Abstract
Colorectal cancer (CRC) screening rates are currently suboptimal. Blood-based screening could improve rates of earlier detection for CRC and adenomatous colorectal polyps. In this study, we evaluated the feasibility of plasma-based detection of early CRC and adenomatous polyps using array-mediated analysis methylation profiling of 56 genes implicated in carcinogenesis. Methylation of 56 genes in patients with stage I and II CRC (N=30) and those with adenomatous polyps (N=30) were compared to individuals who underwent colonoscopy and were found to have neither adenomatous changes nor CRC. Composite biomarkers were developed for adenomatous polyps and CRC, and their sensitivity and specificity was estimated using five-fold cross validation. Six promoters (CYCD2, HIC1, PAX 5, RASSF1A, RB1, and SRBC) were selected for the biomarker, which differentiated CRC patients and controls with 84% sensitivity and 68% specificity. Three promoters (HIC1, MDG1, and RASSF1A) were selected for the biomarker, which differentiated patients with adenomatous polyps and controls with sensitivity of 55% and specificity of 65%. Methylation profiling of plasma DNA can detect early CRC with significant accuracy and shows promise as a methodology to develop biomarkers for CRC screening.
Keywords: cell-free, plasma, methylation, colorectal, polyp, cancer
Introduction
In 2010, it is estimated that over 140,000 Americans will be diagnosed and over 48,000 will die from Colorectal Cancer (CRC). 1 Five year survival is highly dependent on tumor stage at time of detection. Among the unscreened, average risk US population the prevalence of CRC is 0.5%-1%.2 Diagnostic colonoscopy is the most commonly used modality for CRC screening though only a minority or Americans undergo it.3 Colonoscopy quality is variable and depends on a number of factors including difficulty in detection of flat or depressed lesions, right-sided lesions, suboptimal bowel preparations and “interval” cancers with aggressive biology.4–6
Non-invasive screening methods for colorectal neoplasia should have high sensitivity for curable stage CRC as well as advanced precancerous lesions, affordability and specificity.7 Currently commercially available stool tests of hemoccult-based testing (FOBT) and fecal immunochemical testing (FIT) do not detect the majority of advanced adenomas.8 Lesion size, villous histology, and multiplicity of polyps have been correlated with increased risk of future advanced colorectal neoplasia.9
Aberrant methylation of CpG islands at the promoter region of genes leads to transcriptional silencing of tumor suppressor genes.10 Hypermethylation of CpG Islands in tumor suppressor genes has been reported for several cancers including CRC.11–12 Methylation of tumor suppressor genes silenced by hypermethylation and detectable in the plasma or serum of patients with CRC has been shown to hold promise as a potential methodology for the detection of CRC.13–14 Plasma DNA methylation profiles that assess methylation status at multiple sites have been used in biomarker development for detection in cell-free plasma DNA to reflect the primary disease.15–16
In this study, we examined whether patients with early CRC, (stage I and II) and patients with adenomatous polyps could be differentiated from controls using DNA methylation profiling of cell-free circulating plasma. DNA Methylation was evaluated in 56 genes using MethDet56 platform in each sample. The most informative genes selected by a statistical algorithm as genes with the greatest difference in methylation between controls and patients with CRC and with adenomatous polyps respectively were included in the composite biomarker.16–17
Materials and Methods
Patient Enrollment
