correlations with other genes in their respective GEO datasets) and the1000 most expressed genes in the WBC and OO categories (see Section 2.4 for a discussion on the choice of N), we noted that the GO categories to which these 1000 genes belong (a mere 4.5% of the total number of genes measured on the GPL96 platform) had a very high overlap. The distribution of distinct GO categories within each of the three main GO hierarchies for OO and WBC datasets is shown in Figure 3. The hierarchies are ordered by the number of genes annotated for OO and the corresponding number of annotations in WBC shown in the second column of the figure. With the notable exceptions of components of the ribosome, cell adhesion and the immune response, the annotations of these top correlated genes in OO samples are well represented by the most correlated genes in WBC samples. For graphical compactness, Figure 3 only shows the 30 most annotated GO categories in OO samples. If we extend the analysis to all categories, then the GO categories that change the most (as defined by their Change Score described in Section 2) include the ribosomal location and related processes as before, but also includes nucleic acid binding, and spliceosome-related processes as shown in the second column of Table 1.
Fig. 3.
Distribution of GO categories in 1000 most correlated genes in WBC and in OO.
Table 1.
The most changing GO categories from WBC to OO
| GO categories of 1000 most correlated genes | GO categories of 1000 most expressed genes | |
|---|---|---|
| Cell | Ribosome (21 → 121) Cytosolic large ribosomal subunit (sensu Eukaryota) (1 → 34) | Mitochondrion (72 → 129) Extracellular matrix (sensu Metazoa) (7 → 37) |
| MF | Nucleic acid binding (130 → 56) Structural constituent of ribosome (34 → 178) | Transmembrane receptor activity (9 → 1) Sugar binding (16 → 3) |
| BP | Nuclear mRNA splicing, via spliceosome (47 → 9) Protein biosynthesis (62 → 193) | Nucleosome assembly (3 → 14) sensory perception of sound (12 → 2) |
But this focuses on the differences rather than the larger similarities between the WBC and OO transcriptomes suggested by the broad silhouettes in Figure 3. Indeed, 90% of the GO categories change <1.37% for cellular components, 1.83% for molecular function and 2.89% for the biological processes hierarchies, or in average of slightly over 2%. The similarities between WBC and OO are further quantified by examining which GO categories are shared by the 1000 most highly expressed or the 1000 most correlated genes in the two transcriptomes, respectively, as summarized in Table 2. In all instances, the intersection is larger than any of the distinct sets (ranging from 27% larger to 700% larger). The details of the particular GO categories summarized in this table are given in Supplementary Table S2. Of course, if all the genes on the chip were used in this analysis, rather than the top 1000, then all GO annotations would intersect completely across the two transcriptomes, by definition. However, with only 4.5% of the genes measured, the measured degree of overlap is highly improbable (P=0.000499 by using Monte Carlo sampling simulation in χ2 test, see Section 2). Supplementary Figure S1 illustrates the overall stability of numbers of annotation by GO category by plotting the frequency of change scores for each GO category. As shown, the distribution of change score is centered on zero for all three GO hierarchies. The top 1000 most expressed genes have a much tighter distribution (more low change scores) than the 1000 most correlated genes.
A broader examination of which genes change the most in their rank (by entropy or by expression) within the 22 283 genes measured on the GPL96 platform reveals a steep decline in the change in rank of expression as illustrated in Supplementary Figure S2. That is, 10 877 or 48.8% of all measured genes do not change >10% of rank range between WBC and OO. Only 878 (3.9%) change rank >50%. There is somewhat more change in rank of genes by entropy as revealed in Supplementary Figure S3. That is, 5084 (22.8%) of all genes do not change >10% of rank range in entropy, and 4356 (19.5%) by >50%.
