Abstract
MicroRNA (miR) are emerging as important gene expression regulators often involved in a variety of pathogenesis such as cancers and autoimmunity. Signal Transducers and Activators of Transcription (STAT) proteins are the principle signaling proteins for many cytokines and growth factors, thereby play a critical role in regulating immune cell homeostasis, differentiation and cellular functions. In this review, we discuss recent advances in the field demonstrating active interactions between STATs and miRs, with our primary focus on the promotion and inhibition of immune cells and cancer. Additionally, we review the reciprocal regulations between STATs and miR, and discuss how we can use this knowledge in the context of diseases. For example, recent findings related to STAT1 and miR-155 support the presence of a positive feedback loop of miR-155 and STAT1 in response to inflammatory signals or infection. STAT3 is known to play critical roles in tumorigenesis and cancer-induced immunosuppression. There is a growing body of evidence demonstrating that STAT3 directly activates miR-21, one of miRs that promote cancer cell survival and proliferation. While some miRs directly regulate STATs, there are findings demonstrating indirect STAT regulation by miRs also mediate important biological mechanisms. Therefore, further research is warranted to elucidate significant contributions made by direct and indirect miR-STAT mechanisms. As we learn more about miR pathways, we gain the opportunity to manipulate them in cancer cells to slow down growth or increase their susceptibility anti-tumor immunity.
Keywords: miR-17-92, microRNA, STATs, cytokine, chemokine, T cells, B cells, Malignancy
1. Introduction
MicroRNA (miR) are emerging as important gene expression regulators often involved in a variety of pathogenesis such as cancers and autoimmunity. Primary miR transcripts (pri-miR) are transcribed by RNA polymerase II and RNA polymerase III [1], and contain the mature miR in a hairpin structure. The pri-miR hairpin is then cleaved by the class 2 RNase III enzyme, Drosha and the remaining precursor miR (pre-miR) is transported out of the nucleus by Exportin-V. The RNase III superfamily member Dicer then cleaves the hairpin loop structure leaving two single strands of RNA. After one strand is degraded, the other mature miR of about 22 nucleotides is incorporated into an RNA-induced silencing complex (RISC). Binding of the mature miR seed sequence to partial or exact complementary regions of 3′ untranslated region (UTR) on mRNA results in translational inhibition or mRNA degradation, respectively [2, 3]. miR are predicted to regulate up to 90% of human genes making them an important element of cellular processes [4].
Signal Transducers and Activators of Transcription (STAT) proteins are the principle signaling proteins of many cytokines and growth factors in mammals [5]. STATs play a critical role in regulating immune cell homeostasis, differentiation and cellular functions. There are 7 STAT proteins (STAT1, STAT2, STAT3, STAT4, STAT6 and the isoforms, STAT5a and STAT5b), each with established roles in immune cell functioning (Table 1). The key regulation of STATs is mediated by phosphorylation, typically by Janus kinases (JAKs). STAT1 and 2 respond to interferons (IFNs), thereby promote IFN-stimulated genes and anti-viral immunity [6, 7]. STAT3 responds to factors including IL-6, IL-10 and VEGF and is involved in T-helper cell (Th)17 and Treg cell development and tumorigenesis [8, 9]. STAT4 and STAT6 control Th1 and Th2 cell differentiation in response to IL-12 and IL-4/13, respectively [10, 11]. Finally, STAT5a and STAT5b are involved in NK cell activity, IL-2 induced T-cell proliferation and have been suggested to play a role in oncogenesis [12, 13]. Due to the large involvement of STATs in a variety of cell processes, it is critical that their activity is tightly regulated.
Table 1.