The study was approved by the institutional review boards of the Rush University Medical Center, Chicago, Illinois and University of Insubria, Italy. Controls were individuals evaluated and deemed medically appropriate for standard colorectal cancer screening by colonoscopy and had colonoscopy negative for either adenomatous changes or CRC. Patients could have cardiac, pulmonary, renal or liver disease but no chronic medical condition was represented in more than 10% of patients. Patients who had a history of a solid organ tumor were excluded. No cancer patients had neoadjuvant chemoradiotherapy. Colonoscopies in all patients were considered to have adequate preparation and were complete to the cecum. Colorectal cancers included patients with stage I or II disease only. All CRC patients had surgical resections and no evidence of nodal disease. Sample Collection and DNA Isolation: Whole blood was obtained by venipuncture, collected in vacutainer tubes containing EDTA and stored at 4°C for a maximum of 2 hour. Blood was obtained within one week prior to the surgery in patients with CRC. For patients with polyps or controls blood was obtained on the day of colonoscopy. Tubes were centrifuged twice (2600g) for 10 minutes at 4°C to separate plasma from cellular elements. The plasma was then aliquoted and stored at −80°C. DNA was isolated using DNAzol BD and proteinase K as previously described and quantified by fluorimetry.15,18
Microarray Mediated Methylation Assay
The assay was done as previously described.17,19,20, 22 Briefly, DNA samples were split into two equal aliquots, and one was digested with Hin6I (Fermentas, Glen Burnie, MD, USA) while the other was mock digested. Both samples were amplified via a multiplexed, nested polymerase chain reaction (PCR). 5-aminoallyl dUTP (Biotium Inc., Hayward, CA, USA) was added for the second PCR; products of the Hin6I digested DNA were labeled with Cy3, while products of the mock digested DNA were labeled with Cy5. Both labeled products were mixed and hybridized to custom printed DNA microarrays (Microarrays Inc., Huntsville, AL, USA). The slides were then washed and scanned using a Genepix 4000B Microarray Scanner (Molecular Devices, Union City, CA, USA) and the data were analyzed using the Genepix Pro 6.0 software.
Statistical Analysis
Methylation patterns in cfpDNA were determined using the MethDet 56 test in three different cohorts: CRC patients, colorectal adenoma patients and those without any adenomatous changes or CRC on colonoscopy. There was 85% power to detect an effect size of 0.8 in methylation ratio (difference divided by standard deviation) between any two groups.
Data were processed using a fixed cutoff approach as previously described 15,17–19. First, the same PCR product from an undigested sample was divided into two parts, which were labeled with either Cy5 or Cy3 and used for hybridization with the microarray. This PCR product from an undigested DNA presented a model of a completely methylated fragment where the differences in fluorescent signal were dependent solely on the fluorescent dyes. The Cy5/Cy3 ratio (r=4) for this DNA fragment was used as a threshold for binarization (methylated vs. unmethylated) of results. We next applied Fisher’s exact test analysis with p<0.05 to select the most differentially methylated genes. Finally, the most informative combination of genes was selected by naïve Bayes algorithm with 5 fold cross-validation to obtain sensitivity and specificity.
Results
Clinical Specimens
The age of subjects and clinical characteristics are presented in Table 1. The control group free of adenomatous polyps, the adenomatous polyp group and the CRC group each contained 30 patients. Patients in all three groups were similar by age and gender. Eighteen of the patients with polyps had either villous histology or size ≥ 1cm. The presence of right colon neoplasia was 7/30 (23%) in the polyp group and 8/30 (27%) in the CRC group. Among CRC patients 11/30 (37%) had stage I disease and 19/30 (63%) – stage II disease. Table 2 depicts the genes used in the MethDet 56 microarray.
Table 1.
Age (range), extent of dysplasia and tumor location in colorectal clinical specimens in enrolled patients.
| Mean Age | Dysplasia | Location of Neoplasia | |
|---|---|---|---|
| Controls (N=30) | 61.2 (40–80) | None | NA |
| Adenomatous Polyps (N=30) | 61.6 (40–84) | Advanced Adenomas 17/30 (57%) |
Right Sided 7/30 (23%) |
| Colon Cancers (N=30) | 68.3 (49–85) | Adenocarcinoma StageI =11 Stage II=19 |
Right Colon 8/30 (27%) |
Table 2.