The individual genes that change the least in rank across WBC and OO, are itemized in Supplementary Table S3 as are those genes that change the most. In the former list, are included HBB (hemoglobin β chain), H3F3A H3 histone, family 3A, MED6 (mediator of RNA polymerase II transcription, subunit 6 homolog), FTL (ferritin, light polypeptide), ACTG1 (actin, gamma 1), B2M (β-2-microglobulin) several HLA-related genes, several hemoglobin-related genes, S100A9 (S100 calcium binding protein A9—calgranulin B). In the latter list, are included the FCGR3B (Fc fragment of IgG, low affinity IIIb, receptor—CD16b), PSAP (prosaposin—variant Gaucher disease and variant metachromatic leukodystrophy), CA1 (carbonic anhydrase I—most highly expressed in erythrocytes) and UBB (ubiquitin B). In contrast to expression rank, UBB changes the least in entropy rank between WBC and OO. Conversely, GAPDH is highly invariant as viewed by change in rank in expression, but is among the most changed in rank in entropy across WBC and OO. Many more genes have similar rank profiles in expression and entropy: For example, H3F3A (H3 histone) is just as invariant in rank as in expression of HLA genes, ribosomal genes and several of the hemoglobins. The expression of HBB in avascular and non-hematopoietic tissues has been previously documented (e.g. Mansergh et al., 2008).
3.2 Quantitative map between WBC and OO transcriptomes
Having established the broad correlation between OO and WBC expression profiles, we next take the logical step of trying to find a quantitative relationship between the two transcriptomes. The exact question we ask is: can one come up with the expression rank in the OO system given the expression rank in WBC? Or, even more broadly, given the broader expression profile (values and cross-correlations) of a group of WBC genes how can one predict their corresponding rank values and correlations in the OO set, and with what probability? In doing this, we treat WBC expression (or correlation) profile as an independent random variable and determine the conditional probability distribution of the OO expression or correlation coefficients given the WBC ones.
We modeled the OO expression levels and correlation coefficients as a function of their WBC counterparts using linear predictive models. The ribosome-related genes were removed as previously described. The parameters for the resulting linear least squares models as well as the P-values measuring the goodness-of-fit for the expression and for the correlation profiles are summarized in Table 3 together with the PCA model results for the correlation profiles space. The two PCs for the MIC and r correlation spaces show similar structure where a large part of the variance is carried by the first PC.
3.3 Relevance networks and ‘most-connected’ genes
Relevance networks generate graph of connected nodes in which each node represents a gene and each edge represent a correlation metric (e.g. Pearson's) that exceeds a statistically significant threshold (Butte et al., 1999, 2000).
Shown in Supplementary Figure S4 is the relevance network for the WBC transcriptome depleted for ribosomal genes. The most highly connected genes include RRAGB (Ras-related GTP binding B), MAG (myelin-associated glycoprotein, member of the immunoglobulin super-family thought to be involved in the process of myelination and known to mediate certain myelin–neuron cell–cell interactions), BMP7 (bone morphogenetic protein 7), DOPEY1 (dopey family member 1), GTF2H5 (general transcription factor IIH, polypeptide 5, involved with DNA repair mechanisms, and nucleotide excision repair in particular), PURA (purine-rich element binding protein A, deletion of this gene has been associated with myelodysplastic syndrome and acute myelogenous leukemia), EZH1 (enhancer of zeste homolog, a transcriptional regulator and as a component of protein complexes that stably maintain heterochromatin.) and PGDS (prostaglandin D2 synthase, plays a role in the production of prostanoids in the immune system and mast cells). The rank of these 200 top correlated gene pairs in this WBC relevance network are compared with the ranks of those same gene pairs in the OO set, in Supplementary Figure S5. It does reveal a significant concordance in the ranks of correlation in gene pairs from WBC and OO, even though the overall shape of the presented distribution is somewhat peaked—the largest bin (N=50) corresponds to a rank change across WBC to OO of maximum of 11.4%. Ninety percent of all WBC pairs do not change rank by >31 positions or 15.5%. The top shared gene pairs are SF3B1 and DDX5 (the first is in the spliceosome and the other is DEAD domain involved in spliceosome), YME1L1 and FNTA (the first is ATP-dependent metalloprotease specific to mitochondria and the second is a farnesyl transferase, also involved in protein metabolism but in the cytoplasm).