Immunological roles of STATs
| STAT | Functions | Refs |
|---|---|---|
| STAT1 | Type I and II interferon signaling important for host anti-anti tumor response | [15,19] |
| STAT2 | Type II interferon signaling | [15,17,18] |
| STAT3 | IL-6, IL-21 and VEGF signaling | [8,9] |
| STAT4 | Response to IL-12 important for type-1 immune skewing | [10,11] |
| STAT5a | Prolactin signaling and T cell/NK cell proliferation | [66,67,71,72] |
| STAT5b | Hormone signaling and T cell NK cell proliferation | [66,67,71,72] |
| STAT6 | Response to IL-4 or IL-13 important for type-2 immune response | [10,11] |
In this review, we discuss recent advances in the field demonstrating active interactions between STATs and miRs. While providing comprehensive overview of this subject, we address our primary focus to the promotion and inhibition of immune cells and cancer. Additionally, we will review the reciprocal regulations between STATs and miR, and discuss how we can use this knowledge in the context of diseases. Most of the published findings on miR/STAT regulation have occurred over the past few years, and to our knowledge, this is the first review specifically focused on this topic.
2. STATs and miR
2.1 STAT1 and STAT2-Controllers of the IFN network
Interferons (IFNs) are critical cytokines for host defense mechanisms against viral infection [14]. Type I IFNs (IFN-α and IFN-β) signal through the STAT1, STAT2, and STAT3 pathways, whereas the type II interferon, IFN-γ signals uniquely through STAT1 [15]. Consistently, STAT1-deficient mice fail to respond to either IFN-α or IFN-γ and are highly susceptible to viral and bacterial infection [16]. IFNs have been shown to both inhibit tumor growth and promote T cell, NK cell and macrophage activity [17, 18]. Specifically, STAT1 acts in host, non-tumor cells, as IFN-α increases the survival of STAT1 competent but not STAT1 deficient animals inoculated with the STAT1 competent B16F10 melanoma cell line [19]. Furthermore, mice challenged with STAT1 deficient AGS-1 tumor cells or AGS-1 cells reconstituted with STAT1 both exhibit similar survival following IFN-α treatment, indicating the importance of STAT1 in non-tumor host cells for the IFN-α response [19].
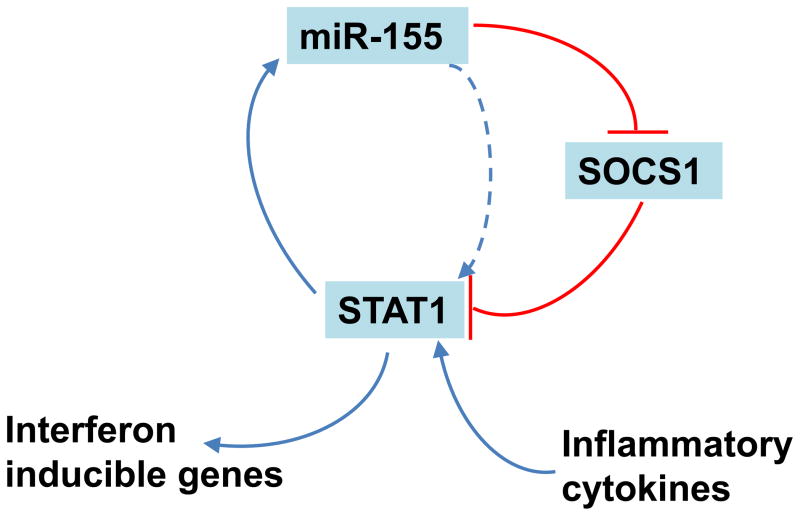
MiR-155 has been shown to regulate STAT1 expression [20, 21] (Figure 1). Ectopic expression of miR-155 in human hepatocellular carcinoma cells (HepG2, H7402) results in suppression of suppressor of cytokine signaling (SOCS)1 [20]. SOCS1 is a known negative regulator of the JAK-STAT pathway [20], and subsequent analyses of miR-155 overexpressing HepG2 cells have revealed increased phosphorylation of STAT1 and STAT3 [20]. This activation of STATs results in 30% and 40% increases of IFN-inducible genes, MxA and ISG15 expression levels, respectively [20]. Additionally, an enhanced anti-HBV effect is observed in HepG2 cells after transfection with a miR-155 mimic [20].
Figure 1.
