Abstract
Autophagy is rapidly developing into a new immunological paradigm. The latest links now include overlaps between autophagy and innate immune signaling via TBK-1 and IKKα/β, and the role of autophagy in inflammation directed by the inflammasome. Autophagy's innate immunity connections include responses to pathogen and damage associated molecular patterns including alarming such as HMGB1 and IL-1β, Toll-like receptors, Nod-like receptors including NLRC4, NLRP3 and NLRP4, and RIG-I-like receptors. Autophagic adaptors referred to as SLRs (sequestosome 1/p62-like receptors) are themselves a category of pattern recognition receptors. SLRs empower autophagy to eliminate intracellular microbes by direct capture and by facilitating generation and delivery of antimicrobial peptides, and also serve as inflammatory signaling platforms. SLRs contribute to autophagic control of intracellular microbes, including Mycobacterium tuberculosis, Salmonella, Listeria, Shigella, HIV-1 and Sindbis viruses, but act as double edged sword and contribute to inflammation and cell death. Autophagy roles in innate immunity continue to expand vertically and laterally, and now include antimicrobial function downstream of vitamin D3 action in tuberculosis and AIDS. Recent data expand the connections between immunity related GTPases and autophagy to include not only IRGM but also several members of the Gbp (guanlyate-binding proteins) family. The efficacy with which autophagy handles microbes, microbial products and sterile endogenous irritants governs whether the outcome will be with suppression of or with excess inflammation, the latter reflected in human diseases that have strong inflammatory components including tuberculosis and Crohn's disease.
Introduction
The sensu stricto autophagy (often referred to as macroautophagy) is a ubiquitous eukaryotic process dependent on Atg factors and internal membrane formation in the cytoplasm that form unique organelles called autophagosomes [1]. Autophagosomes capture diverse cytoplasmic cargo with a variety of end purposes: (a) quality control of disused or defunct organelles such as irreversibly depolarized or leaky mitochondria; (b) removal of toxic macromolecular aggregates too large for handling by smaller capacity or single-molecule-handling proteolytic systems of the cell (e.g. proteasome); (c) digestion of bulk cytoplasm expressly to replenish amino acids and energy during starvation or growth factor withdrawal; (d) acting on or in concert with the molecular machineries and organelles at the interface between cell survival and cell death; and (e) controlling and acting as an effector or a regulator of innate and adaptive immunity and inflammation, which is the focus of this review.
One of the first reviews on this topic, written half a decade ago by the author [2] referred to several studies that have appeared at the time, suggesting rather gingerly a potential role for autophagy in immunity. Five to six years later, the immunological roles of autophagy have become one of the better established physiological functions of autophagy, as summarized in a number of recent comprehensive reviews [3-5]. The anti-inflammatory role of autophagy and its anti-microbial action [4] have not remained “unnoticed” by prospective pathogenic microbes in the process of their adaptation to a potentially susceptible host. Many highly successful pathogens have evolved mechanisms to counter or harness autophagy attesting to the significance of autophagy as a bona fide antimicrobial defense [5]. Autophagy roles in immunity have been established through in vitro, ex vivo, and in vivo models [4,5]. Importantly, immunological autophagy, via a variety of cellular and molecular mechanisms [6-8], shows unequivocal genetic links to inflammatory bowel Crohn's disease [9] and tuberculosis (reviewed in [5]). The genetically established role of autophagy in human immunity is arguably the most critical evidence for the health relevance of immunological autophagy [5,9].
The present review reflects the author's bias that the innate immunity role of autophagy is not just another physiological application of autophagy but is a key evolutionary driver and mainstream effector-regulator of autophagy that may have shaped several facets of this fundamental biological process. Another underlying theme of this opinion article is to consider autophagic adaptors [10], termed in the immunological context as sequestosome 1/p62-like receptors (SLRs) [5], as another category of pattern recognition receptors (PRRs) on par with Toll-like receptors (TLRs), Nod-like receptros (NLRs), and RIG-I-like receptors (RLRs).
Autophagy as an intracellular membrane trafficking pathway with potential links to inflammatory platforms
Autophagosomes are believed to emerge at least in part from the ER membranes [11] via an ER cradle model, with the autophagic phagophore (isolation membrane) being formed from or in the vicinity of a PI3P-possitive (albeit PI3P is atypical for the ER) and DFCP-positive transient omegasome structures (reviewed in [1]) (Fig. 1). This likely occurs with participation of additional compartments and organelles supplying either assembly/signaling platforms, phospholipids, or membrane intermediates including those originating from the biosynthetic secretory pathway [12-14], plasma membrane [15], and ER-mitochondria contact sites [16]. The processes of autophagosomal formation and autophagic flux are under the control of three systems [1]: (i) The first system represents the well known protein conjugation systems (Atg5-Atg12-Atg16L1 and Atg7-Atg3-Atg8/LC3/GABARAP) culminating in the C-terminal lipidation of Atg8 paralogs with phosphatidylethanolamine resulting in, for example, LC3-II, the emblematic marker of mammalian autophagosomes [1]. (ii) The second of these systems is centered upon the Ser/Thr kinase Atg1 (mammalian Ulk1 and Ulk2) controlled by the upstream nutritional regulator mTOR and, perhaps equally if not more importantly, directly by the cellular energy charge sensor and regulator AMPK [17]. (iii) The third system is centered around Atg6 (Beclin 1), which interacts with many proteins including the evolutionarily ancient Class III PI3K Vps34 [18].