It is instructive to note in passing that the results reported here do not depend on the actual permutation thresholds in the two comparison sets. Since the two sets are very different in size (81 for WBC versus 1463 for OO), we did not use a permutation scheme to select subsets for comparison (Kerr, 2009). Instead, we define the number of genes used for the correlation analyses based on global FDR thresholds. We built the relevance network of the WBC correlation profile based on a fixed number (the top 200, as reported in Supplementary Figure S1, most correlated pairs) as it is of illustrative purposes only.
3.4 Transcription factors expression patterns: the Mahoney atlas
The Mahoney is an atlas of transcription factors expressed in the murine brain (Gray et al., 2004). We determined how highly expressed these TF's were in the WBC. First, the homologues of these 2342 human TF's were mapped to their 2168 human homologues by reciprocal best first match (Moreno-Hagelsieb et al., 2008). Of those, 1212 are measured on the HG-U133A microarray. The expression ranks of these 1212 were obtained from the WBC transcriptome and are illustrated in Supplementary Figure S6. As shown, the distribution of ranks is uniform over the range of ranks measured by GPL96.
3.5 Gene ontology enrichment analysis
We used DAVID online tools (Dennis et al., 2003) to perform enrichment analysis of the set of 500 least-changing genes with respect to Gene Ontology (GO) categories. The DAVID functional annotation resulted in the 110 clusters listed in Supplementary Table S4. The top cluster (enrichment score of 20.35) is clearly reflecting the existence of a robust and well-correlated cluster of ribosome-related genes (see discussion about Relevance graphs above).
4 DISCUSSION
If the tools of functional genomics can be applied to peripheral blood cell samples to develop biomarkers for other organs, two questions are apparent: To what extent does expression in WBCs reflect expression in other organ systems? Why should WBCs reflect expression in other organ systems? This investigation focuses on the first question and we review some possibilities with regard to the second at the end of this discussion.
Before a further discussion of our findings, here we comment on the general validity of our approach. Two sets were defined and compared, the WBC and OO. The OO set is composed of a large number of different solid tissues (∼95% of all samples come from such tissue, see Fig. 2). Also, there was no discrimination between diseased or normal samples in OO or the WBC sets (i.e. ∼8% of the samples in OO are from malignant tissues). In a sense, we compare WBC expression to expression of an ‘averaged or generalized’ human tissue. A question might arise about the general validity of such a procedure. To address it, we have redone the bulk of our analyses for the case of an OO set defined as a superposition of five samples taken at random from the solid tissue distribution in Figure 2. The specific differences seen do not affect the quantitative conclusions reported below (results not shown).
A bird's eye view of the correspondence between the WBC and OO transcriptome is provided by the GO (Ashburner, 2000). We have performed a systematic mapping of the three main branches of GO categories in the cases of the most correlated and expressed genes, across the WBC and OO datasets. From the results listed in Figure 1 and Table 1, one is able to characterize the similarities and the difference between the WBC and OO transcriptomes. First, the top most common annotations across the datasets show a tight match, with the notable exception of the ribosome, cell adhesion and the immune response GO codes; the nucleic acid binding and spliceosome-related processes are added to this when one considers all GO categories that change the most between WBC and OO. Overall, 90% of the GO categories change only ∼2% in average across datasets. Next, focusing on the GO categories shared between WBC and OO in the expression and correlation spaces, the intersections between the distinct gene sets across GO branches are consistently larger than the distinct sets themselves, which is a highly improbable for only 4.5% of measured genes (P=0.0004998 by Monte Carlo sampling simulation in the χ2 test; Table 2). This result rules out the enrichment-by-chance hypothesis and suggests the high level of overlap in GO annotation categories across the WBC and OO. Supplementary Figure S2 also demonstrates the overall stability of GO categories change across WBC and OO, in both expression and correlation spaces, as exemplified by their change scores.