The positive feedback cycle between miR155 and STAT1
In converse, STAT1 also regulates miR-155. Treatment of human retinal pigment epithelial (RPE) cells with inflammatory cytokines IFN-γ, TNF-α and IL-1 increases miR-155 expression levels for approximately 8 fold dependent on the JAK/STAT pathway as JAK inhibitor 1 a known blocker of the JAK/STAT pathway abrogated the cytokine-mediated miR-155 up-regulation [21]. STAT1-mediated regulation of miR-155 is supported by: 1) overexpression of STAT1 in RPE cells in response to inflammatory cytokines and 2) The presence of 2 putative STAT1 binding sites in the miR-155 promoter. Electrophoretic mobility shift assay (EMSA) further demonstrates binding ability of STAT1 to the miR-155 promoter [21]. These data support the presence of a positive feedback loop of miR-155 and STAT1 in response to inflammatory signals or infection (Figure 1). The similar mechanism of miR-155-STAT1 regulation in the two unique cell types supports a generalizable role of this pathway where STAT1 activation positively regulates its function by up-regulating miR-155 and down-regulating a STAT inhibitory factor SOCS1.
Expression of miRs-221, 222 (herein referred to as miR-221/222) and miR-145 are up-and down-regulated in tumors, respectively. However, both interact with STAT1 and STAT2 [22]. miRNA-145 is down-regulated in cancers such as prostate cancer, colon cancer, bladder cancer, ovarian cancer and B cell malignancies [22]. miR-221/222, on the other hand, are increased in tumors, such as glioblastoma and human thyroid papillary carcinomas [23, 24]. Differential expression of miR in tumor cells compared with healthy cells is known to promote tumor growth [25, 26].
Overexpression of miR-145 in the DLD-1 colon cancer cell line has lead to discovery of a number of miR binding sites in the 3′ UTR of genes that are down-regulated by miR-145 [22]. By luciferase assay, the Scr kinase family member, YES and STAT1 have been shown to be direct targets of miR-145 in the DLD-1 cells [22]. Further, miR-145 overexpression DLD-1 cells promotes tumor suppressor ability and reduces proliferation potential of cells [22].
Unlike miR-145 which is commonly down-regulated in tumors, miR-221/222 are often up-regulated in cancers [23, 27, 28]. Among the genes whose expression levels are altered by antisense-mediated knockdown of miRs-221/222 in U251 glioma cells, ones in the IFN-α signaling pathway are the most significantly modulated, and this observation is dependent on increased expression of STAT1 and STAT2 [23]. Consistently, overexpression of miR-221/222 in U251 glioma cells interferes with IFN signaling by down-regulating STAT1 and STAT2 [23]. These studies provide a solid foundation for tumor utilization of miRs to promote growth and resistance to immunosurveillance. Tumor use of miR for growth is still partially understood and much research is needed to understand how tumors regulate miR, how this regulation contributes promote tumor survival and how we can manipulate these miR to favor the host response against the tumor [29–33].
2.2 STAT3-diverse roles including promotion of cancer
STAT3 responds to a variety of signals including: growth factors, cytokines and oncogenes. As STAT3 is involved in diverse signaling pathways, it is reasonable that STAT3 deficiency in mice is embryonically lethal [34].
STAT3 is regulated by the miR-17 cluster family members. This family includes 7 miRs (miR-17-5p, miR-17-3p, miR-18a, miR-19a miR-20a, miR-19b and miR92-1) and 2 paralog clusters (miR-106b-25 and miR-106a-363) which include the pertinent miR-20b [35-37]. miR-17–92 cluster is also known as OncomiR-1 for its ability to promote tumorigenesis [36, 38–40]. Our research and others have demonstrated that miR-17 family members promote type-1 immune skewing and enhance T cell functioning by providing resistance to activation induced cell death (AICD) [41, 42]. miRs-17,-20a and-106b directly target STAT3 and MAPK14 [43]. Simultaneous overexpression of STAT3 and MAPK14 mimics the alteration of fibroblast growth factor (FGF)10-induced E-Cadherin distribution observed after miR-17,-20a, and-106b down-regulation, supporting the role of these miR-17 family members in FGF-signaling by specifically targeting STAT3 and MAPK14.
