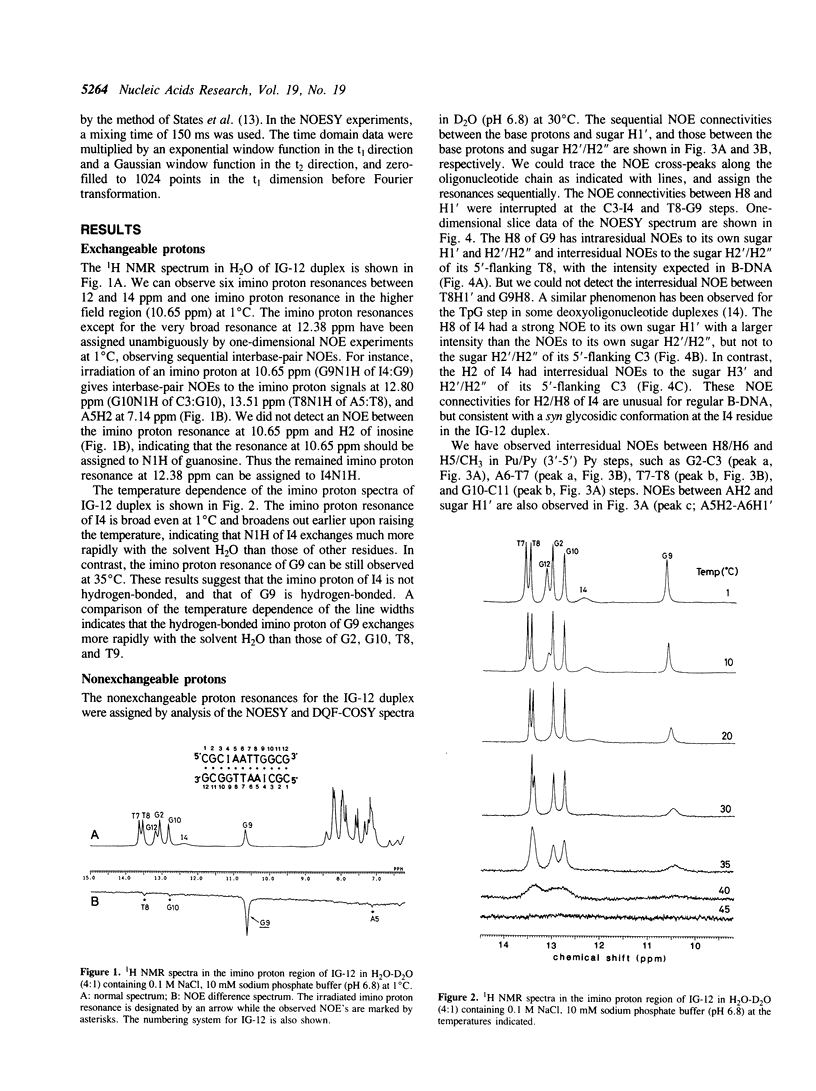
Abstract
One- and two-dimensional NMR experiments have been undertaken to investigate deoxyinosine:deoxyguanosine (dI:dG) base pairing in a self-complementary dodecadeoxyribonucleotide, d(C1-G2-C3-I4-A5-A6-T7-T8-G9-G10-G11-G12) (designated IG-12), duplex. The NMR data indicate formation of a dI(syn):dG(anti) base pair in a B-DNA helix. This unusual base pairing results in altered NOE patterns between the base protons (H8 and H2) of the I4 residue and the sugar protons of its own and the 5'-flanking C3 residues. The dI(syn):dG(anti) base pair is accommodated in the B-DNA duplex with only a subtle distortion of the local conformation. Identification of the dI:dG base pairing in this study confirms that a hypoxanthine base can form hydrogen-bonded base pairs with all of the four normal bases, C, A, T, and G, in DNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carbonnaux C., Fazakerley G. V., Sowers L. C. An NMR structural study of deaminated base pairs in DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4075–4081. doi: 10.1093/nar/18.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield P. W., Hunter W. N., Brown T., Robinson P., Kennard O. Inosine.adenine base pairs in a B-DNA duplex. Nucleic Acids Res. 1987 Oct 12;15(19):7935–7949. doi: 10.1093/nar/15.19.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Aymani J., Kennard O., Brown T., Jack A. G., Leonard G. A. Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs. Nucleic Acids Res. 1989 Jan 11;17(1):55–72. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Quignard E., Teoule R., Guy A., Guschlbauer W. A two-dimensional 1H-NMR study of the dam methylase site: comparison between the hemimethylated GATC sequence, its unmethylated analogue and a hemimethylated CATG sequence. The sequence dependence of methylation upon base-pair lifetimes. Eur J Biochem. 1987 Sep 15;167(3):397–404. doi: 10.1111/j.1432-1033.1987.tb13351.x. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Schroeder S. A., Fu J. M., Metz J. T., Roongta V., Jones C. R. Assignments of 31P NMR resonances in oligodeoxyribonucleotides: origin of sequence-specific variations in the deoxyribose phosphate backbone conformation and the 31P chemical shifts of double-helical nucleic acids. Biochemistry. 1988 Sep 20;27(19):7223–7237. doi: 10.1021/bi00419a009. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hosur R. V., Ravikumar M., Chary K. V., Sheth A., Govil G., Zu-Kun T., Miles H. T. Solution structure of d-GAATTCGAATTC by 2D NMR. A new approach to determination of sugar geometries in DNA segments. FEBS Lett. 1986 Sep 1;205(1):71–76. doi: 10.1016/0014-5793(86)80868-7. [DOI] [PubMed] [Google Scholar]
