Abstract
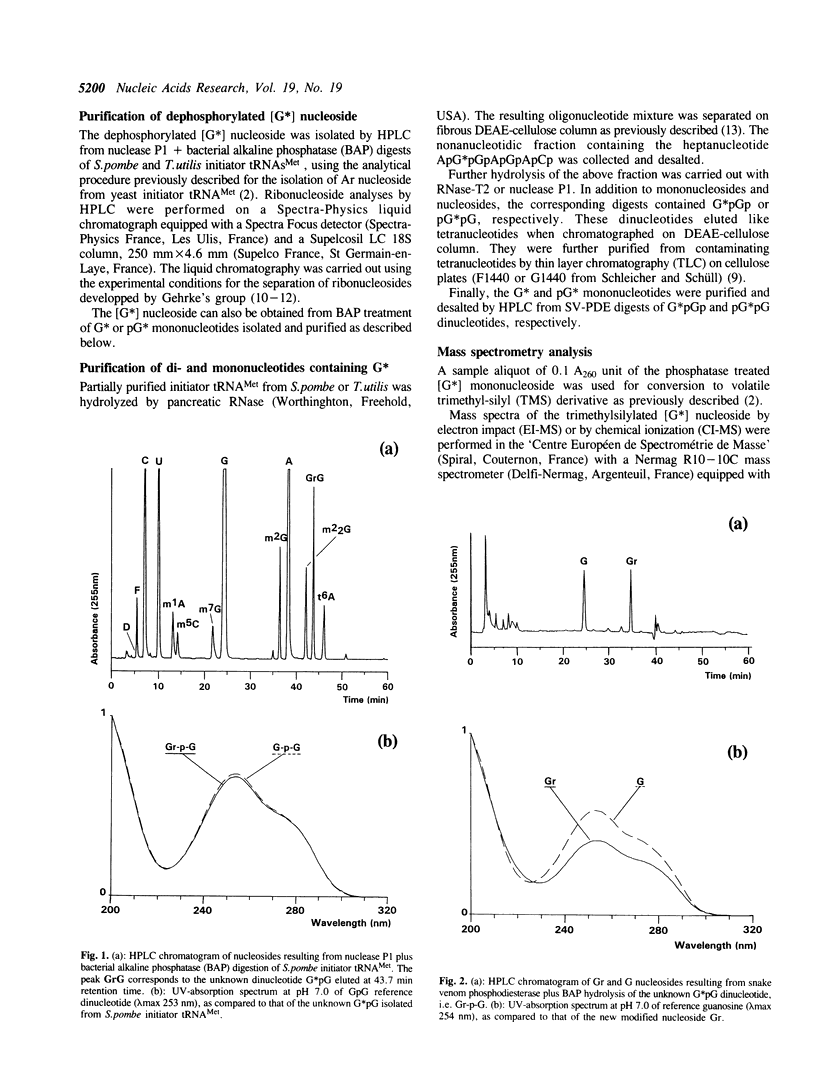
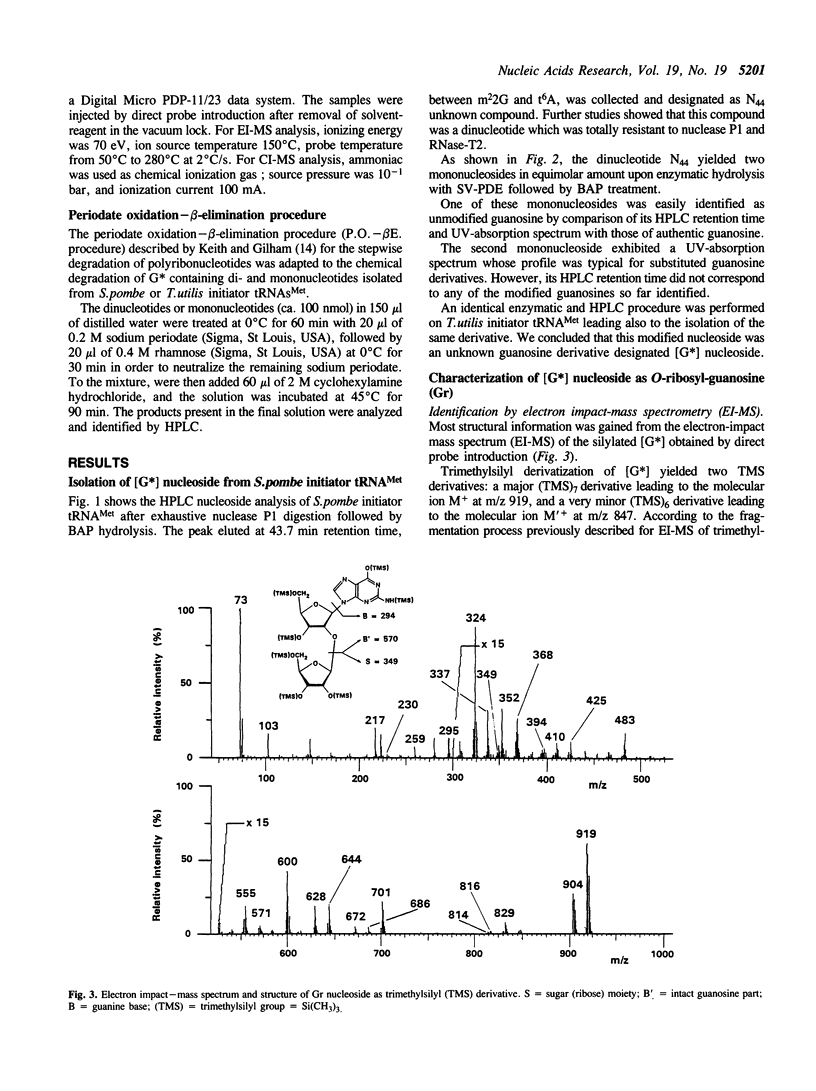
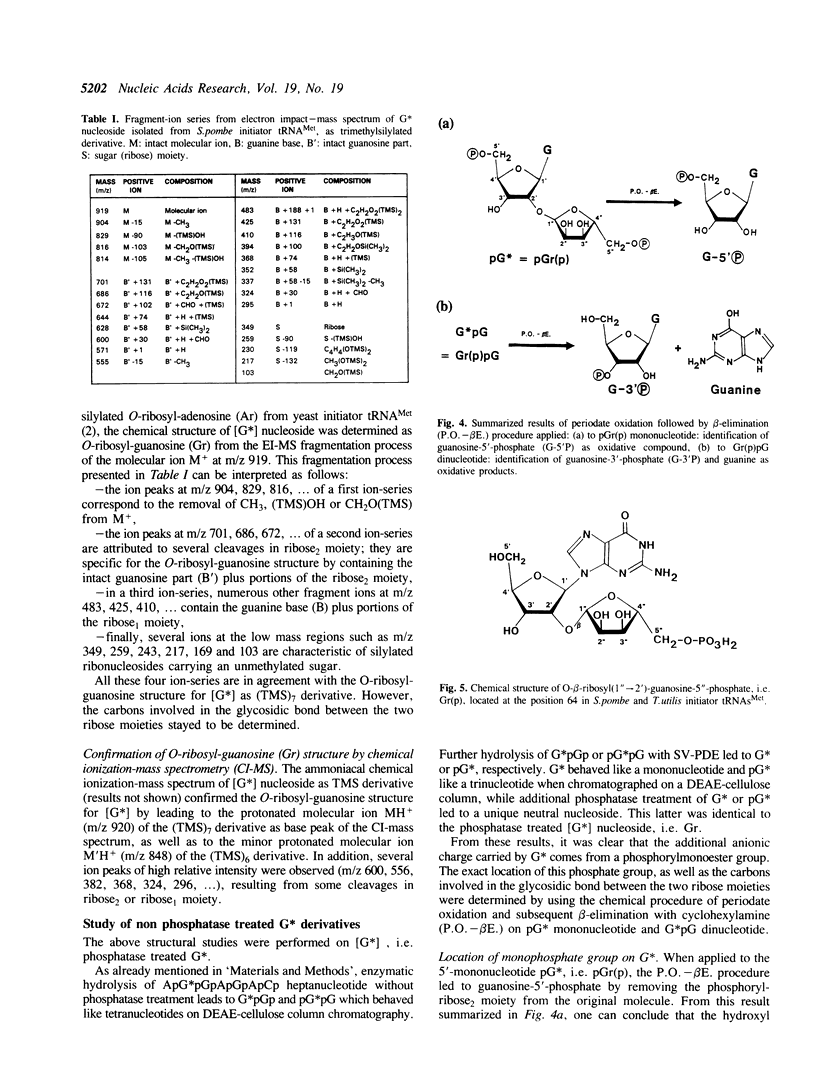
The unknown modified nucleotide G*, isolated from both Schizosaccharomyces pombe and Torulopsis utilis initiator tRNAs(Met), has been identified as an O-ribosyl-(1"----2')-guanosine-5"-phosphate, called Gr(p), by means of HPLC, UV-absorption, mass spectrometry and periodate oxidation procedures. By comparison with the previously published structure of Ar(p) isolated from Saccharomyces cerevisiae initiator tRNA(Met), the (1"----2')-glycosidic bond in Gr(p) has been postulated to have a beta-spatial conformation. The modified nucleotide Gr(p) is located at position 64 in the tRNA(Met) molecules, i.e. at the same position as Ar(p). Since we have also characterized Gr(p) in Candida albicans initiator tRNA(Met), the phosphoribosylation of purine 64 can be considered as a constant nucleotide modification in the cytoplasmic initiator tRNAs(Met) of all yeast species so far sequenced. Precise evidence for the presence of Gr(p) in initiator tRNAs(Met) of several plants is also reported.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amstutz H., Munz P., Heyer W. D., Leupoid U., Kohli J. Concerted evolution of tRNA genes: intergenic conversion among three unlinked serine tRNA genes in S. pombe. Cell. 1985 Apr;40(4):879–886. doi: 10.1016/0092-8674(85)90347-2. [DOI] [PubMed] [Google Scholar]