Two of the most changing (from WBC to OO) GO categories and their corresponding genes are the ‘sensory perception of sound’ (biological process, GO code BP:7605) and ‘sugar binding’ (molecular function, MF:5529) listed in the second column in Table 1. Some of the individual genes in the BP:7605 category are WRD1 (WD repeat domain 1), MYH14 (myosin, heavy polypeptide 14) and DIAPH1 [diaphanous homolog 1 (Drosophila)]. Sugar-binding most frequent genes are LGALS8 [lectin, galactoside-binding, soluble, 8 (galectin 8)], PKD1 [polycystic kidney disease 1 (autosomal dominant)] and BCAN (brevican). Additionally, the ‘nucleosome assembly’ BP:6334 GO category is quite overrepresented in going from OO to the WBC datasets (3 → 14). Some of the genes included in this GO category are H3F3A (H3 histone, family 3A), NAP1L4 (nucleosome assembly protein 1-like 4) and HIST1H1T (histone 1, H1t). The complete lists of the genes included in these two GO categories are listed in Supplementary Table S5.
To cross-validate the enrichment of sugar-binding and sensory perception of sound categories in WBC, we have performed enrichment analysis in DAVID using the ‘Tissue of Expression’ annotation option (see Section 2). For the sugar-binding individual genes, Clusters #3 (enrichment score of 0.59) and Cluster #9 (enrichment score of 0.38) clearly indicate the relative enrichment of this group of genes with respect to WBCs as a tissue of expression. A similar analysis of the hearing genes has yielded a much weaker WBC tissue association results bordering the method's sensitivity (results not shown).
We next took a more detailed look at some of the individual genes in the least-changing group as presented in the Supplementary Table S2. A GO terms enrichment analysis of those genes reveals that two of the most overrepresented types of categories are ‘oxygen transport’ (GO: 15671, BP), and ‘antigen presentation’-related categories (GO:19882, GO:19883 and GO: 19884, BP; Supplementary Material). These antigen presentation categories are of course the genes part of the human leukocyte antigen (HLA) system, the name of the major histocompatibility complex (MHC) in humans and are expected to be widely expressed across most tissues (Shiina, 2009). Another story is the ‘oxygen transport’ category as exemplified by a very stable levels of expression of HBB (hemoglobin β chain) and several other hemoglobin-related genes found in the list between WBC and OO sets. One remote but distinct possibility is the presence of contaminating red blood cells (RBCs) in the samples of both groups. For example, albumin, transferrin and some other major plasma proteins are quite abundant in muscle tissues, but the presence of hemoglobin-related genes in the WBC data is much more counterintuitive. To rule out the possibility of RBC contamination, we have reviewed the annotation for the GEO samples assigned to the WBC dataset (Supplementary Table S6). Although it is difficult to judge the exact sample collection protocol from just the description field in the GEO GSM format, it appears that most studies had followed the proper protocols. Therefore, the finding that HBB (hemoglogin β chain) and other hemoglobin-related genes are widely expressed in WBC is a true, but nevertheless a surprising one. The expression of HBB in avascular and non-hematopoietic tissues has been previously documented thus providing an example of another human tissue (and embryonic development stage) where hemoglobin units might possibly play novel development roles (Mansergh et al., 2008).
Next, we have reviewed the list of 567 human housekeeping genes (Eisenberg et al., 2003) against the two lists presented in Supplementary Table S3. It appears that the housekeeping genes are well represented on both least changing and most changing from WBC to OO lists. Some of the most stable are e.g. HLA-A/B/C (major histocompatibility complex, class I, A/B/C), TPT1 (Tumor protein, translationally controlled 1), VIM (Vimentin); some of the most changing are e.g. TUBB (tubulin, β), PSAP (prosaposin), JUNB (Jun-B oncogene), CREBBP(CREB binding protein).
Finally, we would like to briefly touch upon some of the biological and physiological underpinnings of why the WBC transcriptome might reflect the state of (including disease) another tissue or set of tissues. First, as blood cells contact and interact with all human tissues and transport and convey bioactive molecules (i.e. oxygen, metabolites, nutrient, cytokines and hormones, etc.), there is a distinct possibility that the WBC will themselves reflect the state(s) of those tissues. Conceivably, these subtle changes due to interaction of WBCs with diseased tissues might trigger specific changes in the gene expression of the WBCs paralleling the initial stimulus. A recent study comparing expression in nine human tissues with the WBC transcriptome has found that ∼80% of the WBC expression profile as being shared with any given tissue (Liew, 2006). After all, a ‘cell is a cell’ and the most fundamental characteristics of any cells are expected to be shared across all human tissues. Secondly, the WBC are the ‘etiological organ or tissue’ for some diseases, a good example being asthma which is not a disease of the lungs (Weiss et al., 2009). Thirdly, some diseases affect all tissues, e.g. Pompe's disease affects the function of the lysosomes, which are found in all types of tissue (Vellodi, 2005).