Fig. 1. Autophagy pathway.
Four major contributors proposed as the source of autophagic membrane: 1) ER, endoplasmic reticulum; 2) G/SP, Golgi/secretory pathway; 3) PM, plasma membrane; 4) MT, mitochondria. 5) Autophagic pathway has been stylized into a logo flow chart (crescent, initiation/phagophore formation; double membrane, early/nondegradative autophagosome; single membrane with dense content, degradative autolysosome). SLRs, sequestosome 1/p6-lke receptors. PI3P, phosphatidylinositol 3-phosphate.
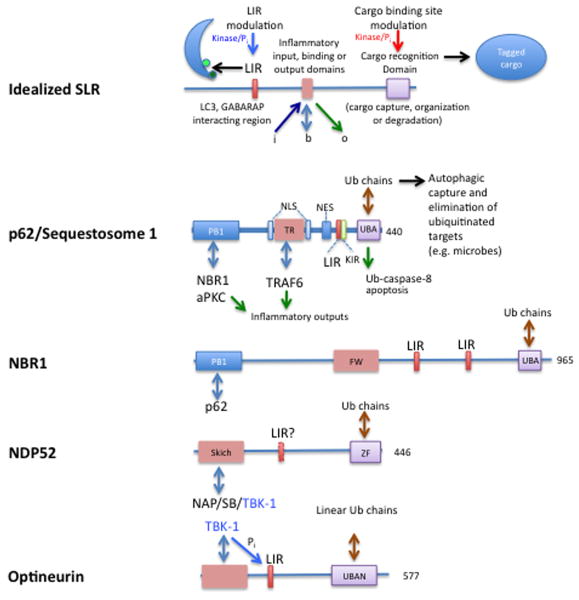
Ulk1/2 and one of the Beclin 1 interactors, Atg14L, mark the sites for the initiation of autophagosome formation on the ER [1]. Significantly, these sites on the ER in mammalian cells receive very early on, at the Ulk1 recruitment step, the autophagic adaptor proteins sequestosome 1/p62 and NBR1 [19]. NBR1 and p62 will be featured in other parts of this review as we explore links with innate immunity and inflammation. These adaptors have been primarily and exceptionally well characterized in the autophagic capture of diverse intracellular cargo [10], but will be considered here in the context of their immunological function (along with several other members) as SLRs (Fig. 2). These two archetypal SLRs are located at the very nidus of autophagosome formation in mammalian cells even before their engagement in cargo recognition and capture [19]. Of note, SLRs have been studied independently of autophagy as complex inflammatory signaling platforms independently of their role as autophagic adaptors [20,21]. This physically connects inflammatory signaling to the very initiation steps of the autophagic pathway. We propose that potential for inflammation may be intimately linked to the earliest stages of autophagy inception in mammalian cells.
Fig. 2. SLRs, sequestome 1/p62-like receptors.
Top, idealized SLR represents no particular member of the group, but is depicted in functional terms. LIR, LC3-interacting region allows an SLR to connect with autophagic organelles via Atg8 paralogs (LC3s and GABARAPs). Cargo recognition domain, so far based on ubiquitin binding: UBA (Ub-associated) and UBAN domains. The inflammatory input, binding, or output domain depicts the ability of SLRs to interact with proinflammatory factors (e.g. TRAF6, TBK-1), and receive signals, e.g. phosphorylation by protein kinases modifying SLR functions (e.g. TBK-1 phosphorylates a Ser residue juxtaposed to the LIR domain core motif (W/FXXL) improving its ability to associate with LC3, or a kinase modifying ability of the UBA domain of p62 to bind different ubiquitin chain subsets. FW, four tryptophans domain. KIR, Keap1-interacting region. NES, nuclear export signal. NLS, nuclear localization signal. PB1, protein-protein interaction (hetero- and homo-oligomerization) domain. ZF, zinc finger domain.
Convergence of pro-inflammatory, immune, and physiological signaling pathways in control of autophagy
Much of the innate immunity signaling occurs through or involves the members of the IκB family of kinases (IKK), which fall into two categories – canonical (IKKα, IKKβ) and IKK-related kinases (IKKε and TBK-1). TBK-1, a key regulator of type I interferon response to viral infections, has been shown to play a role in autophagy [22]. TBK-1 phosphorylates optenurin, a TBK-1-interactor analogous to NEMO (IKK-γ; which serves as a platform for the canonical IKK complex). The phosphorylated optineurin, acting as an SLR, assists through its LC3-interacting region (LIR) in the autophagic uptake and elimination of Salmonella [22].