A separate study evaluating the role of bone morphogenetic protein receptor (BMPR)2 in death of vascular smooth muscle cells in pulmonary hypertension demonstrates a novel miR17-STAT3 pathway [44]. In human pulmonary arterial endothelial cells (HPAECs), HEPG2 and 293 cells, under IL-6 stimulation, activated STAT3 directly binds to the miR-17-92 cluster promoter and facilitates mIR-17-92 cluster expression. The Increase of miR-17-5p and miR-20a in HPAECs directly target 3′-UTR of HEPG2 and suppresses BMPR2 protein expression. This suggests a new pathway for down-regulation of BMPR2 mediated by IL-6-STAT3-miR-17-92 [44]. While the miR-17-STAT3 pathway has not been reported in the context of cancer, FGF signaling and BMPR2 have been known for their roles in cancer [45–47]. Further examination of these pathways in cancers has potential to lead to miR-modulation treatment strategies.
Vascular endothelial growth factor (VEGF) is one of the most well recognized angiogenic factors promoting tumor growth, a promising target for cancer therapies and thought to be regulated by hypoxia [48, 49]. Cascio and colleagues examined the mechanism of VEGF regulation in MCF-7 breast cancer cells cultured in the presence of CoCl2. In this hypoxia-mimicking condition, miR-20b was down-regulated and ectopic overexpression of pre-miR-20b in MCF-7 cells inhibited VEGF. As HIF-1α and STAT3 are verified targets of miR-20b, binding of HIF-1α and STAT3 to the VEGF promoter was examined by chromatin immuno-precipitation (ChIP) assay. Although HIF-1α but not STAT3 directly binds to the VEGF promoter, siRNA targeting of STAT3 revealed that STAT3 is necessary for the HIF-1α induced VEGF expression. These findings reveal critical mechanisms of miR-17 family member interaction with STAT3 which may be targeted in cancers [49].
Arguably the most extensively studied miR/STAT interaction is the miR-21/STAT3 pathway [50–53]. It is well established that STAT3 directly activates miR-21 [51, 53]. In myeloma cells, miR-21 transcription is controlled directly by an upstream enhancer containing 2 STAT3 binding sites [50]. Further, miR-21 induction by IL-6 is strictly STAT3-dependent, as knocking down of STAT3 prevents the IL-6 mediated up-regulation of miR-21 [51].
MCF-10A-ER-Src cells undergo malignant transformation after tamoxifen treatment making them ideal for studying factors that influence the epigenetic switch and cancer. During transformation of MCF-10A-ER-Src cells, miRs-21,-181b-1 and-210 are rapidly and dramatically up-regulated [50]. IL-6 activation of STAT3 directly promotes miR-21 and miR-181b-1 expression. These miR then directly target the tumor suppressor genes, phosphatase and tensin homolog (PTEN) and cylindomatosis (CYLD), thereby promote NF-κB activity which is required to maintain the transformed state [50]. Importantly, overexpression of either miR-21 or 181b-1 is sufficient to cause transformation, indicating that these miRs act as oncogenes. As miR have oncogenic properties, targeting miR in tumors has potential to inhibit tumor growth and development.
Unlike the IL-6-STAT3 pathway, the IFN-STAT3 pathway has yielded conflicting results in its regulation of miR. Human glioma cells treated with IFN-β demonstrate increased STAT3 which down-regulated miR-21. This was further supported by evaluation of miR-21 following overexpression or knocking down of STAT3 [52]. On the other hand, when PC3 prostate cancer cells are exposed to type I IFNs in vitro, those cells up-regulate miR-21, in a STAT3 dependent manner [53]. While there seems to be miR systems that are generalizable to other cell types, these conflicting o highlight the importance of possible cell type variation and the tight control in regulation of miR.
