Abstract
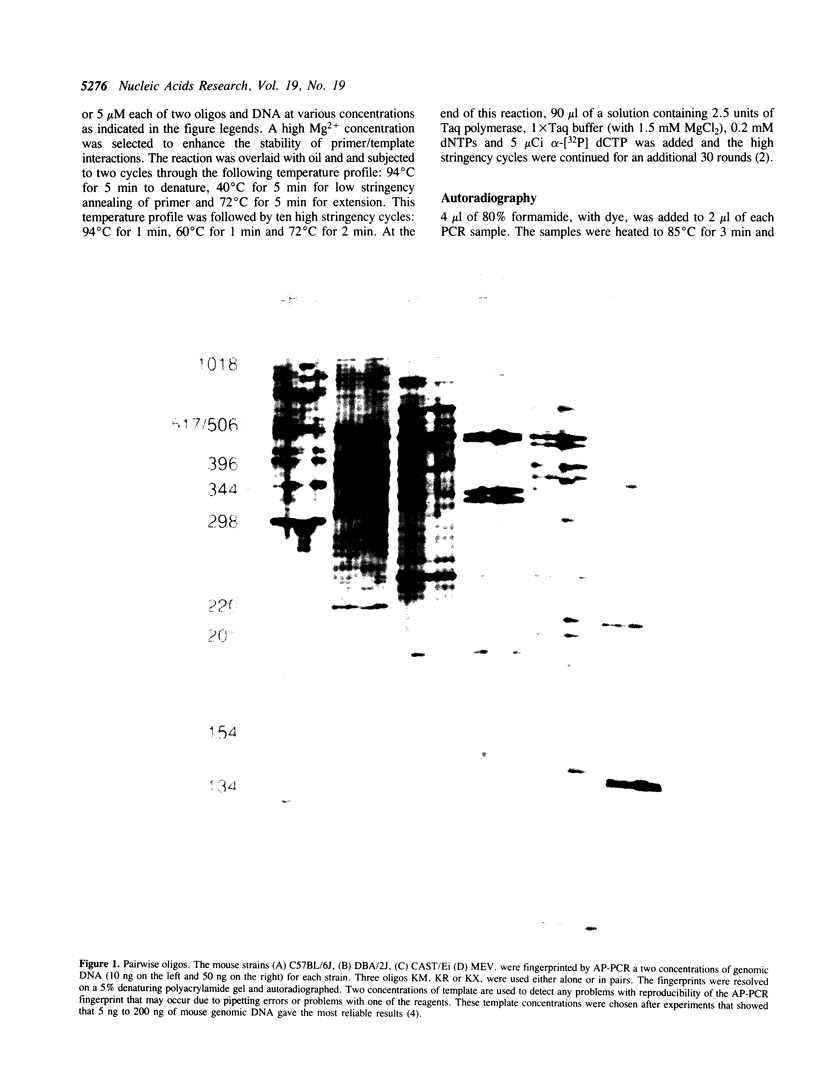
Polymorphisms in genomic fingerprints generated by arbitrarily primed PCR (AP-PCR) can distinguish between slightly divergent strains of any organism. Single oligodeoxyribonucleotide (oligo) primers have been used to generate such fingerprints, with the same primer being present at the 5' end of both strands for every PCR product. We used three arbitrary oligos, individually and in pairs, to generate six different genomic fingerprints of the same mouse genomic DNAs. Fewer than half of the products in genomic fingerprints generated using the oligos in pairs were the same as those produced by AP-PCR using one of the three oligos alone. Thus, a few oligos could be used in a very large number of single and pairwise combinations, each producing a distinct AP-PCR fingerprint with the potential to identify new polymorphisms. For example, 50 oligos can be used in a matrix of pairwise combinations to produce 2,500 fingerprints, in which at least half the data can be expected to be unique to each pair. We demonstrate this principle by using two oligos, alone and together, to generate three sets of fingerprints and map thirteen polymorphisms in the C57BL/6J x DBA/2J set of recombinant inbred mice.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cox R. D., Copeland N. G., Jenkins N. A., Lehrach H. Interspersed repetitive element polymerase chain reaction product mapping using a mouse interspecific backcross. Genomics. 1991 Jun;10(2):375–384. doi: 10.1016/0888-7543(91)90322-6. [DOI] [PubMed] [Google Scholar]
- Ledbetter S. A., Nelson D. L., Warren S. T., Ledbetter D. H. Rapid isolation of DNA probes within specific chromosome regions by interspersed repetitive sequence polymerase chain reaction. Genomics. 1990 Mar;6(3):475–481. doi: 10.1016/0888-7543(90)90477-c. [DOI] [PubMed] [Google Scholar]
- Nelson D. L., Ledbetter S. A., Corbo L., Victoria M. F., Ramírez-Solis R., Webster T. D., Ledbetter D. H., Caskey C. T. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991 Feb 25;19(4):861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Petersen C., McClelland M. Polymorphisms generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res. 1991 Jan 25;19(2):303–306. doi: 10.1093/nar/19.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]