The canonical IKKs, IKKα and IKKβ turned out to participate in transducing the classical signal for autophagy induction – starvation [23,24]. This signaling is not based on NF-κB [23,24]. It appears that IKK induces autophagy in response to starvation by engaging 4 systems [23]: IKK-dependent depletion of cytosolic p53 (cytosolic p53 suppresses autophagy); IKK pathway-associated AMPK activation, likely engaging TAK1 signaling upstream of IKK since TAK1 is a known kinase activating AMPK [25], with AMPK directly phosphorylating and activating Ulk1 [17] and inhibiting mTOR complex 1 via Raptor and TSC2; and IKK-sponsored activation of JNK-1, the details of which are not known, with JNK-1 then phosphorylating Bcl-2 to release Beclin 1 (reviewed in [18]). These relationships between autophagy and IKK signaling underscore the links between autophagy, innate immunity, and inflammatory signaling.
Autophagy and conventional PRRs: TLRs
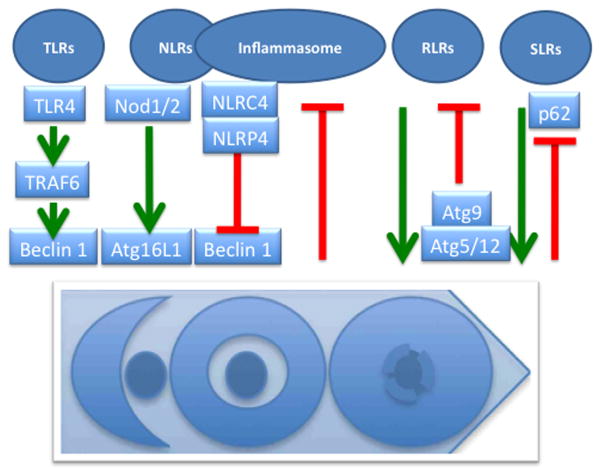
Continuing along with the theme of connections between autophagy and innate immunity responses, autophagy pathway and machinery shows physical, signaling, and regulatory interactions with pattern recognition receptors (PRR), such as Toll-like receptors (TLR), RIG-I like receptors (RLR), Nod-like receptors (NLR), and inflammasomes [5] (Fig. 3).
Fig. 3. Pattern recognition receptors, inflammasomes, and autophagy.
NLRs, Nod-like receptors. RLRs, RIG-I-like receptors. SLRs, Sequestome 1/p62-like receptors. TLRs, Toll-like receptors. All are collectively referred to as pattern recognition receptors (PRRs). Positive regulation of autophagy by PRRs can occur via TRAF6 downstream of TLR4 (TRAF6 ubiquitinates Beclin 1 and releases it from inhibitory complexes with Bcl-2), or via direct interactions with Atg factors (e.g. Nod1/2-Atg16L1). Negative regulation of inflammasome by autophagy is presently ascribed to autophagic prevention of formation of the endogenous sources of inflammasome agonists (e.g. reactive oxygen species or DNA released from unkempt mitochondria). Atg5-Atg12 can negatively regulate RLR signaling by binding and interfering with RLR signaling complex formation (e.g. Atg5-Atg12 conjugate binding to VISA/IPS-1/Cardif/MAVS located on mitochondria). SLRs as autophagic adaptors play a positive role in autophagic elimination, whereas autophagy degrades (and thus downregulates) SLRs in the process of autophagy.
TLRs were historically the first class of PRRs to be connected with autophagy [5]. Autophagy is induced by signaling from TLRs as recently summarized or discussed [3-5]. Importantly, the details of how TLRs connect to autophagy have been elegantly delineated [26]. TLR4 triggers immunological autophagy via TRAF6 E3 ligase that ubiquitinates Beclin 1, followed by Bcl-2 dissociation from the BH3 domain of Beclin 1 [26]. Incidentally, processes following induction by NF-κB downstream of TRAF6 may lead to increase in A20 deubiquitinating enzyme [26], potentially explaining the reported sporadically negative results with TLR stimulation and autophagy induction as well as a the initially observed negative role of NF-κB on autophagy (reviewed in [3]).
Autophagy and NLRs
Connections between NLR signaling and autophagy (Fig. 3) exist in species from Drosophila to mammals (reviewed in [5]). Murine Nod1 and Nod2 have been reported to interact with Atg16L1 and may modulate autophagy in the context of Crohn's disease by several mechanisms [4] including influencing the localization of Atg16L1 at the point of microbial entry at the plasma membrane [27]. Intriguingly, this independently supports the model of plasma membrane as a source of Atg16L1 vesicular precursors leading to LC3-positive autophagosomal profiles [15]. Polymorphisms in the ATG16L1 gene have been linked with predisposition to Crohn's disease mammals with the caveat of a low penetrance of the risk allele ATG16L1*300A. This has been explained in a recent study indicating that phenotypic expression of ATG16L1*300A depends on an “immunological trifecta”: genetic predisposition, endogenous flora in the gut, and viral infection [6]. The reader is directed to a recent review in which these aspects have been extensively analyzed [4].