Sézary cell lymphoma, a cutaneous CD4+ T cell lymphoma displays constitutively activated STAT3 [54]. Treatment of Sézary cells with IL-21 highly activates STAT3 and subsequently increases miR-21 expression [54]. ChIP assay demonstrated that STAT3 can bind to the miR-21 promoter. Further, inhibition of miR-21 results in increased apoptosis of Sézary cells [54]. These data support miR inhibitors for Sézary cell lymphoma, but the findings may extend to miR-21-mediated reduced apoptosis in anti-tumor T cells.
In addition to miR-17-92 family members and miR-21, a few other miR have been demonstrated to be involved in the STAT3 signaling [54]. miR-125b, which has been shown to interfere with granulocyte-colony stimulating factor (G-CSF)-induced granulocytic differentiation, can directly target both Bcl-2 homologous antagonist/killer (BAK1) and STAT3. However, knockdown of STAT3 or BAK1 is not sufficient to interfere with G-CSF [55]. Jun family member D (JUND), however, is suggested as another miR-125b target and reduction of JUND partially mimicked the miR-125b interference with G-CSF [55]. Hepatitis B virus X protein (HBx) downregulates miR let-7a, which directly targets STAT3 in HepG2 cells [56]. The viral targeting of let-7a and the subsequent increase in STAT3 further supported proliferation of HBV infected cells [56]. In the context of cancer, let-7a might be used to target STAT3 in cancers and reduce tumor cell proliferation. Additionally, knocking down of let-7a may improve anti-tumor immune cell proliferation [57–59].
The following 2 studies provide insights into miR-STAT interactions despite the indirect nature of their interactions. In response to TGF-β1 and inflammatory cytokines, human bronchial cells (HBECs) undergo apoptosis, which is blocked by etopic miR-146a expression. HBECs overexpressing miR-146a also display high levels of phosphorylated STAT3 (Tyr 705) [60]. Finally, lentiviral vector-mediated STAT3-knockdown in rat cardiomyocytes lead to an increase in miR-199a-5p expression, suggesting a suppressive role of STAT3 on miR-199 transcription [61]. While there are multiple findings on the miR-21/STAT3 interactions, further studies are needed to gain more insights for future application of miR-engineering in each cell types for treatment of human diseases.
2.3 STAT4 and STAT6-Controlling type-1 and type-2 immune skewing
In CD4+ T helper cell development, the primary cytokines that influence type-1 and type-2 polarization are IL-12 and IL-4, respectively. For type-1 polarization, IL-12 activates STAT4 and T-bet, while IL-4 (and the closely related IL-13) stimulates STAT6 and GATA3 as the master regulators for type-2 polarization. Mice Lacking STAT4 or STAT6 are unable to mount effective type-1 or type-2 immunity, respectively [62, 63].Importantly, type I skewing is necessary for effective anti-tumor immunity [64, 65]. Additionally, STAT6 is necessary for B-cell antibody class switching to IgE and IgG1 [66, 67].
Despite the importance of STAT4 and STAT6 and the high likelihood of miR crosstalk, there are few reports demonstrating the role of miR in regulating those STAT proteins. We have previously reported that IL-4 suppresses the miR-17-92 cluster in CD4+ T cells, and that the suppression is dependent on STAT6 [41]. Additionally, while CD4+ T cells in tumor bearing mice exhibit a decrease of miR-17 cluster expression compared with those in non-tumor bearing mice, such decrease is not seen in STAT6 deficient mice [41]. As miR-17 cluster promotes T-cell survival and IFN-γ production [41], our data suggest a novel IL-4 induced mechanism in suppression of type-1 T-cell functions.
miR-155 is also important in immune regulation and the pro Th1/and type-1 polarization of macrophages (M1) [68–70]. miR-155 has recently been shown to directly inhibit IL-13Rα1, a component of the type II IL-4 receptor. Down regulation of IL-13Rα1 further diminishes the activation of STAT6 in human macrophages [70]. Although these findings demonstrate merely indirect STAT regulation by miRs, such indirect regulations through direct targets can mediate important biological mechanisms. Therefore, further research is warranted to elucidate significant contributions made by indirect miR-STAT mechanisms.