- Kawase Y., Iwai S., Inoue H., Miura K., Ohtsuka E. Studies on nucleic acid interactions. I. Stabilities of mini-duplexes (dG2A4XA4G2-dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC) containing deoxyinosine and other mismatched bases. Nucleic Acids Res. 1986 Oct 10;14(19):7727–7736. doi: 10.1093/nar/14.19.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Matsuki S., Ikehara M., Takahashi Y., Matsubara K. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J Biol Chem. 1985 Mar 10;260(5):2605–2608. [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Nordheim A., Rich A. Right-handed and left-handed DNA: studies of B- and Z-DNA by using proton nuclear Overhauser effect and P NMR. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1413–1417. doi: 10.1073/pnas.79.5.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Hare D. Sequence-dependent conformations of DNA duplexes: the TATA segment of the d(G-G-T-A-T-A-C-C) duplex in aqueous solution. Biopolymers. 1986 Apr;25(4):693–706. doi: 10.1002/bip.360250412. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kato K., Hayashizaki Y., Wakabayashi T., Ohtsuka E., Matsuki S., Ikehara M., Matsubara K. Molecular cloning of the human cholecystokinin gene by use of a synthetic probe containing deoxyinosine. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1931–1935. doi: 10.1073/pnas.82.7.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi S., Oda Y., Ikehara M., Kawase Y., Ohtsuka E. Identification of I:A mismatch base-pairing structure in DNA. J Biol Chem. 1987 May 25;262(15):6965–6968. [PubMed] [Google Scholar]
- Westerink H. P., van der Marel G. A., van Boom J. H., Haasnoot C. A. Conformational analysis of r(CGCGCG) in aqueous solution: an A-type double helical conformation studied by two-dimensional nuclear Overhauser effect spectroscopy. Nucleic Acids Res. 1984 May 25;12(10):4323–4338. doi: 10.1093/nar/12.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos C., Kouchakdjian M., Yarema K., Basu A., Essigmann J., Patel D. J. NMR studies of the exocyclic 1,N6-ethenodeoxyadenosine adduct (epsilon dA) opposite deoxyguanosine in a DNA duplex. Epsilon dA(syn).dG(anti) pairing at the lesion site. Biochemistry. 1991 Feb 19;30(7):1828–1835. doi: 10.1021/bi00221a015. [DOI] [PubMed] [Google Scholar]