In contrast to Nod2, NLRC4 (a.k.a. Ipaf) and NLRP4 have inhibitory effects on autophagy [28]. They both inhibit autophagic initiation, whereas NLRP4 additionally inhibits autophagic maturation [28]. Both NLRC4 and NLRP4 interact with Beclin 1 complexes apparently via their NACHT nucleotide-binding domains interacting with ECD (evolutionarily conserved domain) of Beclin 1. In addition, NLRP4 shows some affinity for VPS class C proteins (recall that VPS class C factors play a role in endosomal and autophagosomal maturation). NLRP1, NLRP10, and importantly NLRP3 (a key component of the principal form of inflammasome reacting to diverse agonist), also interact with Beclin 1 but the functional consequences of these interactions are yet to be reported [28]. Interestingly, NALP4 is found together with Beclin 1 in large cytoplasmic macromolecular complexes (500-700 kDa) [28], which is of further interest given that NLRC4-, NLRP1 and NLRP3 form inflammasome complexes in that size range. The emerging interrelationships between the inflammasome and autophagy will be treated in a separate section below.
Autophagy and RLRs
RLRs connections with autophagy (Fig. 3) are notable for the usual emphasis in published reports on the negative regulation of RLR signaling by autophagy including autophagy factors Atg5-Atg12 and Atg9 (reviewed in [3]). Atg9 has been reported to negatively regulate trafficking, assembly and activation of TBK-1 in type I interferon response elicited by intracellular double stranded DNA (dsDNA) [3]. However, in the study showing that Atg9 negatively regulates assembly of STING (stimulator of IFN genes), TBK-1, and interferon responses, the core autophagy conjugation system did not follow this pattern, since Atg7 and Atg16L1 knockout MEFs were not derepressed for type I interferon response to dsDNA and the STING puncta assembled normally in the absence of Atg7 and Atg16L1 [29]. As a forerunner of yet to be fully explored positive interactions between RLRs and autophagy, it has been demonstrated that RLRs can activate autophagy with biologically significant effects [30]. In these experiments, dsRNA (polyinosine-polycytidylic acid) in conjunction with MDA-5, an RLR, induced autophagy. It is worth noting here that there is much room for expansion of the standalone role of autophagy upon RLR agonist stimulation.
Autophagic adaptors SLRs, as a new class of PRRs, link autophagy and innate immunity signaling
Autophagic targets in the cytoplasm, ranging in nature, size, and complexity from protein aggregates to whole organelles are recognized and collected by proteins that act as autophagic adaptors [10]. The two principal adaptors, p62 and NBR1 (Fig. 2), have been well characterized and show domain and sequence similarity [10]. These autophagic adaptors have been termed SLRs [5] to emphasize that they represent a new family of innate immunity receptors, as another category of PRRs engaged in recognition and capture of intracellular microbes. Broadly defined, SLRs feature a cargo recognition and capture domain, contain an LC3 interacting region (LIR) [31-34], and invariably contain additional protein interaction modules that cause or participate in inflammatory processes [21,22,32,35]. This latter feature of SLRs has the potential to trigger inflammatory signaling, perhaps concomitantly with microbial target capture or when the process of autophagic elimination does not proceed smoothly [21,22,32,34,35].
The intracellular pathogen recognition and capture occurs in the case of Salmonella via multiple SLRs (p62, NDP52, optineurin) [22,33,35] recognizing either classical or branched ubiquitin chains in association with or in the vicinity of cytosolic salmonellae. Although ubiquitin binding domains of different SLRs involved may recognize different ubiquitin chains (Fig. 2), it is possible that they act along the same pathway, involving cooperation of p62, NDP52 and optineurin, albeit p62 domains on cytosolic bacteria appears to be physically segregated from the NDP52/optineurin domains [22,33,35]. Other microorganism utilizing these SLRs and parts of this pathway are Shigella via p62 [32] and NDP52 [36], Streptococci via NDP52 [35], Listeria via p62 [31] and NDP52 [36] and possibly Sindbis virus via p62 [34]. Through one or more LIRs, SLRs loaded with their cargo connect to nascent autophagosome [22,31-35]. SLRs' ability to recognize microbial targets is controlled by kinases, as recently revealed in the case of TBK-1-dependent phosphorylation of a Ser residue juxtaposed to the LIR domain of optineurin (Fig. 2); this modification enhances both optineurin binding to LC3 and autophagic capture of Salmonella via a pathway that involves optineurin along with p62 and NDP52 [22]. Although it seems that microbial targets are predominantly earmarked for autophagy by ubiquitin tags recognized by SLRs, additional lipid signals or tags (e.g. diacylglycerol) have been invoked in these processes with Salmonella [37].
SLRs act as dual-function innate immunity devices. As described above, SLRs endow autophagic machinery with the capacity to find its microbial targets in the cytoplasm, whereas they can via proinflammatory signaling induce other innate immunity mechanisms of the cell (e.g. p62 via TRAF6; Fig. 2). Since autophagic adaptors are consumed during autophagy, their proinflammatory signaling should be reduced when autophagy and microbial elimination proceed efficiently. However, if autophagic clearance of microbial targets does not proceed efficiently, the innate immunity signaling from SLRs and their interacting components (e.g. in the case of p62, TRAF6 and atypical PKC, as reviewed in [21]) feature may serve to expand the scope of pathogen containment by summoning other cellular and tissue innate immunity mechanisms.