2.4 STAT5a and STAT5b-Homologous but unique roles
STAT5a and STA5b are paralogous STATS and despite having high homology, each STAT5 has its own unique function [71]. In mice, STAT5a is important for mammary gland development and prolactin signaling, which controls lactation [66, 67]. STAT5b controls growth hormone pathways and regulates expression of sexually dimorphic genes that controls the male/female size difference in mice. In addition to the unique functions of each STAT5 as discussed above, they also have redundant functions as mice deficient of STAT5a/5b have additional phenotypes. These include: a defect in T-cell proliferation and NK cell survival. STAT5 has further been suggested to regulate IL-3, IL-5, IL-7, thrombopoietin (TPO), erythropoietin (Epo), as well as granulocyte macrophage-colony stimulating factor (GM-CSF), and also implicated in cellular transformation [66, 67, 72].
STAT5a mediates the effects of T-cell-derived IL-3 and FGF in endothelial cells [73]. Following treatment with IL-3 and FGF, endothelial cells down-regulate miR-296, miR-126 and miR-221/222, thereby promote proliferation, migration and inflammation-mediated vascular remodeling of the endothelial cells [73]. Using gain-of-function studies, overexpression of pre-miR-222, but not miRs-296,-126 and-221 blocks the IL-3 + FGF promoted responses. Furthermore, direct targeting of STAT5a by miR-222 was demonstrated by luciferase reporter assays, making miR-222 a potential target for modulation of STAT5a [73].
Dicer deficient animals exhibit a 2 to 3 fold decrease of regulatory T-(Treg) cells compared to control animals, posing the question about the role of miR in Tregs [74]. miR-155 is up-regulated in Treg cells and its expression is controlled by Foxp3 [75]. Furthermore, elevated Foxp3 is required for maintaining Treg cell proliferative activity, and miR-155 deficient Tregs demonstrate increased SOCS1 and impaired STAT5 in response to IL-2 [75]. As IL-2 is also important in anti-tumor immune cells such as helper T cells, cytotoxic T cells and NK cells, this finding further supports a role for miR-155 in enhancing immune cells, and suggests a strategy of inhibiting miR-155 in immune-suppressor cells in cancer immunotherapy.
STAT5 can also promote B-cell lymphoma 2 (bcl-2), an anti-apoptotic factor often overexpressed in cancers [76]. Reconstitution of STAT5ab deficient fetal liver cells with STAT5a promotes BCL-2 expression. Furthermore, IL-3 and stem cell factor (SCF) promote STAT5 expression in mast cells, and STAT5 directly induces BCL-2 mRNA and inhibits miR-15b/16 cluster expression. Further the miR-15b/16 cluster directly suppresses BCL-2 [76]. Further understanding of mechanisms underlying these observations may lead to development of novel cancer therapy, as miR-15b/16 cluster overexpression in tumor cells can deplete BCL-2 and promote tumor cell apoptosis.
3. Conclusion
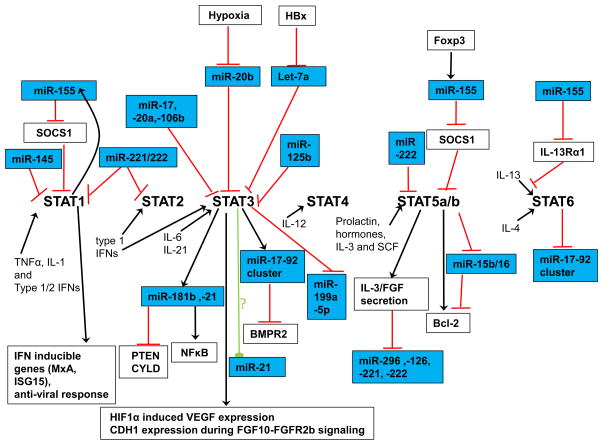
We reviewed the literature regarding interactions between miR and STAT pathways (Figure 2). The original observations discussed are not always in cancer settings, but provide us with potentially valuable insights for a variety of aspects of cancer biology including oncogenesis, angiogenesis and immunity. The field of miR-STAT interactions is still very new. As we learn more about miR pathways, we gain the opportunity to manipulate them in cancer cells to slow down growth or increase their susceptibility anti-tumor immunity. Cancer is equipped with a variety of immune escape mechanisms. Modulation of miR in immune cells has potentials to overcome the immuno-suppressive tumor environment. It is possible to carefully utilize “OncomiRs” (miR that promote cancer cell survival and/or proliferation) in immune cells to promote their survival. Although we are still far from completely understanding all the critical miR pathways in cells, the described pathways provide us with a foundation to continue to build upon. Many miRs we discussed have attractive biological properties that may be used to enhance our ability to fight against cancer. We recognize concerns with miR-engineering therapies, such as saturation of the cell machinery for miRNA processing, effects on endogenous miR expression and the truncation of 3′ UTR target sites in cancer cells. Nevertheless, we believe that miRs have tremendous potential for cancer immunotherapy.