5 CONCLUSION
We study the similarity between the WBC and OO transcriptomes using public repository expression data (GEO). The OO sets has a wide representation of solid tissues (∼95%) and normal (∼92%) tissue samples.
We utilize GO mapping and ranking across correlation and expression level profiles selected by FDR thresholds to quantify similarity between comparison sets. We first identify that components of the ribosome, cell adhesion and immune response are among the most pronounced differences between the transcriptomes.
We next build predictive models of the rank in OO given the rank in WBC, across correlation (using linear least squares) and expression level spaces (using general least squares and PCA), after removing the ribosome-related genes. Two trans-tissue gene lists were also defined, the most- and least-changing in expression genes. We also consider the individual genes on both ends of the spectrum of WBC-OO change and gain further insight using ad hoc analyses, including looking at TF expression in WBC from Mahoney atlas as well as GO enrichment in tissue of expression.
We report on an overall very tight, quantitative match between the WBC human transcriptome and the transcriptome of a generalized set of solid tissues, across both correlation and expression level spaces. These findings are essentially independent on exact subset of tissues in the generalized OO set indicating a shared fundamental biology connection between the WBCs and OOs in the human body. Our results underscore the utility of the peripheral blood cells in applying the functional genomics tools for discovering biomarkers for other organs.
ACKNOWLEDGEMENTS
We are thankful to the PHS HPC Cluster staff for computational support.
Funding: Library of Medicine grant number (R01 LM 010125 to I.S.K.).
Conflict of Interest: none declared.
Footnotes
†Also at: New Atlantic Technology Group, LLC, Newton, MA 02468, USA.
REFERENCES
- Ashburner M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., et al. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Butte A.J., Kohane I.S. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pacific Symposium on Biocomputing. 1999;5:415–426. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- Butte A.J., et al. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc. Natl Acad. Sci. USA. 2000;97:121826. doi: 10.1073/pnas.220392197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G., et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann. Neurol. 2008;64:92–96. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Eisenberg E., Levanon E.Y. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Golub T.R., et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Gray P.A., et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Kathleen K. Comments on the analysis of unbalanced microarray data. Bioinformatics. 2009;25:2035–2041. doi: 10.1093/bioinformatics/btp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo W.P., et al. Analysis of matched mRNA measurements from different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- Liew C.C., et al. The peripheral blood transcriptome dynamically reflects system-wide biology: a potential diagnostic tool. J. Lab. Clin. Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Mansergh F.C., et al. Developmentally regulated expression of hemoglobin subunits in avascular tissues. Int. J. Dev. Biol. 2008;52:873–886. doi: 10.1387/ijdb.082597fm. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Nimgaonkar A., et al. Reproducibility of gene expression across generation of Affymetrix microarrays. BMC Bioinformatics. 2003;4:26. doi: 10.1186/1471-2105-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmos R.C., et al. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch. Gen. Psychiatry. 2008;65:395–407. doi: 10.1001/archpsyc.65.4.395. [DOI] [PubMed] [Google Scholar]
- Scherzer C.R., et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc. Natl Acad. Sci. USA. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweder T., Spjøtvoll E. Plots of p-values to evaluate many tests simultaneously. Biometrika. 1982;69:493–502. [Google Scholar]
- Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., et al. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Vellodi A. Lysosomal storage disorders. Br. J. Haematol. 2005;128:413–431. doi: 10.1111/j.1365-2141.2004.05293.x. [DOI] [PubMed] [Google Scholar]
- Washizuka S., et al. Expression of mitochondrial complex I subunit gene NDUFV2 in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. Neurosci. Res. 2009;63:199–204. doi: 10.1016/j.neures.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Weiss S.T., et al. Asthma genetics and genomics 2009. Curr. Opin. Genet. Dev. 2009;19:279–282. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]