The above and additional features of SLRs, e.g. that p62 can promote NF-kB induction and caspase-8 aggregation (Fig. 2), activation and cell death (reviewed in [21]), need to be further explored in the context of infection. We envision that SLRs have the potential to act as multi-stage regulators capable of orchestrating gradually escalating levels of innate defenses with the ultimate purpose of limiting microbial spread albeit at increasing inflammatory damage cost. Important examples of this duality are the role of p62 in elimination and/or inflammatory signaling with Shigella [32] and Sindbis virus [34]. The duality of SLR functions in pathogen clearance and pro-inflammatory signaling may further be reflected in pathologies and NBR1's potential relationship to asthma [20]. NBR1 along with p62 plays a role in bone formation and remodeling [38,39], whereas mutations in the SQSTM gene encoding p62 are associated with activation of NF-κB in Paget's disease of the bone. NBR1 is also important in NFATc1 activation required for Th2 differentiation [20]. The association between TRIM5α and p62 in the context of HIV-1 infection [40] may engender yet to be explored inflammatory effects and autophagy connections. The elements outlined above primarily based on p62 and partially on NBR1, may nevertheless apply to other SLRs since for example NDP52 and optineurin link up with TBK-1 [22,35], and should depend in principle on the presence or absence of domains enabling interactions with proinflammatory signaling partners.
SLRs, antimicrobial peptides, and vitamin D3
SLRs can also act in a completely different manner to promote autophagic killing of intracellular microbes. In the case of p62, this SLR can collect cytoplasmic precursors to be converted in autolysosomes into neo-antimicrobial products [41]. The antimicrobial peptides generated in autolysosomes or autophagolysosomes can be derived by limited proteolysis from cytosolic proteins such as ubiquitin [42] and ribosomal proteins [41]. Thus, autophagolysosomes can acquire additional microbicidal properties by converting innocuous cytoplasmic proteins into products that have been shown to kill M. tuberculosis [41,42] and likely will apply to other microbes.
There are also indications of a relationship between autophagy and conventional antimicrobial peptides, i.e. cathelicidin (LL-37) [43] an antimycobacterial and antiviral peptide that is derived by conventional proteolysis from larger precursors. Although the details of the interplay between cathelicidin and autophagy are not clear at present (published work indicates that cathelicidin affects autophagy and not the other way around [43]), this is of interest both in the context of bacterial (tuberculosis) [43] and viral (HIV-1) [44] infections. Cathelicidin expression and its antimycobacterial action are induced by vitamin D3 [43], a known activator of autophagy. Significantly, vitamin D receptor gene variations are well know to predispose to tuberculosis in human populations, whereas low vitamin D3 levels are found in individuals with AIDS.
In the context of HIV-1 infection, the interplay between autophagy, vitamin D3 and cathelicidin appears to be complex. Cathelicidin has been shown to boost anti-HIV-1 properties of blood mononuclear cells but it failed to induce autophagy [44]. Thus, at least in the context of HIV-1 the published work indicates that cathelicidin may not be upstream of autophagic processes. Vitamin D3-induced autophagy nevertheless suppressed HIV-1 [44] in keeping with prior observations that autophagy induction can inhibit HIV-1 in macrophages [45]. As a postscript to these considerations, it should be noted that autophagy is countered by HIV-1 proteins Nef [45], which interferes with autophagic maturation, and Env [46], which interferes with autophagy initiation. In this context, it is of potential interest that some autophagy-activating agonists, such as vitamin D3, can bypass the inhibitory effects of HIV-1 Nef and Env on autophagy [44].
DAMP signaling and autophagy
Alarmins or damage associated molecular patterns (DAMP) represent a number of diverse cellular components that undergo a change in their intracellular localization or are released from damaged cells serving as reporters of a need to contain cell or tissue injury under sterile or septic conditions. DAMPs induce autophagy [47,48]. HMGB1, an alarmin [49], undergoes a stepwise displacement from the nucleus (where it is a chromatin component) into the cytoplasm with eventual extracellular release, at each stage inducing autophagy thorough intracytoplasmic events or extracellular receptor signaling [48]. As HMGB1 translocates from the nucleus to the cytoplasm it displaces Bcl-2 from Beclin 1 thus inducing autophagy [48]. As damage continues and HMGB1 ends up being released extracellularly, HMGB1 action widens to a tissue level where it can act both in an autocrine and paracrine fashion via the RAGE receptor and induce autophagy or cell death, an outcome modulated by a number of factors including HMGB1 oxidation state [47]. In addition to HMGB1, DAMPs such as ATP, IL-1β, and DNA complexes are known to induce autophagy (reviewed in [5]).