The genes used in the MethDet 56 Microarray. Negative controls include coding sequences of three genes (ACTB, GADPH, TUBA3) and heterologous DNA from Arabidopsis thaliana
| ABCB1 | ACTB | APAF1 | BRCA1 | CALCA | CASP8 | CCND2 | CDH1 |
| CDKN1A | CDKN1B | CDKN1C | CDKN2A | CDKN2B | DAPK1 | DNAJC15 | EDNRB |
| EP300 | ESR1prA | ESR1prB | FABP3 | FAS | FHIT | GPC3 | GSTP |
| HIC1 | HLTF | ICAM1 | MCTS1 | MGMT | MLH1 | MSH2 | MUC2 |
| MYOD1 | NR3C1 | PAX5 | PGK1 | PGRdist | PGRprox | PLAU | PRDM2 |
| PRKCDBP | PYCARD | RARB | RASSF1A | RB1 | RPL15 | S100A2 | SCGB3A1 |
| SLC19A1 | SOCS1 | SYK | TES | THBS1 | TNSF11 | TP73 | VHL |
Methylation data by the fixed cutoff approach to differentiate Patients with CRC from Healthy Controls
Using the fixed cutoff approach, differentiation between patients with CRC versus controls (Fig. 1A) was more accurate than differentiation of patients with adenomatous polyps versus controls (Fig. 1B). Using the Fisher’s Exact test between paired samples, 7 genes were informative with a p<0.05. The informative genes were (CYCD2, HIC, MDR1, PAX5, RASSF1A, RB1, SRBC). Next, naïve Bayes algorithm with 5 fold cross-validation was used to obtain the most informative panel of genes. This yielded a 6 gene panel (CYCD2, HIC1, PAX5, RASSF1A, RB1, SRBC). The sensitivity of this panel was 83.7% and the specificity was 67.9% to differentiate CRC patients from those without polyps (figure 1C). In all comparisons, methylation status of informative genes was higher in CRC patients than in controls.
Figure 1.
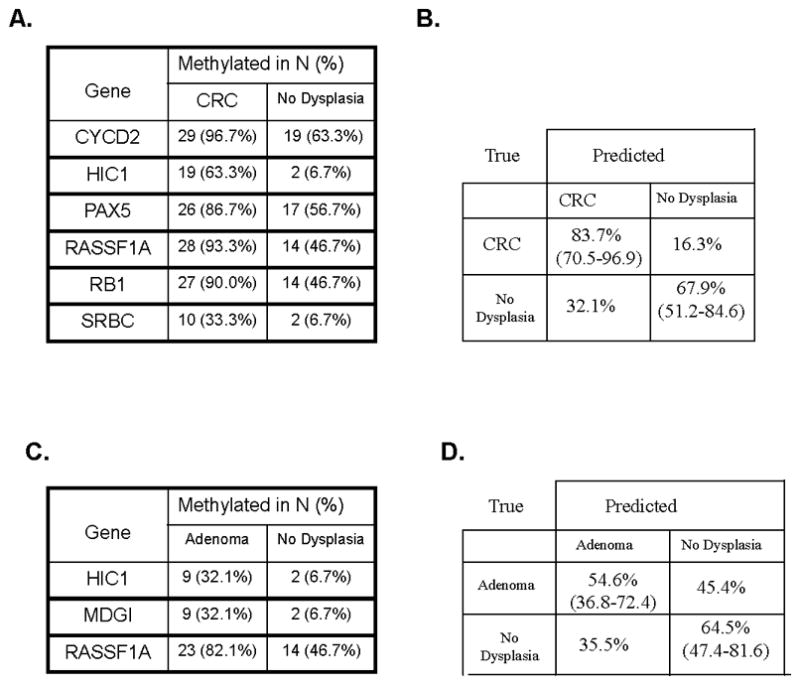
Analysis of results using the fixed cutoff approach following 5 fold cross validation. For each comparison the percentage of the sample that was methylated by binary fixed cut off approach is listed A) Individual informative genes that comprise the panel to differentiate patients with CRC from controls. (B) The informative genes panel yielded a sensitivity of 83.7% and specificity of 67.9% to differentiate CRC from controls C) Individual informative genes that comprise the panel to differentiate patients with adenoma from controls. D) The informative genes as a panel yielded sensitivity of 54.6% and specificity of 64.5% to differentiate patients with adenomatous polyps from controls.