- Desgrès J., Keith G., Kuo K. C., Gehrke C. W. Presence of phosphorylated O-ribosyl-adenosine in T-psi-stem of yeast methionine initiator tRNA. Nucleic Acids Res. 1989 Feb 11;17(3):865–882. doi: 10.1093/nar/17.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin A., Gruhl H., Feldmann H. Mapping of yeast tRNAs by two-dimensional electrophoresis on polyacrylamide gels. FEBS Lett. 1975 Feb 1;50(2):185–189. doi: 10.1016/0014-5793(75)80485-6. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., Davis G. E., Suits R. D., Waalkes T. P., Borek E. Quantitative high-performance liquid chromatography of nucleosides in biological materials. J Chromatogr. 1978 Mar 21;150(2):455–476. doi: 10.1016/s0021-9673(00)88205-9. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., McCune R. A., Gerhardt K. O., Agris P. F. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J Chromatogr. 1982 Jul 9;230(2):297–308. [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr. 1989 Jun 2;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- Ghosh H. P., Ghosh K., Simsek M., RajBhandary U. L. Nucleotide sequence of wheat germ cytoplasmic initiator methionine transfer ribonucleic acid. Nucleic Acids Res. 1982 May 25;10(10):3241–3247. doi: 10.1093/nar/10.10.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam I., Blew D., Warrington R. C., von Tigerstrom M., Tener G. M. A general procedure for the isolation of specific transfer ribonucleic acids. Biochemistry. 1968 Oct;7(10):3459–3468. doi: 10.1021/bi00850a022. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Hurd R. E., Reid B. R., Rimerman R. A., Hatfield G. W. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1068–1071. doi: 10.1073/pnas.72.3.