Figure 2.
Summary of available information in the literature with respect to miR-STAT interactions.
Acknowledgments
Grant Support from: the National Institutes of Health [2R01NS055140, 2P01 NS40923, 1P01CA132714, the Cancer Center Support Grant P3CA047904] and Musella Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winter J, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 3.Ying SY, Lin SL. Intron-mediated RNA interference and microRNA biogenesis. Methods Mol Biol. 2009;487:387–413. doi: 10.1007/978-1-60327-547-7_19. [DOI] [PubMed] [Google Scholar]
- 4.Provost P. Interpretation and applicability of microRNA data to the context of Alzheimer’s and age-related diseases. Aging (Albany NY) 2010;2(3):166–9. doi: 10.18632/aging.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(Pt 8):1281–3. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 6.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2(9):675–87. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 7.Li X, et al. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271(10):5790–4. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- 8.He G, Karin M. NF-kappaB and STAT3-key players in liver inflammation and cancer. Cell Res. 2011;21(1):159–68. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egwuagu CE. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine. 2009;47(3):149–56. doi: 10.1016/j.cyto.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19(21):2577–2584. doi: 10.1038/sj.onc.1203485. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32(6):840–51. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriggl R, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10(2):249–59. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 13.Eckelhart E, et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117(5):1565–73. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 14.Stark GR, et al. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 15.Platanias LC. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 16.Meraz MA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84(3):431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 17.Belardelli F, et al. Interferon-alpha in tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002;13(2):119–34. doi: 10.1016/s1359-6101(01)00022-3. [DOI] [PubMed] [Google Scholar]
- 18.Fujita M, et al. Role of type 1 IFNs in antiglioma immunosurveillance--using mouse studies to guide examination of novel prognostic markers in humans. Clin Cancer Res. 2010;16(13):3409–19. doi: 10.1158/1078-0432.CCR-10-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesinski GB, et al. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J Clin Invest. 2003;112(2):170–80. doi: 10.1172/JCI16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su C, et al. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8(1):354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutty RK, et al. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010;402(2):390–5. doi: 10.1016/j.bbrc.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregersen LH, et al. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5(1):e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, et al. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol. 2010;36(6):1503–12. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- 24.Visone R, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14(3):791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 25.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 27.Ueda R, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A. 2009;106(26):10746–51. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation--opportunities for cancer immunotherapy. Int J Biochem Cell Biol. 2010;42(8):1256–61. doi: 10.1016/j.biocel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzo M, et al. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. J Cell Mol Med. 2010;14(11):2633–40. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krutovskikh VA, Herceg Z. Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. Bioessays. 2010;32(10):894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 31.Gotte M. MicroRNAs in breast cancer pathogenesis. Minerva Ginecol. 2010;62(6):559–71. [PubMed] [Google Scholar]
- 32.Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai) 2010;42(6):363–9. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 33.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21(3):470–9. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Akira S. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells. 1999;17(3):138–46. doi: 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- 35.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309(1):62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Hong L, et al. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70(21):8547–57. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond SM. RNAi, microRNAs, and human disease. Cancer Chemother Pharmacol. 2006;58(Suppl 1):s63–8. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki K, et al. miR-17-92 expression in differentiated T cells-implications for cancer immunotherapy. J Transl Med. 2010;8:17. doi: 10.1186/1479-5876-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lykken EA, Li QJ. microRNAs at the regulatory frontier: an investigation into how microRNAs impact the development and effector functions of CD4 T cells. Immunol Res. 2011;49(1–3):87–96. doi: 10.1007/s12026-010-8196-4. [DOI] [PubMed] [Google Scholar]