Basal autophagy prevents spurious inflammasome activation
Autophagy and inflammasome show complex functional interactions (Fig. 3). DAMPs such as ATP that signal to induce K+ efflux, pore-forming toxins and ionophores (e.g. streptolysin O and nigericin), structurally diverse particulates such as silica, alum and asbestos, salt precipitates calcium pyrophosphate dehydrate and monosodium urate (linked to gout), and protein aggregates associated with inflammatory pathologies such as fibrillar amyloid β (Amβ), can activate inflammasome. These inflammasome agonists can induce potassium fluxes, ROS, and/or cathepsin B leakage from lysosomal compartments and activate inflammasomes leading to release of IL-1β, a key pro-inflammatory cytokine that in turn can induce autophagy [26,50].
The basal autophagy, at rates when cells are not exogenously stimulated, prevents spurious activation of inflammasome by ROS and DNA released from unkempt mitochondria or through additional mechanisms [51-53]. Leaky or depolarized mitochondria are continuously removed by mitophagy, a form of housekeeping autophagy acting in all cells at all times. If this basal autophagy is impaired, ROS and mitochondrial DNA cause unscheduled inflammasome activation [51,52]. Where might the negative/suppressive control of inflammasome by autophagy discussed above be of relevance for a known disease? The connections between autophagy and IL-1β could be of relevance for Crohn's disease. For example, the Atg16L1 mouse model of inflammatory colitis shows elevated IL-1β (reviewed in [3]). However, issues concerning the role of autophagy in Crohn's disease are complex, and may not involve only the inflammasome or even not primarily be expressed through inflammasome. For example, altered responses to multiple pro-inflammatory inputs are likely to be at play [4,6].
Is autophagy's role in inflammation limited to CD? This is unlikely. To take the central nervous system as one example, IL-1β levels are increased both in the brains and the serum of Alzheimer's patients and both neurons and glia have the capacity to activate inflammasome (with microglia relying on NLRP3 and neurons on NLRP1); both cell types are being subject to Amβ (an inflammasome agonist) action, with potential cell death outcomes known as pyroptosis (caspase 1-dependent “apoptosis-with-inflammation”) or pyronecrosis (caspase 1-independent cell death) in neurons and microglia, respectively. It should be explored whether autophagy prevents spurious inflammasome activation in the sterile environment of the nervous tissues, of potential relevance for Alzheimer's disease and other neurodegenerative disorders with inflammatory components. When the CNS is not sterile, e.g. in HIV-associated dementia, autophagy has been implicated in pathology-inducing relationship between the HIV-infected glia and HIV-uninfected neurons [54]. This was shown in an SIV encephalitis as a model and in brains of individuals with HIV-associated dementia, suggesting that retrovirally infected microglia produces mediators that suppress autophagy in neurons and negate is neuroprotective role [54]. This area deserves more study.
Induced autophagy enhances inflammasome output
In contrast to the anti-inflammatory effects of basal autophagy, which suppresses unscheduled inflammasome activation [51-53], induced autophagy promotes IL-1β secretion [55]. This turned out to be related to the fundamental issue of how IL-1β gets secreted outside of the cell. IL-1β is a cytosolic protein without a leader peptide and thus cannot utilize the conventional secretory pathway (export via ER lumen, Golgi, post-Golgi carriers, and their fusion with plasma membrane). It has now been shown [55] that IL-1β, following inflammasome activation and processing of pro-IL-1β into mature IL-1β by caspase-1, utilizes the autophagy-based unconventional secretion pathway to exit the cytosol into the extracellular milieu where it acts in pro-inflammatory signaling. Significantly, the autophagy-based unconventional secretion pathway contributes to secretion of other alarmins, e.g. HMGB1, from the cells [55].
Other innate immunity systems and autophagy: IRGM and Gbp
In the context of inflammatory bowel disease, IRGM, a human autophagy factor and a member of the family of immunity related GTPases (IRG), has been genetically linked with Crohn's disease as a locus of polymorphisms associated with increased risk [9]. IRGM is also a risk locus for tuberculosis in human populations (reviewed in [5]). IRGM is important for physiologically (starvation) pharmacologically (rapamycin), and immunologically (IFN-γ) induced autophagy [8]. IRGM impacts autophagic maturation [7,56] and exerts its effects on autophagy via mitochondria where it binds to cardiolpin [8] (Fig. 5). Autophagy links with the immunity related GTPases such as IRGM have now been expanded to interferon-induced guanlyate-binding proteins Gbp1, Gbp2 and Gbp7, until recently orphaned for function [42,56]. The details of Gbp connections to autophagy include Gbp1 interaction with an SLR, p62/sequestosome 1, whereas Gbp7 acts in concert with Atg4 [42].