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Gilham P. T. Stepwise degradation of polyribonucleotides. Biochemistry. 1974 Aug 13;13(17):3601–3606. doi: 10.1021/bi00714a031. [DOI] [PubMed] [Google Scholar]
- Keith G., Glasser A. L., Desgrès J., Kuo K. C., Gehrke C. W. Identification and structural characterization of O-beta-ribosyl-(1"----2')-adenosine-5"-phosphate in yeast methionine initiator tRNA. Nucleic Acids Res. 1990 Oct 25;18(20):5989–5993. doi: 10.1093/nar/18.20.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Pixa G., Fix C., Dirheimer G. Primary structure of three tRNAs from brewer's yeast: tRNAPro2, tRNAHis1 and tRNAHis2. Biochimie. 1983 Nov-Dec;65(11-12):661–672. doi: 10.1016/s0300-9084(84)80030-9. [DOI] [PubMed] [Google Scholar]
- Keith G., Roy A., Ebel J. P., Dirheimer G. The primary structure of tRNA trp from brewer's yeast. I. Complete digestion with pancreatic ribonuclease and T 1 ribonuclease. Biochimie. 1972;54(11):1405–1415. doi: 10.1016/s0300-9084(72)80082-8. [DOI] [PubMed] [Google Scholar]
- Kiesewetter S., Ott G., Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNA(iMet) from yeast and wheat germ. Nucleic Acids Res. 1990 Aug 25;18(16):4677–4682. doi: 10.1093/nar/18.16.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro-Matsumura S., Takemura S. The primary structure of cytoplasmic initiator transfer ribonucleic acid from Torulopsis utilis. J Biochem. 1979 Aug;86(2):335–346. doi: 10.1093/oxfordjournals.jbchem.a132531. [DOI] [PubMed] [Google Scholar]