Methylation data by the fixed cutoff approach to differentiate patients with adenomatous Polyps from controls
Differentiation by the fixed cut off approach of patients with adenomatous polyps versus those without any polyps showed three informative genes by Fisher’s Exact test with a p<0.05 (HIC1, MDGI, RASSF1A). Naïve Bayesian analysis included all three genes (HIC1, MDG1, RASSF1A) which yielded a 54.6% sensitivity with a 64.5% specificity (Fig 1B, 1D).
Table 3 lists the informative genes of the composite biomarker for CRC differentiation from controls (RASSF1A, HIC1, CYCD2, PAX5, RB1, SRBC) by the fixed cut-off approach. Using this binary assessment of methylation, all of the above six informative genes likewise had a higher frequency of methylation status in patients with adenomatous polyps than in healthy controls though all 6 genes were methylated in CRC patients. Two genes (HIC1; RASSF1A) were informative for differentiation of both adenomatous polyps and likewise CRC patients from controls though these cohorts were analyzed separately. This observation suggests that patients with adenomatous polyps and CRC have common genes that tend to be methylated at greater frequency than seen with healthy controls. The sensitivity and specificities of the six individual genes to differentiate controls without adenomatous lesions from CRC patients are also listed in table 3.
Table 3.
Informative (Methylated) Genes Selected for Composite Biomarker for Colorectal Cancer and for Colonic Adenoma Detection by the Fixed Cut-Off Approach. The number (and percentage) of samples with methylated gene is shown. Sensitivity (Se) and Specificity (Sp) for the informative genes that differentiated colorectal cancer from controls are listed.
| Gene | Informative for CRC | Informative For Adenoma | Controls N=30 | Adenoma N=30 | Colorectal Cancer N=30 |
|---|---|---|---|---|---|
| RASSF1A | + | + | 14 (47%) | 23 (82%) | 28 (Se=93%, Sp=53%) |
| HIC1 | + | + | 2 (7%) | 9 (30%) | 19 (Se=63%, Sp=93%) |
| CYCD2 | + | − | 19 (63%) | 23 (82%) | 29 (Se=97%, Sp=37%) |
| PAX5 | + | − | 17 (57%) | 21 (75%) | 26 (Se=87%, Sp=43%) |
| RB1 | + | − | 14 (47%) | 19 (68%) | 27 (Se=90%, Sp=53%) |
| SRBC | + | − | 2 (7%) | 3 (11%) | 10 (Se=33%, Sp=93%) |
Discussion
Mortality from CRC is reduced by early detection and removal of adenomatous polyps 20–21,23–24. Currently commercially available stool tests of FOBT and FIT have shown some reduction in mortality in CRC. (20) Rates of screening for CRC are significantly lower than for other malignancies including breast (mammography), prostate (PSA) and cervix (pap smear) and most Americans do not undergo age appropriate screening for CRC by any methodology.3,22 Failure to screen for CRC stems from multiple reasons including time, cost, privacy concerns as well as perceptions of procedural and bowel preparation related-pain and aversion to stool based non-invasive modalities. Sensitivity of the above mentioned noninvasive modalities is reduced in early stage cancers and adenomas.23
Promoter methylation of varied genes can differentiate patients with adenomatous polyps and CRC from controls with some accuracy in tissue and stool.24–26 Plasma markers have been investigated for early detection of CRC and numerous individual genes including SEPT9 and ALX4 have been identified as potential biomarkers with limited sensitivity for detection of precancerous polyps.27–28,32–34 Other individual promoters including TMEFF2, NGFR and MGMT, CDKN2A, RARB2, RASSF1A, and APC have been suggested as potential biomarkers of CRC in plasma, but their accuracy remains limited.29–30
As methylation status of single genes have limited accuracy we sought to develop a methodology for the analysis of multiple different methylation sites simultaneously. This technology should allow identification of the most informative methylation sites or methylation profiling that can distinguish patients with colorectal malignant and premalignant lesions from controls to show promise as a potential screening modality. In this study we used the proof-of-principle platform for methylation analysis of 56 genes (MethDet-56) in order to compare methylation patterns in plasma from controls and patients with CRC and adenomatous polyps.