Concluding remarks
Autophagy is intimately intertwined with nearly all aspects of innate immunity, attesting to the contention that immunological functions are one of autophagy's mainstream roles. In this opinion article we have focused on the interrelationship between autophagy and conventional innate immunity systems including TLRs, RLRs, NLRs, and inflammasomes. Additionally, we have ascribed a PRR function to the autophagic adaptors (SLRs), such as p62/sequestosome 1, NBR1, NDP52, and optineurin, characterized by the presence of LC3-interacting region, ability to recognize and target for autophagy the invading intracellular microbes, and propensity to connect with proinflammatory signaling. A common theme emerges whereby different PRRs, now including a new category, the SLRs, coordinate cell-autonomous capture of intracellular microbes with other aspects of innate immunity responses. We propose that, among the PRRs, what sets apart the SLRs is that they have a unique capability of executing their antimicrobial function in full without having to trigger inflammation, but can switch to a pro-inflammatory mode when needed (Fig. 5). If the autophagic clearance of microbes fails, SLRs can trigger pro-inflammatory signaling at cellular and tissue levels to recruit additional forces and limit the spread of infection. In keeping with the dual anti-inflammatory and pro-inflammatory capabilities of autophagy as an innate immunity mechanism, basal autophagy keeps inflammasomes from being spuriously activated by intracellular triggers, but induced autophagy can augment inflammasome-dependent processing and secretion of potent pro-inflammatory DAMPs/alarmins IL-1β and HMGB1.
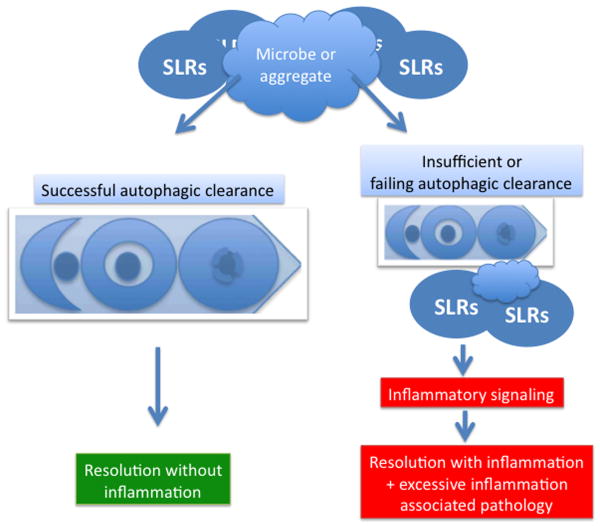
Fig. 4. A model of autophagic clearance with and without inflammation.
Efficacy of autophagic elimination of insults (microbes, endogenous and exogenous particulate irritants such as inflammasome agonists) and SLRs dictates clearance of insult and resolution with and without inflammation. Left, efficacious clearance of insult along with removal of attached SLRs prevents inflammation. A stalled or inefficient autophagic removal of the insult (aggregates, microbes, etc) may lead to accumulation of SLRs and signaling through their pro-inflammatory partners. A realistic process in vivo falls somewhere within the spectrum between the two extremes, and may be skewed by direct or indirect pathological inputs and genetics.
Highlights.
Autophagy is a developing immunological paradigm with vertical and lateral connections in innate immunity
Sequestosome 1/p62-like adaptors (SLRs) serve both as autophagic adaptors and innate immunity signaling platforms
Balance between autophagic clearance via SLRs and their and other inflammatory signaling determines degree of collateral inflammatory damage
Interactions between autophagy and inflammasome involve both suppression of inflammasome activation and enhancement through the autophagy-based unconventional secretion of inflammasome substrates
Acknowledgments
The author apologies to all researchers active in this field for not being able to include all worthy references and in some cases having to resort to referencing reviews instead of primary articles although this was avoided to the maximum extent possible. This work was supported by grants R01 AI069345, RC1AI086845, and R01 AI042999 from National Institutes of Health and a Bill and Melinda Gates Grand Challenge Explorations grant OPP1024376.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annual review of cell and developmental biology. 2010 doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **6.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. This study explains low penterance of Crohn's disease risk alleles, including those involved in autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- **8.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. This study has determined how IRGM controls autophagy, via binding to cardiolipin, affecting mitochondrial function and polarization, autophagy induction, and in extreme cases apoptotic cell death with some necrotic features including release of HMGB1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7 doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009 doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 12.van der Vaart A, Griffith J, Reggiori F. Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **15.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. This study shows that a set of Atg16L1 precursor profiles emanate from the plasma membrane and undegro homotypic fusion in the formation of early autophagic organelles; of interest is that this potentially connects with immunological studies (see ref. [27]) in the context of Crohn's disease, where Atg16L1 has been shown to be recruited to the plasma membrane at the microbial entry site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **19.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. The Journal of cell biology. 2011;192:17–27. doi: 10.1083/jcb.201009067. This study shows that autophagic adaptors p62 and NBR1 are present at the earlies sites on the ER leading to the initiation of autophagosome formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JQ, Liu H, Diaz-Meco MT, Moscat J. NBR1 is a new PB1 signalling adapter in Th2 differentiation and allergic airway inflammation in vivo. The EMBO journal. 2010;29:3421–3433. doi: 10.1038/emboj.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. This study shows that optineurin is an SLR enhancing autophagic uptake of Salmonella. Most importantly, this study shows that SLRs need activation regulated by phosphorylation; in the case of optineurin the regulatory kinase is the non-canonical IKK, TBK-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. This study establishes that canonical IKK, often associated with inflammatory signaling, are involved in initiating autophagy in response to classical stimuli such as starvation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comb WC, Cogswell P, Sitcheran R, Baldwin AS. IKK-dependent, NF-kappaB-independent control of autophagic gene expression. Oncogene. 2011;30:1727–1732. doi: 10.1038/onc.2010.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. The EMBO journal. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. This study shows how TLR4 stimulation leads to autophagy activation. This occurs by TRAF6-dependent ubiquitination of Beclin 1, immediately downstream of TLR4 stimulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2009 doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- **28.Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin1. Journal of immunology. 2011;186:1646–1655. doi: 10.4049/jimmunol.1001654. This study estabalishes that, unexpectedly, NLRs, including those essential for inflammasome assembly and activation such as NRLP3, are found in complexes with Beclin 1. Additionally, NLRC1 (Ipaf) and NLRP4 act to inhibit autophagy, NLRC1 at the initiation stage whereas NLRP4 acts negatively at both initiation and maturation stages. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–114. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 32.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- **34.Orvedahl A, Macpherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy Protects against Sindbis Virus Infection of the Central Nervous System. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. This study is one fo the most effective demonstrations that SLRs such as p62, while acting in pathogen clearance, perhaps even more improtantly are key to suppressing inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 36.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 Proteins Target Intracytosolic Shigella and Listeria to Different Autophagy Pathways. The Journal of biological chemistry. 2011;286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A Diacylglycerol-Dependent Signaling Pathway Contributes to Regulation of Antibacterial Autophagy. Cell Host Microbe. 2010 doi: 10.1016/j.chom.2010.07.002. This study shows that signals in addition to ubiquitin tags are needed or can independently promote antimicrobial autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, Xavier GM, Kashima TG, Cobourne MT, Richards GO, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12913–12918. doi: 10.1073/pnas.0913058107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters S, Marchbank K, Solomon E, Whitehouse CA. Autophagic receptors Nbr1 and p62 coregulate skeletal remodeling. Autophagy. 2010;6:981–983. doi: 10.4161/auto.6.7.13155. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor C, Pertel T, Gray S, Robia SL, Bakowska JC, Luban J, Campbell EM. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **41.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWt, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. This study shows that SLRs don't act only by bringing microbes into the autophagosomes but that SLRs such as p62 are responsible for bringing cytoplasmic precursors of antimicrobial peptides into the autolysosomes or autophagolysosomes containing microbes. These antimicorbial peptides (termed cryptides) can be derived from ribosomal proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **42.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. This sudy expands the connections between autophagy and immunity related GTPases, and adds a new class (Gbp) of such innate immunity factors previously orphaned for function; Gbp1 interacts with the SLR p62, and Gbp7 interacts with Atg4. [DOI] [PubMed] [Google Scholar]
- *43.Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, Lee ZW, Lee SH, Kim JM, Jo EK. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–243. doi: 10.1016/j.chom.2009.08.004. This study expands connections between autophagy and antimicrobial peptides, to incldue cathelicidin in conjunction with vitamin D3-induced autophagy. [DOI] [PubMed] [Google Scholar]
- **44.Campbell G, Spector S. Hormonally Active Vitamin D3 (1,25-Dihydroxycholecalciferol) Triggers Autophagy in Human Macrophages That Inhibits HIV-1 Infection*. Journal of Biological Chemistry. 2011;268:18890–18902. doi: 10.1074/jbc.M110.206110. This study expands on the findings by Kyei et al [45] and combines the observaions by Yuk et al [43] to HIV control by autophagy. It furthermore points to a connection to low vitamin D3 levels in AIDS patients as a contributor to diminished autophagic defenses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. This study establishes how cytoplasmic HMGB1, an alarmin, regulates autophagy: upon HMGB1 translocation form the nucleus into the cytoplasm, HMGB1 displaces Bcl-2 from Baclin 1 thus inducing autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 50.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009 doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **51.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. This study is a landmark work showing that basal autophagy prevents inflamamsome activation and thus acts as an improtant anti-inflammatory mechanism. The proposed mechanism of action is that autophagy, known to remove depolarized mitochondria, keeps ROS sources to a minimum thus preventing ROS-dependent inflammasome activation. [DOI] [PubMed] [Google Scholar]
- **52.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2010 doi: 10.1038/ni.1980. Similarly to the work by Tschopp and colleagues [51], these authors show that mitophagy keeps inflammasome activation low but atribute the effects of impaired autophagy to DNA released from mitochondria; mitochondrial DNA (released when leaky mitochondria are not removed by autophagy) in turn activates inflammasome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, et al. Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J Biol Chem. 2011 doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS ONE. 2008;3:e2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **55.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011 doi: 10.1038/emboj.2011.398. In press. This study shows that unlike basal autophagy, which keeps inflammasome activity low, induction of autophagy promotes secretion of IL-1β, the product of inflammasome activation. The autophagy-based unconventional secretion explains how cytosolic IL-1β is brought to the outside of the cell. More broadly, this pathway applies to other alarmins such as HMGB1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, Beer S, Pfeffer K, Coers J, Taylor GA. Immunity-related gtpase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]