- 43.Carraro G, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333(2):238–50. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brock M, et al. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104(10):1184–91. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- 45.Schwertfeger KL. Fibroblast growth factors in development and cancer: insights from the mammary and prostate glands. Curr Drug Targets. 2009;10(7):632–44. doi: 10.2174/138945009788680419. [DOI] [PubMed] [Google Scholar]
- 46.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 47.Yoshikawa H, et al. Bone morphogenetic proteins in bone tumors. J Orthop Sci. 2004;9(3):334–40. doi: 10.1007/s00776-004-0764-9. [DOI] [PubMed] [Google Scholar]
- 48.Harmey JH, et al. Regulation of macrophage production of vascular endothelial growth factor (VEGF) by hypoxia and transforming growth factor beta-1. Ann Surg Oncol. 1998;5(3):271–8. doi: 10.1007/BF02303785. [DOI] [PubMed] [Google Scholar]
- 49.Cascio S, et al. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224(1):242–9. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 50.Iliopoulos D, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loffler D, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 52.Ohno M, et al. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res. 2009;7(12):2022–30. doi: 10.1158/1541-7786.MCR-09-0319. [DOI] [PubMed] [Google Scholar]
- 53.Yang CH, et al. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70(20):8108–16. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Fits L, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol. 2011;131(3):762–8. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 55.Surdziel E, et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011;117(16):4338–48. doi: 10.1182/blood-2010-06-289058. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53(1):57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 57.Dranoff G, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A. 1993;90(8):3539–43. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leong SP, et al. Recombinant human granulocyte macrophage-colony stimulating factor (rhGM-CSF) and autologous melanoma vaccine mediate tumor regression in patients with metastatic melanoma. J Immunother. 1999;22(2):166–74. doi: 10.1097/00002371-199903000-00008. [DOI] [PubMed] [Google Scholar]
- 59.Dong Q, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 2010;5(4):e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, et al. MicroRNA-146a modulates human bronchial epithelial cell survival in response to the cytokine-induced apoptosis. Biochem Biophys Res Commun. 2009;380(1):177–82. doi: 10.1016/j.bbrc.2009.01.066. [DOI] [PubMed] [Google Scholar]
- 61.Haghikia A, et al. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J. 2011;32(10):1287–97. doi: 10.1093/eurheartj/ehq369. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan MH, et al. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382(6587):174–7. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 63.Kaplan MH, et al. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4(3):313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 64.Okada H, et al. Immunotherapeutic approaches for glioma. Crit Rev Immunol. 2009;29(1):1–42. doi: 10.1615/critrevimmunol.v29.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sasaki K, et al. IL-4 suppresses very late antigen-4 expression which is required for therapeutic Th1 T-cell trafficking into tumors. J Immunother. 2009;32(8):793–802. doi: 10.1097/CJI.0b013e3181acec1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13(2):211–7. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 67.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25(10):496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 70.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286(3):1786–94. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crispi S, et al. Characterization of the human STAT5A and STAT5B promoters: evidence of a positive and negative mechanism of transcriptional regulation. FEBS Lett. 2004;562(1–3):27–34. doi: 10.1016/S0014-5793(04)00166-8. [DOI] [PubMed] [Google Scholar]
- 72.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 73.Dentelli P, et al. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562–8. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 74.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–7. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 75.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li G, et al. STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 2010;115(7):1416–24. doi: 10.1182/blood-2009-07-234963. [DOI] [PMC free article] [PubMed] [Google Scholar]