MethDet is a technique developed to produce a multigene methylation signature in each sample, so that selection of the most informative genes and their combinations becomes possible.18 A set of six genes (CYCD2, HIC1, PAX5, RB1, SRBC; Fig. 3C) selected by a fixed cut-off approach provides a sensitivity of 84% and a specificity of 68% to discriminate CRC from healthy controls. Patients with adenomatous polyps could also be differentiated from healthy controls but to a lesser extent. This suggests that different and likely larger sets of genes are required to improve differential detection of patients with adenomatous polyp from controls.
HIC1 and RASSF1A, the two sites with preferential methylation in both the CRC and polyp plasma cohorts in this series have been described as informative genes for detection of CRC and polyps in tissue.25,31,32 Increase in frequency of methylated RB1 in CRC has been described but is reported in plasma for the first time.33 Preferential methylation of SRBC and PAX 5 in plasma of CRC patients is reported for the first time.34,35 RASSF1A and PAX5 have been shown to be preferentially methylated in other solid organ malignancies. While patients with known malignancies in other organs were excluded, it is possible that occult malignancies may have impacted on our results.36, 37
In this proof-of-principle study we demonstrate that patients with adenomatous polyps and CRC share common preferentially methylated sites (HIC1, RASSF1A) when compared separately to controls without adenomatous change. While this proof-of-principle study is not intended as a clinical-grade biomarker discovery, we have nonetheless identified genes – RASSF1A and HIC1 – as preferentially methylated and informative in plasma in two separate cohorts (CRC and adenomatous polyps) compared to healthy controls. These common features suggest that differences in specific methylation sites may be present from lower grade dysplasia to malignant lesions. It is intriguing that the accuracy of detection of early CRC is higher than that of adenomatous polyps. This suggests that clinically higher grades of dysplasia might be detected better using this approach.
We intentionally sought after early cancers (stages I, II) that would be of most benefit to detect by a screening modality. There are, however, several limitations to this proof-of-principle study, including homogeneity of the test population (only European Caucasian) and a lack of several genes that may have added to the accuracy of detection, e.g. NDRG4, SEPT9, RUNX3 and ALX4. 28,29,38 The methylation patterns developed in this study should not be considered actionable clinical-grade biomarkers. Nonetheless, they suggest that accurate biomarkers could be developed once larger, preferably genome-wide analysis of blood-based methylation profiles is completed.
Acknowledgments
This work was supported in part by NIH R21RR024420 to VVL. Assistance of Ms. Sushma Shrestha and Mr. Brian Danzer, Michael Greenspan is gratefully acknowledged.
Abbreviations
- CRC
colorectal cancer
- FOBT
fecal occult blood testing
- FIT
fecal immunochemical testing
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010 Jul 7; doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Mariotto AB, Rowland JH, Ries LA, Scoppa S, Feuer EJ. Multiple cancer prevalence: A growing challenge in long-term survivorship. Cancer Epidemiol Biomarkers Prev. 2007 Mar;16(3):566–71. doi: 10.1158/1055-9965.EPI-06-0782. [DOI] [PubMed] [Google Scholar]
- 3.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the united states. Cancer Epidemiol Biomarkers Prev. 2006 Feb;15(2):389–94. doi: 10.1158/1055-9965.EPI-05-0678. [DOI] [PubMed] [Google Scholar]
- 4.Citarda F, Tomaselli G, Capocaccia R, Barcherini S, Crespi M Italian Multicentre Study Group. Efficacy in standard clinical practice of colonoscopic polypectomy in reducing colorectal cancer incidence. Gut. 2001 Jun;48(6):812–5. doi: 10.1136/gut.48.6.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rembacken BJ, Fujii T, Cairns A, Dixon MF, Yoshida S, Chalmers DM, Axon AT. Flat and depressed colonic neoplasms: A prospective study of 1000 colonoscopies in the UK. Lancet. 2000 Apr 8;355(9211):1211–4. doi: 10.1016/s0140-6736(00)02086-9. [DOI] [PubMed] [Google Scholar]
- 6.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009 Jan 6;150(1):1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 7.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010 Jun;138(6):2127–39. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004 Dec 23;351(26):2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 9.Martinez ME. Primary prevention of colorectal cancer: Lifestyle, nutrition, exercise. Recent Results Cancer Res. 2005;166:177–211. doi: 10.1007/3-540-26980-0_13. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Epigenetic lesions causing genetic lesions in human cancer: Promoter hypermethylation of DNA repair genes. Eur J Cancer. 2000 Dec;36(18):2294–300. doi: 10.1016/s0959-8049(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Harrington JJ, Shire AM, Rego RL, Wang L, Campbell ME, Oberg AL, Ahlquist DA. Highly methylated genes in colorectal neoplasia: Implications for screening. Cancer Epidemiol Biomarkers Prev. 2007 Dec;16(12):2686–96. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 12.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010 Mar;29(1):181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H. High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett. 2003 May 8;194(1):99–105. doi: 10.1016/s0304-3835(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 14.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH. Aberrant methylation of APC, MGMT, RASSF2A, and wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009 Oct 1;15(19):6185–91. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 15.Melnikov A, Scholtens D, Godwin A, Levenson V. Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn. 2009 Jan;11(1):60–5. doi: 10.2353/jmoldx.2009.080072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melnikov AA, Scholtens D, Talamonti MS, Bentrem DJ, Levenson VV. Methylation profile of circulating plasma DNA in patients with pancreatic cancer. J Surg Oncol. 2009 Feb 1;99(2):119–22. doi: 10.1002/jso.21208. [DOI] [PubMed] [Google Scholar]
- 17.Liggett TE, Melnikov A, Yi Q, Replogle C, Hu W, Rotmensch J, Kamat A, Sood AK, Levenson V. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol Oncol. 2010 Nov 5; doi: 10.1016/j.ygyno.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melnikov AA, Scholtens DM, Wiley EL, Khan SA, Levenson VV. Array-based multiplex analysis of DNA methylation in breast cancer tissues. J Mol Diagn. 2008 Jan;10(1):93–101. doi: 10.2353/jmoldx.2008.070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liggett T, Melnikov A, Yi QL, Replogle C, Brand R, Kaul K, Talamonti M, Abrams RA, Levenson V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer. 2010 Apr 1;116(7):1674–80. doi: 10.1002/cncr.24893. [DOI] [PubMed] [Google Scholar]
- 20.Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000 Nov 30;343(22):1603–7. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen OD, Kronborg O, Fenger C. A randomised study of screening for colorectal cancer using faecal occult blood testing: Results after 13 years and seven biennial screening rounds. Gut. 2002 Jan;50(1):29–32. doi: 10.1136/gut.50.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith RA, von Eschenbach AC, Wender R, Levin B, Byers T, Rothenberger D, Brooks D, Creasman W, Cohen C, Runowicz C, Saslow D, Cokkinides V, et al. American cancer society guidelines for the early detection of cancer: Update of early detection guidelines for prostate, colorectal, and endometrial cancers. also: Update 2001--testing for early lung cancer detection. CA Cancer J Clin. 2001 Jan-Feb;51(1):38, 75. doi: 10.3322/canjclin.51.1.38. quiz 77–80. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005 Aug;129(2):422–8. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 24.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001 Sep;159(3):1129–35. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pehlivan S, Artac M, Sever T, Bozcuk H, Kilincarslan C, Pehlivan M. Gene methylation of SFRP2, P16, DAPK1, HIC1, and MGMT and KRAS mutations in sporadic colorectal cancer. Cancer Genet Cytogenet. 2010 Sep;201(2):128–32. doi: 10.1016/j.cancergencyto.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Azuara D, Rodriguez-Moranta F, de Oca J, Soriano-Izquierdo A, Mora J, Guardiola J, Biondo S, Blanco I, Peinado MA, Moreno V, Esteller M, Capella G. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer. 2010 Jul;9(3):168–76. doi: 10.3816/CCC.2010.n.023. [DOI] [PubMed] [Google Scholar]
- 27.Grutzmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F, Roblick UJ, Bruch HP, Koch R, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3(11):e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, Grutzmann R, Pilarsky C, Habermann JK, Fleshner PR, Oubre BM, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009 Jul;55(7):1337–46. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 29.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, Molnar B, Grutzmann R, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008 Feb;54(2):414–23. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 30.Taback B, Saha S, Hoon DS. Comparative analysis of mesenteric and peripheral blood circulating tumor DNA in colorectal cancer patients. Ann N Y Acad Sci. 2006 Sep;1075:197–203. doi: 10.1196/annals.1368.027. [DOI] [PubMed] [Google Scholar]
- 31.Wagner KJ, Cooper WN, Grundy RG, Caldwell G, Jones C, Wadey RB, Morton D, Schofield PN, Reik W, Latif F, Maher ER. Frequent RASSF1A tumour suppressor gene promoter methylation in wilms’ tumour and colorectal cancer. Oncogene. 2002 Oct 17;21(47):7277–82. doi: 10.1038/sj.onc.1205922. [DOI] [PubMed] [Google Scholar]
- 32.Sakamoto N, Terai T, Ajioka Y, Abe S, Kobayasi O, Hirai S, Hino O, Watanabe H, Sato N, Shimoda T, Fujii H. Frequent hypermethylation of RASSF1A in early flat-type colorectal tumors. Oncogene. 2004 Nov 25;23(55):8900–7. doi: 10.1038/sj.onc.1207993. [DOI] [PubMed] [Google Scholar]
- 33.Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol. 2005 Feb;18(2):170–8. doi: 10.1038/modpathol.3800261. [DOI] [PubMed] [Google Scholar]
- 34.Palmisano WA, Crume KP, Grimes MJ, Winters SA, Toyota M, Esteller M, Joste N, Baylin SB, Belinsky SA. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res. 2003 Aug 1;63(15):4620–5. [PubMed] [Google Scholar]
- 35.Xu XL, Wu LC, Du F, Davis A, Peyton M, Tomizawa Y, Maitra A, Tomlinson G, Gazdar AF, Weissman BE, Bowcock AM, Baer R, et al. Inactivation of human SRBC, located within the 11p15.5-p15.4 tumor suppressor region, in breast and lung cancers. Cancer Res. 2001 Nov 1;61(21):7943–9. [PubMed] [Google Scholar]
- 36.Yazici H, Terry MB, Cho YH, Senie RT, Liao Y, Andrulis I, Santella RM. Aberrant methylation of RASSF1A in plasma DNA before breast cancer diagnosis in the breast cancer family registry. Cancer Epidemiol Biomarkers Prev. 2009 Oct;18(10):2723–5. doi: 10.1158/1055-9965.EPI-08-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liggett TE, Melnikov AA, Marks JR, Levenson VV. Methylation patterns in cell-free plasma DNA reflect removal of the primary tumor and drug treatment of breast cancer patients. Int J Cancer. 2011 Jan 15;128(2):492–9. doi: 10.1002/ijc.25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007 Nov;18(5):1225–30. [PubMed] [Google Scholar]


